Abstract
Usher syndrome type 3 (USH3) is an autosomal recessive disorder characterized by progressive hearing loss, severe retinal degeneration, and variably present vestibular dysfunction, assigned to 3q21-q25. Here, we report on the positional cloning of the USH3 gene. By haplotype and linkage-disequilibrium analyses in Finnish carriers of a putative founder mutation, the critical region was narrowed to 250 kb, of which we sequenced, assembled, and annotated 207 kb. Two novel genes—NOPAR and UCRP—and one previously identified gene—H963—were excluded as USH3, on the basis of mutational analysis. USH3, the candidate gene that we identified, encodes a 120-amino-acid protein. Fifty-two Finnish patients were homozygous for a termination mutation, Y100X; patients in two Finnish families were compound heterozygous for Y100X and for a missense mutation, M44K, whereas patients in an Italian family were homozygous for a 3-bp deletion leading to an amino acid deletion and substitution. USH3 has two predicted transmembrane domains, and it shows no homology to known genes. As revealed by northern blotting and reverse-transcriptase PCR, it is expressed in many tissues, including the retina.
Introduction
The Usher syndromes (USHs) are characterized by variable degrees of loss of hearing and of loss of eyesight. Three distinct phenotypes have been described. Patients with USH1 have congenital severe-to-profound hearing loss and absence of vestibular function, whereas patients with USH2 have congenital moderate-to-severe hearing loss and normal vestibular function. The onset of retinitis pigmentosa (RP) is prepubertal in patients with USH1 and occurs during the 2d decade of life in patients with USH2 (Smith et al. 1994). Patients with USH3 have postlingual, progressive hearing loss, and the onset of retinitis pigmentosa (RP) symptoms—nyctalopia, progressive constriction of visual fields, and reduction of central visual acuity—usually occurs by the 2d decade of life (Pakarinen et al. 1996). At least 10 loci account for subtypes of USH (see the Hereditary Hearing Loss Homepage). Five genes have so far been identified: myosin VIIA (MYO7A), for USH1B (MIM 276903) (Weil et al. 1995); usherin, for USH2A (MIM 276901) (Eudy et al. 1998); harmonin, for USH1C (MIM 276904) (Bitner-Glindzicz et al. 2000; Verpy et al. 2000); cadherin 23 (CDH23), for USH1D (MIM 601067) (Bolz et al. 2001; Bork et al. 2001); and protocadherin 15 (PCDH15), for USH1F (MIM 602083) (Ahmed et al. 2001).
We previously assigned USH3 to 3q21-q25 (Sankila et al. 1995). Significant linkage disequilibrium between USH3 and the linked markers suggested enrichment of a founder mutation in Finland and allowed us to refine the localization of USH3 to a 250-kb genomic interval between markers 107G19CA7 and D3S3625 (Joensuu et al. 1996, 2000). Haplotype analysis indicated 107G19CA7 to be the proximal historic recombination breakpoint, and a recombination in one family with USH3 placed the USH3 gene proximal to D3S3625. In this study, we (a) sequenced, assembled, and annotated 207 kb of the USH3 critical region, (b) excluded two novel genes—NOPAR and UCRP—and one previously identified gene—H963—as candidates for USH3, and, finally, (c) positionally cloned USH3.
Subjects and Methods
Families with USH3 and Controls
Clinical and genealogical data on the Finnish and Italian families with USH3 that are studied here also have been described elsewhere (Pakarinen et al. 1995; Sankila et al. 1995; Joensuu et al. 1996, 2000; Gasparini et al. 1998). Control DNA samples from 100 anonymous blood donors from eastern Finland (where there is a high prevalence of USH3) and from 100 donors from western and southern Finland (where there is a low prevalence of USH3) were provided by the Finnish Red Cross Blood Transfusion Service. A total of 51 grandparents in families from the Centre d'Étude du Polymorphisme Humain (CEPH) were also studied as controls.
Bacterial Artificial Chromosome (BAC) DNA and Shotgun Sequencing
We shotgun sequenced partly overlapping BACs—specifically, 355L4 and 545J16. Shotgun libraries were prepared and plasmid DNA was extracted by GATC Shotgun Library Construction service. DNA fragments were ligated to the PCR-Blunt II-Topo cloning vector (Invitrogen), and we sequenced the subclones, from both directions, with custom vector primers designed by GATC and with BigDye-terminator chemistry (PE Applied Biosystems), by use of a PE9700 Thermal Cycler (Perkin Elmer). Sequence readings were transferred to a UNIX-based system; edited and filtered by removal of matches to the cloning and sequencing vectors, as well as to Escherichia coli, by the GASP software package (University of Washington Genome Center); and assembled by Phil Green's Phred, Phrap, and Consed sequence-assembly programs. The orientation of contigs, from centromere to telomere, was determined on the basis of the marker order of the physical map described elsewhere (Joensuu et al. 2000).
Computational Analyses and Sequence Annotation by Integrated Gene-Prediction Programs
The complete genomic sequence assembled was masked for repetitive elements, by the RepeatMasker2 program (University of Washington Genome Center). Sequence-similarity searches were performed using BLAST programs (Altschul et al. 1997) against different databases, Human Index for Nonredundant Transcripts (HINT) (Zhuo et al. 2001), UNIGENE (Boguski and Schuler 1995), and the TIGR Human Gene Index (Liang et al. 2000). The sequences were compared with the High-Throughput Genome Sequence and Genomic Survey Sequence databases (National Center for Biotechnology Information), to identify BAC clones or P1-derived artificial clones mapping to the same region. To compare orthologous mouse genomic sequences, homology searches were performed by use of the Celera Assembled and Annotated Mouse Genome. Complete sequence annotation was performed by a modified computational sequence annotation tool, Genotator (Harris 1997), and by the NIX program (UK Human Genome Mapping Project Resource Centre). Transcription-factor and promoter predictions at the 5′-flanking genomic sequences of identified genes were performed by MatInspector/TRANSFAC and Neural Network Promoter Input programs (Baylor College of Medicine Search Launcher). Protein predictions were performed by the PIX program (UK Human Genome Mapping Project Resource Centre), LASERGENE (DNASTAR), TMpred (Swiss Institute of Bioinformatics), SignalP (Center for Biological Sequence Analysis), and PSORT II (Human Genome Center).
DNA and cDNA
We extracted BAC DNA as described elsewhere (Joensuu et al. 2000). Integrated Molecular Analysis of Genomes and Their Expression (IMAGE) cDNA clones for expressed-sequence tags (ESTs) homologous to the USH3 region were obtained from Research Genetics. To obtain the complete sequence of the inserts, cDNA clones were sequenced on both strands by vector-specific primers.
Human-placenta RNA and Marathon Ready Human Retina cDNA were obtained from Clontech. The first-strand cDNA synthesis was performed with 1 μg of total RNA, according to the supplied protocol for the SMART PCR cDNA Synthesis kit (Clontech). Subsequently, 2 μl of the 100-μl reaction mixture was PCR amplified by gene-specific primers. To isolate full-length cDNAs of the predicted genes, we performed rapid amplification of cDNA ends (RACE) PCR with the Marathon Ready Human Retina cDNA libraries, according to the manufacturer's instructions, by a modified PCR with annealing at 65°C for 1 min 30 s, for 20 cycles, followed by nested PCR with annealing at 60°C for 1 min 30 s, for 40 cycles, in a total volume of 50 μl.
We designed combinations of forward and reverse first-amplification primers and nested primers with the program Primer3 (Whitehead Institute for Biomedical Research/Massachusetts Institute of Technology Center for Genome Research). When possible, the primer pairs were designed to span introns in order to minimize false-positive results by contaminating DNA in the reverse-transcriptase PCRs (RT-PCRs). PCR primer sequences and conditions are available on request.
PCR and RT-PCR
We analyzed candidate genes by PCR amplification and direct sequencing of all exon regions with adjacent splice sites, from genomic DNA and, when possible, from total RNA and poly(A) RNA—using the Qiagen Total RNA kit and the poly(A) RNA–extraction kit provided by Amersham, respectively—from Epstein-Barr virus–transformed lymphoblasts extracted from two patients with USH3, two carriers, and two controls. Amplification of genomic DNA was performed as described elsewhere (Joensuu et al. 1996). For RT-PCR, either 200 ng of lymphoblast poly(A) RNA or 1 μg of total RNA was primed by random hexamers and was reverse transcribed for the first-strand synthesis of cDNA. Subsequent nested PCR from cDNA was essentially performed with 2 μl of the 20-μl cDNA reaction mix in a total volume of 25 μl with 0.2 mM of each dNTP, 20 pmol of each primer, and 0.5 U of AmpliTaq Gold DNA polymerase (PE Applied Biosystems). Control reactions containing human genomic DNA and water, without cDNA, were performed. Amplification of GC-rich regions was attempted with 1.2 mM betaine or 7% dimethyl sulfoxide.
We electrophoresed PCR products on either 1%–2% SeaKem agarose gel or 0.7 × MDE SSCP (FMC) polyacrylamide gels and visualized them by ethidium-bromide or silver staining (Bassam et al. 1991). Direct sequencing of the purified amplification products (PCR purification kit; Qiagen) was performed by automated sequencing (ABI 377-sequencer; PE Applied Biosystems) by use of the PCR primers.
Analysis of Novel Single-Nucleotide Polymorphisms (SNPs)
By genomic sequencing of NOPAR, in families with USH3, we identified novel SNPs—RSNP2 and TSNP1, in NOPAR introns 1 and 9, respectively. Intronic forward and reverse primers used in the genotyping of the families with USH3 were as follows: TSNP1 (forward, 5′-TGGTAGGTAATCTTAGTCCAACAA-3′; reverse, 5′-TTTTGCCTCACATAGAATGGTG-3′), 390 bp; and RSNP2 (forward, 5′-CATGCACCAAGCTCTCACAT-3′; reverse, 5′-AAGCTGAGGGAAGAAACTGC-3′), 264 bp. The alleles were detected either by direct sequencing or by SSCP analysis, in 0.7 × MDE gel, for 18 h at 5 W, at room temperature. The SSCP alleles were visualized by silver staining. Haplotypes were manually constructed under the assumption that there were a minimum number of recombinations.
Isolation of the Candidate Gene USH3
The interexon 5′- and 3′-UTR primers used in combining the exons from cDNA synthesized, by PCR (conditions were as described above), from human-retina total RNA or the Marathon Ready Human Retina cDNA library (Clontech) were as follows: E3F (5′-CTCCTGTGGCTGTCTTGTCA-3′) and W27577R (5′-TGGTGGGTTTGCCTCTTAGT-3′); E2F (5′-AAGCAATCCCAGTGAGCATC-3′) and E3R (5′-TGACAAGACAGCCACAGGAG-3′); E1F (5′-TCACTATCTGAAACTATCTTGTTGT-3′) and E2R (5′-TAGGGGACCATGCAGAGTTT-3′); and E1F2 (5′-TGCCTCCCCACCATTCACCA–3′) and E2R2 (5′-CATGAAGAAGGCTGTCCCCACCA-3′).
Mutation Detection
Using genomic DNA, we amplified genomic fragments encompassing five USH3 exons, by use of the following intronic primers: E1F (see above) and P1R (5′-AAGCCCCTGAACTTTATAGG-3′), 910 bp; P1BF (5′-TTGTGGCCATTTTTGGAGAT-3′) and P1BR (5′-CCCCAAACATGTATCAAGTGC-3′), 207 bp; P2F (5′-TCAGAAGGATTTTAGTGATGTTTGA-3′) and P2R (5′-TCTTTTTGACATATTGAAAAGCACA-3′), 352 bp; P3F (5′-ATGTCAATGGGGATGATGGT-3′) and P3R (5′-GGAGCCCATTCAGAAAATGA-3′), 291 bp; and P4F (5′-TTCCCCTGAATTACCCATCA-3′) and P4R (5′-AGCATCTGGAAACTCGGTGT-3′), 339 bp. PCR products were purified and sequenced as described above.
The Finmajor mutation and the Italian 3-bp deletion were screened by direct sequencing of the 291-bp product amplified by primers P3F and P3R. For the Finminor mutation, we used SSCP; a 198-bp fragment was PCR amplified by primers P2F2 (5′-TCCCAGTGAGCATCCACGTC-3′) and P2R2 (5′-TGAAAAGCACATTTGTCTTCAGAGG-3′), and the alleles were separated on 0.7 × MDE gels, for 18 h at 5 W, at room temperature and were visualized by silver staining.
Northern Blot and In Situ Hybridization
A human multiple-tissue northern blot from 12 adult tissues was hybridized by use of an α[32P]-dCTP random-labeled cDNA probe (USH3 nucleotides −33–321; 354 bp) comprising exons 1–3 and part of the 5′ UTR. The membrane was washed at 65°C with 2 × SSC (saline sodium citrate) and 0.05% SDS, then exposed to x-ray film at −80°C for 2 d. In situ hybridization and the production of cRNA probes were performed as described by Wilkinson and Green (1990). In short, 4% paraformaldehyde–fixed paraffin sections from embryonic 16-d-old mouse embryos and from dissected adult eyes and inner ears were used for analysis. A 262-bp fragment from the Marathon Ready Mouse Brain cDNA library (Clontech) was produced by primers mouse/p1 (5′-ATCCCCGTAAGCATCCACAT-3′) and mouse/p2 (5′-TTCTGTAGGCATAGGTCCCTTC-3′). The amplified fragment was cloned into the pGEM-T Easy vector (Promega) according to the manufacturer's instructions. Subsequently, [35S]-labeled antisense and control-sense cRNA probes were synthesized by in vitro transcription. The control-sense probe did not produce any hybridization signal above background level.
Results
Construction of the Genomic Contigs and Transcript Map
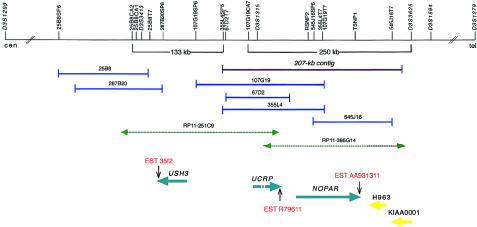
We chose two partly overlapping BAC clones (355L4 and 545J16) flanking both 107G19CA7 and most of the previously defined USH3 critical region toward D3S3625, for shotgun sequencing. Database-derived BAC clone RP11-385G14 sequence was aligned with genomic contigs of BAC clone 545J16, to extend the distal sequences. Gap filling was performed by primer walking from the contig end sequences and by PCR across the gaps. A final sequence contig of 207 kb, which is depicted in figure 1, was assembled.
Figure 1.
Schematic representation of physical and transcript maps covering the USH3 region, depicting the initial 250-kb USH3 region and the extended 133-kb USH3 region, with partly overlapping BAC clones. Polymorphic markers and sequence-tagged sites are indicated by vertical lines. Previously mapped BAC clones are shown in dark blue, and the 207-kb sequence contig assembled is indicated by a purple line. Database-derived and partly assembled BAC sequences from clones RP11-385G14 and RP11-251C9 are indicated by green dashed arrows. The centromere is on the left. A pair of previously known genes—KIAA0001 and H963—and a group of three novel genes—NOPAR, UCRP, and USH3—are indicated by yellow and light-blue arrows, respectively, showing the orientation of transcription. ESTs representing parts of the 3′ UTRs of the transcripts are indicated by vertical arrows. H963, NOPAR, UCRP, and USH3 were subjected to mutation analyses in this study.
Use of nucleotide-nucleotide BLAST for searches of the contig identified 30 distinct or partly overlapping human ESTs and 11 orthologous mouse or rat ESTs (table 1). Starting from the identified ESTs and exons predicted by GRAIL and GENESCAN, we used a combination of RT-PCR, RACE experiments, and direct PCR of cDNA libraries, to assemble two novel genes: NOPAR (for no opposite-paired repeat) and UCRP (for USH critical region pseudogene) (fig. 1). We identified two alternatively spliced transcripts—NOPAR and NOPAR2—which encode putative polypeptide isoforms, of 721 and 756 amino acids, respectively, showing 61% identity to the amino terminus of TRAP230 (Ito et al. 1999), also called the “human OPA-containing gene,” or “HOPA” (Philibert et al. 1999). The NOPAR coding region comprises approximately one-third of the length of TRAP230/HOPA and lacks the 3′ OPA repeat contained in TRAP230/HOPA. Northern blot analysis of NOPAR showed widespread expression in adult tissues (data not shown). Transcription in human retina was shown by RT-PCR.
Table 1.
Location of ESTs in the 207-kb Contig
| EST | Location(s) | GenBankAccession Number | dbEST Details |
| 1 | 24264–24640 | R75628 | Homo sapiens cDNA clone IMAGE: 143509 3′ |
| 2 | 65074–65380 | R79610 | H. sapiens cDNA clone IMAGE: 146349 5′ |
| 3 | 65715–65825 | T39157 | H. sapiens cDNA clone IMAGE: 60197 5′ |
| 4 | 66204–66483 | T40442 | H. sapiens cDNA clone IMAGE: 60197 3′ |
| 5 | 66139–66502 | R79611 | H. sapiens cDNA clone IMAGE: 146349 3′ |
| 6 | 78227–78422, 107727–107829, 114184–114230 | AI501968 | Rattus norvegicus cDNA clone UI-R-C0-jb-b-08-0-UI 3′ |
| 7 | 78227–78422, 107727–107829, 114184–114251 | AW920576 | EST351880 Bento Soares R. norvegicus cDNA clone RGIGZ01 5′ |
| 8 | 79115–79401 | AW897677 | CM1-NN0062-280400-202-d05 NN0062 H. sapiens cDNA |
| 9 | 79118–79401 | AW897669 | CM1-NN0062-280400-202-b05 NN0062 H. sapiens cDNA |
| 10 | 99994–100283 | AA776659 | H. sapiens cDNA clone IMAGE: 970436 3′ |
| 11 | 78227–78422, 107727–107829, 114184–114241 | BE948099 | UI-M-BH3-aws-h-03-0-UI.s1 Mus musculus cDNA clone UI-M-BH3-aws-h-03-0-UI 3′ |
| 12 | 136681–136758 | AF062710 | AF062710 H. sapiens cDNA clone HA0028 |
| 13 | 136681–136758 | AI110602 | H. sapiens cDNA HA0028 |
| 14 | 181289–181421 | AA931311 | H. sapiens cDNA clone IMAGE: 1565326 3′ |
| 15 | 186110–186218 | BE843843 | RC0-TN0079-310700-021-b10 TN0079 H. sapiens cDNA |
| 16 | 186131–186641 | AA170490 | M. musculus cDNA clone IMAGE: 618166 5′ |
| 17 | 186185–186518 | AW591154 | H. sapiens cDNA clone IMAGE: 2703652 3′ |
| 18 | 186193–186446 | BB209769 | BB209769 M. musculus cDNA clone A430094C06 3′ |
| 19 | 186193–186773 | BE283279 | 601101050F1 M. musculus cDNA clone IMAGE: 3493212 5′ |
| 20 | 186235–186446 | AI172577 | R. norvegicus cDNA clone UI-R-C2p-nz-h-03-0-UI 3′ |
| 21 | 186313–186789 | AA162789 | M. musculus cDNA clone IMAGE: 598871 5′ |
| 22 | 186665–187193, 190857–190910 | AV653286 | AV653286 GLC H. sapiens cDNA clone GLCDJC11 3′ |
| 23 | 186660–187193, 190857–190910 | AV653266 | AV653266 GLC H. sapiens cDNA clone GLCDJB03 3′ |
| 24 | 186676–187016 | AV757663 | H. sapiens cDNA clone: BMFBHF02, 5′ end |
| 25 | 186903–187075 | AA184698 | M. musculus cDNA clone IMAGE: 634121 3′ |
| 26 | 186952–187148 | AI592667 | M. musculus cDNA clone IMAGE: 634121 5′ |
| 27 | 187086–187195, 190857– 190954 | AA353758 | EST61951 H. sapiens cDNA 5′ |
| 28 | 200484– 201179 | AU138895 | H. sapiens cDNA clone: PLACE1009521, 5′ |
| 29 | 204146– 204843 | AL042755 | H. sapiens mRNA; EST DKFZp434C0522_r1 |
| 30 | 199885– 200422 | AU157802 | H. sapiens cDNA clone: PLACE1009521, 3′ |
| 31 | 205968– 206348 | AL042756 | H. sapiens mRNA; EST DKFZp434C0522_s1 |
| 32 | 199885–200231 | AI167543 | Soares_NhHMPu_S1 H. sapiens cDNA clone IMAGE: 1661439 3′ |
| 33 | 199883–200242 | AA027011 | Soares_pregnant_uterus_NbHPU H. sapiens cDNA clone IMAGE: 469358 3′ |
| 34 | 199885–200183 | AI277229 | Soares_placenta_8to9weeks_2NbHP8to9W H. sapiens cDNA clone IMAGE: 1893709 3′ |
| 35 | 206135–206348 | AI889440 | H. sapiens cDNA clone IMAGE: 2444461 3′ |
| 36 | 200575–200753 | AA026830 | H. sapiens cDNA clone IMAGE: 469358 5′ |
| 37 | 200005–200189 | AI686699 | H. sapiens cDNA clone IMAGE: 2268648 3′ |
| 38 | 200580–200783 | R35752 | Soares placenta Nb2HP H. sapiens cDNA clone IMAGE: 136983 5′ |
| 39 | 202796–202998 | AA056078 | Soares retina N2b4HR H. sapiens cDNA clone IMAGE: 381530 3′ |
| 40 | 201215–201642 | AI050350 | M. musculus cDNA clone IMAGE: 1379171 5′ |
| 41 | 199880–200139 | R35645 | H. sapiens cDNA clone IMAGE: 136983 3′ |
The 2,045 bp of UCRP cDNA obtained consists of five exons spanning >70 kb genomic DNA. However, the open reading frame (ORF) is disrupted, and the gene thus is probably an unprocessed pseudogene. The region also contained two previously identified genes: KIAA0001 (Nomura et al. 1994), which was previously excluded as USH3 (Joensuu et al. 2000), and platelet-activating–receptor homolog mRNA H963 (Jacobs et al. 1997).
Refinement of the USH3 Critical Region
The exons and flanking intronic sequences of NOPAR, UCRP, and H963 were examined for mutations in patients with USH3; however, no potential disease-causing mutations were detected. Genomic sequencing of the NOPAR introns in patients with USH3 indicated the presence of two novel SNPs, RSNP2 (nucleotide C/G) and TSNP1 (nucleotide T/C), in introns 1 and 9, respectively (fig. 1). Haplotype analysis revealed conservation of the segment RSNP2–TSNP1 in all 48 USH3 chromosomes studied (data not shown). This supported our previous haplotype analysis placing USH3 distal to 107G19CA7 (Joensuu et al. 2000); however, a recombination event in one family excluded the RSNP2-TSNP1-tel segment as the site of the causative mutation (fig. 2). No additional transcripts in the remaining 60-kb critical region between markers 107G19CA7 and RSNP2 were found. This implicated the region immediately centromeric to 107G19CA7 and extending to 25B8CA2, as part of the USH3 critical region.
Figure 2.
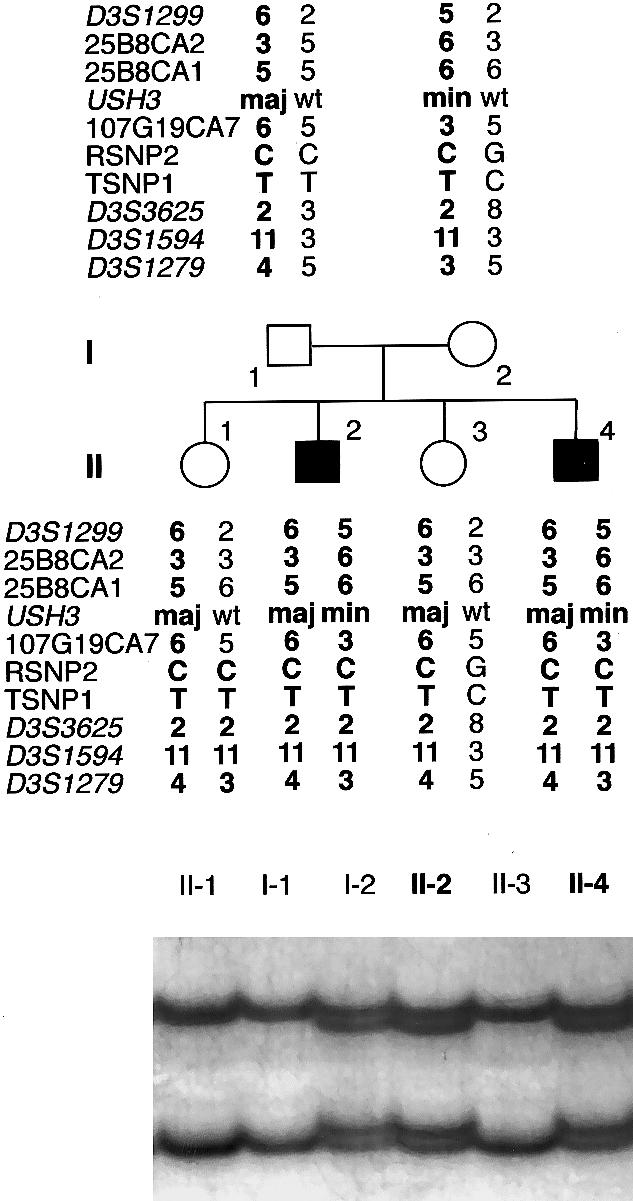
Pedigree of a Finnish family with USH3, segregating a paternal Finmajor mutation (maj)—c.300T→G—and a maternal Finminor mutation (min)—c.131T→A. Alleles at seven microsatellite markers, two SNPs, and USH3 are shown; wt = wild type. The chromosomes assumed to carry the disease allele are in boldface type. The father carries a conserved ancestral haplotype, whereas the mother’s disease-associated haplotype shares alleles at the RSNP2–D3S1594 segment only. A recombination in the maternal chromosome of individual II-1 excludes the segment RSNP2–D3S1279 as the site of the USH3 mutation. At the bottom is the result of an SSCP analysis used in the detection of the Finminor mutation. A mobility shift caused by the mutation was detected in individuals I-2, II-2, and II-4.
Identification of the USH3 Gene
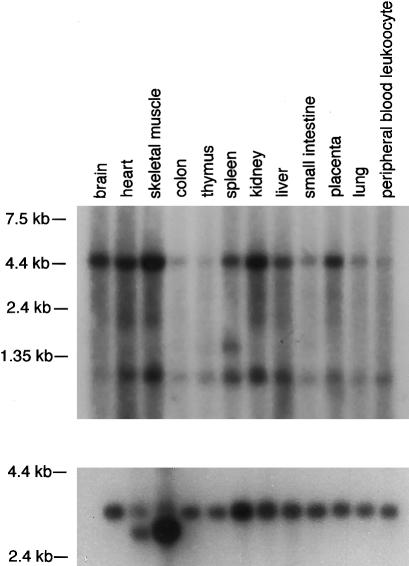
We assembled database-derived RP11-251C9 sequences between 107G19CA7 and 25B8CA2. BLAST analyses of the newly constructed 133-kb contig revealed 11 ESTs (data not shown). Starting from a retina-derived EST 35f2 and exons predicted in its vicinity, a contiguous cDNA sequence of 1,444 bp was obtained by RT-PCR of human retina-derived total RNA (a gift from F. P. M. Cremers) and by PCR of Marathon Ready Human Retina cDNA. Sequence analysis suggested a 360-bp ORF comprising four exons that span ∼18 kb at the genomic level. By comparison of the genomic sequence with the USH3 cDNA sequence, the exon-intron boundaries and their flanking sequences were confirmed and were in agreement with the consensus GT-AG splicing rule. A splice variant, USH3 isoform b, comprising an additional 87-bp exon (1b) with an in-frame stop codon between exons 1 and 2 was also detected. The first in-frame ATG codon in exon 1 conforms with the Kozak motif (Kozak 1987). No splice acceptor was found at the 5′ end of exon 1, suggesting that this exon is the first. There is a 5′ UTR of ⩾392 bp and a 3′ UTR of ⩾692 bp. Computational promoter analysis revealed five possible promoters for USH3. The putative promoter regions lack TATA boxes but include CAAT and GC boxes. Three alternative polyadenylation signals were found, at 156 bp, 764 bp, and 3,568 bp downstream of the termination codon, consistent with both the northern blot analysis showing three transcripts, of 4.5 kb, 1.5 kb, and 1.0 kb, respectively, and expression in many adult-human tissues (fig. 3). Expression was also detected in adult-human retina, by RT-PCR. Only human lymphoblasts did not show a signal, as confirmed by both RT-PCR and northern blotting.
Figure 3.
USH3 expression. Northern analysis of an adult-human multiple-tissue blot was performed by use of a probe comprising the coding region of USH3. Two signals, of ∼4.5 kb and ∼1.0 kb, are detected in all tissues. In addition, a weak signal, of ∼1.5 kb, is detected in spleen mRNA. The blot was hybridized with a β-actin probe as a control (bottom).
The analysis with the Celera Assembled and Annotated Mouse Genome indicated several genomic fragments highly homologous to exons 2 and 3 of USH3. In situ hybridization with [35S]-labeled riboprobes corresponding to the orthologous mouse sequence showed a low-level, ubiquitous expression in several mouse tissues (data not shown).
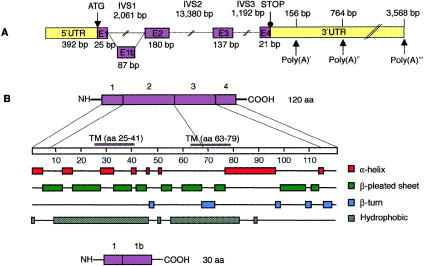
The candidate gene USH3 encodes a novel, putative transmembrane protein of 120 amino acids with a calculated molecular mass of 13.4 kD (fig. 4). It shows no homology to previously known genes or proteins. Bioinformatics analyses suggest a cytosolic amino terminus, two helical transmembrane domains (residues 25–41 and 63–79), and a putative endoplasmic-reticulum membrane–retention signal, TKGH, in the carboxy terminus.
Figure 4.
Predicted schematic structure and regions of secondary structure in the human USH3 gene and in USH3 protein. A, The five exons of USH3, depicted by numbered boxes (E1–E4). The lengths of the exons are shown below the boxes, and those of the three separating introns are shown above the lines. The translation-initiation site and the first stop codon are indicated. Three polyadenylation signals (Poly(A)) and their predicted locations downstream of the termination codon are indicated by arrows. B, One-hundred-twenty-amino-acid protein, encoded by USH3, with two predicted transmembrane domains, at residues 25–41 and 63–79. The combination of putative secondary structures, such as α-helices, β-pleated sheets, and β-turns, features possible functional protein domains. An alternatively spliced transcript (bottom) of USH3 predicts a 30-amino-acid protein.
USH3 Mutations
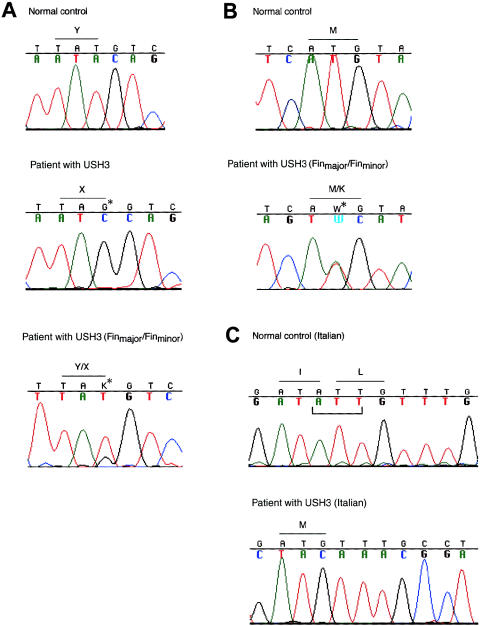
We identified three causative mutations in Finnish and Italian patients with USH3 (fig. 5). The Finnish founder mutation is a nonsense mutation, c.300T→G in exon 3 (Y100X, or Finmajor), predicting the truncation of 21 amino acids. Finmajor was found in 52 patients with USH3 who were homozygous for a conserved ancestral haplotype, whereas all 36 parents analyzed and 26 of 45 unaffected siblings were heterozygous for the mutation. The second Finnish mutation is a c.131T→A transversion at codon 44 in exon 2, resulting in an amino acid substitution of lysine for methionine (M44K, or Finminor). In two Finnish families, four patients are compound heterozygous for Finmajor and Finminor. These patients have inherited the common ancestral haplotype, from one parent, and a partly distinctive haplotype, from the other parent (fig. 2). The Finminor mutation originates from the same region in eastern Finland where USH3 is known to be prevalent. In a consanguineous Italian family with USH3 showing linkage to the 3q21-q25 markers (Gasparini et al. 1998), we detected a homozygous 3-bp deletion, c.231–233delATT in exon 3, resulting in the substitution of one methionine for isoleucine and leucine at codons 77 and 78.
Figure 5.
Detection of three different mutations segregating in families with USH3. A, Sequence analysis of a genomic PCR fragment comprising exon 3 in a normal control, in a patient with USH3 who is homozygous for the ancestral Finmajor mutation—c.300T→G (resulting in Y100X)—in a heterozygous carrier, and in a compound heterozygous patient (Finmajor/Finminor). Positions of mutated nucleotides are indicated by arrows and asterisks. B, Finnish mutation Finminor—c.131T→A (resulting in M44K) in exon 2: sequence chromatograms of a control and of a compound-heterozygous patient with USH3 (Finmajor/Finminor). C, Sequence chromatograms of a control, of a homozygous patient, and of a heterozygous carrier, representing the Italian mutation—a 3-bp deletion, c.231–233delATT—in exon 3. The deletion results in the substitution of one methionine for isoleucine and leucine. The deleted nucleotides are indicated below the normal control sequence.
By direct sequencing (in the case of Finmajor and the Italian mutation, c.231–233delATT) and by SSCP analysis (in the case of Finminor), none of either 100 anonymous blood donors from southern and western Finland or 51 grandparents in families from the CEPH were found to carry any of the three mutant alleles. Of 100 anonymous blood donors from eastern Finland, 1 was heterozygous for Finmajor. This was expected, because USH3 is clustered in eastern Finland (Pakarinen et al. 1995).
Discussion
The association of RP with sensorineural deafness is named “USH.” More than 60 genes responsible for RP and for related disorders have been identified (Retnet), and, to date, ∼20 genes for nonsyndromic deafness are known. Products of RP genes include phototransduction proteins, photoreceptor structural proteins, transcription factors, and proteins involved in metabolism of the photoreceptor and of the retinal pigment epithelium. The recent identification of several deafness genes by molecular genetic studies has enabled the investigation of the molecular basis of normal and pathological auditory function. The genes encode proteins that have a role in hair-cell transduction, in ionic homeostasis in the cochlear duct, and in integrity of the tectorial membrane (Steel and Kros 2001).
Five genes underlying USH have been identified in previous studies. Mutations of myosin VIIA cause nonsyndromic deafness as well as USH1B (Weil et al. 1995; Liu et al. 1997a, 1997b). The shaker-1 mice with myo7a mutations show progressive disorganization of the stereocilia bundle (Self et al. 1998). Although there is no retinal degeneration in shaker-1 mice, abnormal accumulation of opsin in the connecting cilium of photoreceptors and a failure of melanosome transport in the retinal pigment epithelium have been detected (Liu et al. 1998, 1999; Liu and Williams 2001). It is thought that harmonin interacts with myosin VIIA in the sensory cells of the inner ear, and harmonin mutations underlie USH1C as well as autosomal recessive nonsyndromic deafness DFNB18 (Verpy et al. 2000). Surprisingly, no harmonin expression was detected in neonatal-mouse eyes (Verpy et al. 2000), whereas anti-harmonin staining was found in portions of the developing-human eyes (Bitner-Glindzicz et al. 2000). Mutations in two novel cadherin-related genes—CDH23 and PCDH15—are responsible for USH1D and USH1F, respectively (Ahmed et al. 2001; Bolz et al. 2001; Bork et al. 2001). Disorganized stereocilia in the corresponding deaf-mouse mutants waltzer (Di Palma et al. 2001) and Ames waltzer (Alagramam et al. 2001) indicate that cadherins are involved in either the lateral links or the tip links that join adjacent stereocilia, although no gross retinal pathology has been detected in these mouse models. Humans with CDH23 mutations show variable retinal symptoms or, as in autosomal recessive nonsyndromic deafness DFNB12, no retinal phenotype at all (Bolz et al. 2001; Bork et al. 2001). USH2A, as well as nonsyndromic RP, is caused by mutations in usherin, which encodes a putative extracellular-matrix protein (Eudy et al. 1998; Rivolta et al. 2000).
Our results demonstrate that a gene encoding a 120-amino-acid protein with two predicted transmembrane domains and no homology to previously known proteins underlies USH3. In contrast to previously characterized USH genes that show distinct expression patterns in the sensory structures of the inner ear and of the retina, USH3 shows ubiquitous expression in several tissues. However, two genes involved in nonsyndromic X-linked RP—RP2 and RPGR—are also ubiquitously expressed but, nonetheless, have an important and specific role in the retina (Roepman et al. 1996; Schwahn et al. 1998). Patients with USH3 are born with normal hearing and vision, and the progressive sensory defects start later in life, leading to profound hearing loss, variable vestibular dysfunction, and severe RP with nyctalopia, constriction of the visual fields, and loss of central visual acuity.
Previously, we assigned, by linkage, a USH3 locus to 3q21-q25 and established the existence of a USH subtype characterized by postlingual, progressive hearing loss (Sankila et al. 1995). That cases of USH3 comprise ∼40% of all cases of USH in Finland has been explained by a founder effect. Families with USH showing linkage to the USH3 region have also been reported in the United States and Sweden (Kimberling et al. 1995), Spain (Espinos et al. 1998), Israel (Adato et al. 1999), and Italy (Gasparini et al. 1998). Using the significant linkage disequilibrium observed in the Finnish families with USH3, we were able to refine the USH3 critical region to 250 kb (Joensuu et al. 2000). The entire segment was subcloned, sequenced, and assembled, resulting in a 207-kb–sequence contig with 30 human ESTs. Three genes (NOPAR, UCRP, and H963)—two of which were novel—were identified and excluded as candidates, by sequencing. Of these, UCRP is probably an unprocessed pseudogene; however, interestingly, NOPAR shows significant homology to a thyroid-hormone receptor–associated protein, TRAP230 (Ito et al. 1999). The candidate gene that we identified, USH3, lies outside the initial critical region defined by haplotype analyses. This is explained by the fact that two mutations instead of the expected one founder mutation were found to segregate in the Finnish families with USH3. The Finnish founder mutation, c.300T→G in exon 3, was found in all USH3 chromosomes with the Finnish ancestral haplotype. It leads to a premature stop codon, which most probably affects mRNA stability or produces a truncated protein. In two Finnish families, patients were heterozygous for Finmajor and a c.131T→A transversion resulting in an amino acid substitution of lysine for methionine. In an Italian family with USH3, a homozygous 3-bp deletion, c.231–233delATT in exon 3, was detected. Among 200 Finnish controls and 51 individuals from the CEPH families, 1 Finnish individual was, as expected, heterozygous for the Finmajor mutation, whereas neither of the other two mutations was detected. Although the biochemical mechanisms by which USH3 causes retinal and cochlear disease remain to be elucidated, the identification of USH3 will already enable exact and even presymptomatic diagnosis of the disease.
Acknowledgments
We thank the patients and their families for cooperation, Ms. Sinikka Lindh for collecting the Finnish USH3 samples, Dr. Pertti Sistonen for providing the Finnish control samples, Dr. Angelo Notarangelo for preparing the Italian USH3 cell lines, Drs. Robert Chadwick and Lars Paulin for sequencing assistance, and Dr. Jose Dieguez for technical and computational advice and helpful discussions. This study was supported by the Ulla Hjelt Fund, the Finnish Eye and Tissue Bank Foundation, the Finnish Eye Foundation, Finnish State grants TYH8222 and TYH9235, the Academy of Finland, National Institutes of Health grant CA16058, and Telethon grant E.813.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Baylor College of Medicine Search Launcher, http://searchlauncher.bcm.tmc.edu/ (for MatInspector/TRANSFAC and Neural Network Promoter Input programs)
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Celera Publication Site, http://publication.celera.com/ (for Assembled and Annotated Mouse Genome)
- Expressed Sequence Tags Database, http://www.ncbi.nlm.nih.gov/dbEST/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for 207-kb contig sequence [accession number AF388363], BAC clones RP11-385G14 [accession number AC011103] and RP11-251C9 [accession number AC020636], human NOPAR coding sequence [accession number AF388364], human NOPAR2 coding sequence [accession number AF388365], human UCRP putative pseudogene sequence [accession number AF388367], human HOPA coding sequence [accession number AF132033], human H963 coding sequence [accession number AF002986], human TRAP230 mRNA [accession number NM_005120], human KIAA0001 coding sequence [accession number NM_014879], EST 35f2 sequence [accession number W27577], and cDNA of USH3 [accession number AF388366] and of a splice variant, USH3 isoform b [accession number AF388368], of USH3)
- Genome Survey Sequences Database, http://www.ncbi.nlm.nih.gov/dbGSS/
- Hereditary Hearing Loss Homepage, http://www.uia.ac.be/dnalab/hhh/ (for loci accounting for subtypes of USH)
- High-Throughput Genomic Sequences, http://www.ncbi.nlm.nih.gov/HTGS/
- Human Cancer Genetics BioInformatics at the Ohio State University, http://pandora.med.ohio-state.edu/HINT/ (for HINT database)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for USH1B [MIM 276903], USH1C [MIM 276904], USH1D [MIM 601067], USH1F [MIM 602083], and USH2A [MIM 276901])
- Phred/Phrap/Consed System Home Page, The, http://www.phrap.org/ (for Phred, Phrap, and Consed sequence-assembly programs)
- Primer3 Software Distribution, http://www-genome.wi.mit.edu/genome_software/other/primer3.html
- PSORT WWW Server, http://psort.nibb.ac.jp/ (for PSORT II)
- RepeatMasker Web Server, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker (for RepeatMasker2)
- RetNet: Retinal Information Network, http://www.sph.uth.tmc.edu/Retnet/ (for genes responsible for RP and related disorders)
- SignalP V1.1 World Wide Web Prediction Server, http://www.cbs.dtu.dk/services/SignalP/
- TIGR Human Gene Index, http://www.tigr.org/tdb/hgi/
- TMpred: Prediction of Transmembrane Regions and Orientation, http://www.ch.embnet.org/software/TMPRED_form.html
- UK Human Genome Mapping Project Resource Centre, http://www.hgmp.mrc.ac.uk/ (for NIX and PIX program packages)
- University of Washington Genome Center, http://www.genome.washington.edu/UWGC/ (for GASP software package)
References
- Adato A, Kalinski H, Weil D, Chaib H, Korostishevsky M, Bonne-Tamir B (1999) Possible interaction between USH1B and USH3 gene products as implied by apparent digenic deafness inheritance. Am J Hum Genet 65:261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Riazuddin S, Wilcox ER (2001) Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet 69:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP (2001) The mouse Ames Waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet 27:99–102 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83 [DOI] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, Hussain K, Furth-Lavi J, Cosgrove KE, Shepherd RM, Barnes PD, O'Brien RE, Farndon PA, Sowden J, Liu XZ, Scanlan MJ, Malcolm S, Dunne MJ, Aynsley-Green A, Glaser B (2000) A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet 26:56–60 [DOI] [PubMed] [Google Scholar]
- Boguski MS, Schuler GD (1995) ESTablishing a human transcript map. Nat Genet 10:369–371 [DOI] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del C-Salcedó Cabrera M, Vila MC, Molina OP, Gal A, Kubisch C (2001) Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet 27:108–112 [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, et al (2001) Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet 68:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Palma F, Holme RH, Bryda EC, Belyantseva IA, Pellegrino R, Kachar B, Steel KP, Noben-Trauth K (2001) Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat Genet 27:103–107 [DOI] [PubMed] [Google Scholar]
- Espinos C, Najera C, Millan JM, Ayuso C, Baiget M, Perez-Garrigues H, Rodrigo O, Vilela C, Beneyto M (1998) Linkage analysis in Usher syndrome type I (USH1) families from Spain. J Med Genet 35:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao SF, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayoso C, Cremers C, Davenport S, Moller C, Talmadge CB, Beisel KW, Tamayo M, Morton CC, Swaroop A, Kimberling WJ, Sumegi J (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 280:1753–1757 [DOI] [PubMed] [Google Scholar]
- Gasparini P, De Fazio A, Croce AI, Stanziale P, Zelante L (1998) Usher syndrome type III (USH3) linked to chromosome 3q in an Italian family. J Med Genet 35:666–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris NL (1997) Genotator: a workbench for sequence annotation. Genome Res 7:754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell 3:361–370 [DOI] [PubMed] [Google Scholar]
- Jacobs KA, Collins-Racie LA, Colbert M, Duckett M, Golden-Fleet M, Kelleher K, Kriz R, LaVallie ER, Merberg D, Spaulding V, Stover J, Williamson MJ, McCoy JM (1997) A genetic selection for isolating cDNAs encoding secreted proteins. Gene 198:289–296 [DOI] [PubMed] [Google Scholar]
- Joensuu T, Blanco G, Pakarinen L, Sistonen P, Kääriäinen H, Brown S, Chapelle A, Sankila EM (1996) Refined mapping of the Usher syndrome type III locus on chromosome 3, exclusion of candidate genes, and identification of the putative mouse homologous region. Genomics 38:255–263 [DOI] [PubMed] [Google Scholar]
- Joensuu T, Hämäläinen R, Lehesjoki A-E, de la Chapelle A, Sankila E-M (2000) A sequence-ready map of the Usher syndrome type III critical region on chromosome 3q. Genomics 63:409–416 [DOI] [PubMed] [Google Scholar]
- Kimberling WJ, Weston MD, Moller C, van Aarem A, Cremers CW, Sumegi J, Ing PS, Connolly C, Martini A, Milani M, Tamayo ML, Bernal J, Greenberg J, Ayuso C (1995) Gene mapping of Usher syndrome type IIa: localization of the gene to a 2.1-cM segment on chromosome 1q41. Am J Hum Genet 56:216–223 [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15:8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J (2000) Gene index analysis of the human genome estimates approximately 120,000 genes. Nat Genet 25:239–240 [DOI] [PubMed] [Google Scholar]
- Liu X, Ondek B, Williams DS (1998) Mutant myosin VIIa causes defective melanosome distribution in the RPE of shaker-1 mice. Nat Genet 19:117–118 [DOI] [PubMed] [Google Scholar]
- Liu X, Udovichenko IP, Brown SD, Steel KP, Williams DS (1999) Myosin VIIA participates in opsin transport through the photoreceptor cilium. J Neurosci 19:6267–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Mburu P, Kendric-Jones J, Cope MJ, Steel KP, Brown SD (1997a) Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet 16:188–190 [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD (1997b) Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet 17:268–269 [DOI] [PubMed] [Google Scholar]
- Liu X, Williams DS (2001) Coincident onset of expression of myosin VIIa and opsin in the cilium of the developing photoreceptor cell. Exp Eye Res 72:351–355 [DOI] [PubMed] [Google Scholar]
- Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S (1994) Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001–KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res 1:47–56 [DOI] [PubMed] [Google Scholar]
- Pakarinen L, Sankila E-M, Tuppurainen K, Karjalainen S, Kääriäinen H (1995) Usher syndrome type III (USH3): the clinical manifestations in 42 patients. Scand J Logoped Phoniatr 20:141–150 [Google Scholar]
- Pakarinen L, Tuppurainen K, Laippala P, Mäntyjärvi M, Puhakka H (1996) The ophthalmological course of Usher syndrome type III. Intl Ophthalmol 19:307–311 [DOI] [PubMed] [Google Scholar]
- Philibert RA, Winfield SL, Damschroder-Williams P, Tengstrom C, Martin BM, Ginns EI (1999) The genomic structure and developmental expression patterns of the human OPA-containing gene (HOPA). Hum Genet 105:174–178 [DOI] [PubMed] [Google Scholar]
- Rivolta C, Sweklo EA, Berson EL, Dryja TP (2000) Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet 66:1975–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepman R, van Duijnhoven G, Rosenberg T, Pinckers AJ, Bleeker-Wagemakers LM, Bergen AA, Post J, Beck A, Reinhardt R, Ropers HH, Cremers FP, Berger W (1996) Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum Mol Genet 5:1035–1041 [DOI] [PubMed] [Google Scholar]
- Sankila EM, Pakarinen L, Kääriäinen H, Aittomäki K, Karjalainen S, Sistonen P, de la Chapelle A (1995) Assignment of an Usher syndrome type III (USH3) gene to chromosome 3q. Hum Mol Genet 4:93–98 [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, Pinckers AJ, Fundele R, Rosenthal A, Cremers FP, Ropers HH, Berger W (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet 19:327–332 [DOI] [PubMed] [Google Scholar]
- Self T, Mahony M, Fleming J, Walsh J, Brown SD, Steel KP (1998) Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development 125:557–566 [DOI] [PubMed] [Google Scholar]
- Smith RJ, Berlin CI, Hejtmancik JK, Keats BJ, Kimberling WJ, Lewis RA, Moller CG, Pelias MZ, Tranebjaerg L, Usher Syndrome Consortium (1994) Clinical diagnosis of the Usher syndromes. Am J Med Genet 50:32–38 [DOI] [PubMed] [Google Scholar]
- Steel KP, Kros CJ (2001) A genetic approach to understanding auditory function. Nat Genet 27:143–149 [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C (2000) A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet 26:51–55 [DOI] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, Kelley PM, Kimberling WJ, Wagenaar M, Levi-Acobas F, Larget-Piet D, Munnich A, Steel KP, Brown SDM, Petit C (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374:60–61 [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Green DK (1990) In situ hybridization and three-dimensional reconstruction of serial sections. In: Copp AJ, Cockroft DL (eds) Postimplantation mammalian embryos: a practical approach. IRL press, Oxford, pp 151–171 [Google Scholar]
- Zhuo D, Zhao WD, Wright FA, Yang H-Y, Wang J-P, Sears R, Baer T, Kwon D-H, Gordon D, Gibbs S, Dai D, Yang Q, Spitzner J, Krahe R, Stredney D, Stutz A, Yuan B (2001) Assembly, annotation, and integration of UNIGENE clusters into the Human Genome Draft. Genome Res 11:904–918 [DOI] [PMC free article] [PubMed] [Google Scholar]