Abstract
To better understand the pathogenetics of pseudoxanthoma elasticum (PXE), we performed a mutational analysis of ATP-binding cassette subfamily C member 6 (ABCC6) in 122 unrelated patients with PXE, the largest cohort of patients yet studied. Thirty-six mutations were characterized, and, among these, 28 were novel variants (for a total of 43 PXE mutations known to date). Twenty-one alleles were missense variants, six were small insertions or deletions, five were nonsense, two were alleles likely to result in aberrant mRNA splicing, and two were large deletions involving ABCC6. Although most mutations appeared to be unique variants, two disease-causing alleles occurred frequently in apparently unrelated individuals. R1141X was found in our patient cohort at a frequency of 18.8% and was preponderant in European patients. ABCC6del23–29 occurred at a frequency of 12.9% and was prevalent in patients from the United States. These results suggested that R1141X and ABCC6del23–29 might have been derived regionally from founder alleles. Putative disease-causing mutations were identified in ∼64% of the 244 chromosomes studied, and 85.2% of the 122 patients were found to have at least one disease-causing allele. Our results suggest that a fraction of the undetected mutant alleles could be either genomic rearrangements or mutations occurring in noncoding regions of the ABCC6 gene. The distribution pattern of ABCC6 mutations revealed a cluster of disease-causing variants within exons encoding a large C-terminal cytoplasmic loop and in the C-terminal nucleotide-binding domain (NBD2). We discuss the potential structural and functional significance of this mutation pattern within the context of the complex relationship between the PXE phenotype and the function of ABCC6.
Introduction
Pseudoxanthoma elasticum (PXE [MIM 177850 and MIM 264800]) is a heritable disorder characterized by dermal, vascular, and ocular lesions that result from the accumulation of morphologically abnormal and mineralized elastic fibers in these tissues (Uitto and Shamban 1987). The skin manifestations are the most prevalent characteristic of PXE and generally are the first physical signs of the developing disorder. These skin changes are often detected during childhood or adolescence, and they progress slowly and unpredictably during adulthood. The accumulation of abnormal calcified elastic fibers in the middermis produces typical skin lesions, which consist of yellowish papules and plaques and of laxity with loss of elasticity. These lesions are primarily seen on the neck, axilla, antecubital fossa, groin, and periumbilical area (Uitto and Shamban 1987; Neldner 1988; Uitto et al. 1998). The diagnosis of PXE is confirmed histologically by the demonstration, in biopsies of lesional skin, of fragmented calcified elastic fibers, by use of the Verhoeff–van Gieson and von Kossa stains. Similar elastic fiber changes within the internal elastic lamina of medium-sized arteries are most frequently associated with intimal fibroplasia and early onset of peripheral vascular occlusive disease (Nishida et al. 1990; Lebwohl et al. 1993). Retinal angioid streaks, the other hallmark of PXE, result from fragmentation and calcification of the elastic component of the Bruch’s membrane. Angioid streaks are associated with subretinal neovascularization and hemorrhage, which can lead to disciform scarring and severe loss of central vision (Weenink et al. 1996). In addition to elastic fiber alterations, the abnormal deposition of several other extracellular matrix components, such as collagen fibrils and proteoglycan, has also been demonstrated in tissue from patients with PXE (Baccarani-Contri et al. 1996; Passi et al. 1996).
PXE was first observed in 1881 (Rigal 1881) but was not formally identified as a heritable disorder until 1896 (Darier 1896). Despite an abundant literature after these initial reports, particularly in the last two decades, the genetic basis of PXE remained entirely unknown until recently. The PXE gene was first mapped to a 3–5-cM locus at 16p13.1 (Struk et al. 1997; van Soest et al. 1997). This locus was then refined to a region of ∼500 kb (Le Saux et al. 1999; Cai et al. 2000) that contains five candidate genes. Mutational analysis of all these candidate genes led several groups to identify the first mutations responsible for PXE in a gene that encodes an ATP-binding cassette (ABC) subfamly C member 6 (ABCC6) transporter, also known as “multidrug resistance–associated protein 6” or MRP6 (Bergen et al. 2000; Germain et al. 2000; Le Saux et al. 2000; Ringpfeil et al. 2000; Struk et al. 2000). ABCC6 consists of 31 exons spanning ∼75 kb and encodes a 165-kD transmembrane protein. The ABCC6 gene is predominantly expressed in both the liver and kidney. In a recent study, ABCC6 has been localized to the basolateral surface of hepatocytes (Madon et al. 2000). Although ABCC6 is a typical ABC transporter, we do not yet know what molecule(s) this transmembrane protein transports. Similarly, the relationship between aberrant ABCC6 activity and extracellular changes in PXE tissues remains to be elucidated.
To date, a limited number of apparently recurrent and unique mutations of ABCC6 have been identified in unrelated patients with PXE. To ascertain whether these preliminary findings are indicative of a larger spectrum of ABCC6 mutations and to determine, in particular, whether the nature of these mutations may provide any indication of the relationship of ABCC6 function to the PXE phenotype, we performed a mutational analysis (described in the present article) of ABCC6 in 122 unrelated patients with PXE, the largest cohort of such patients yet studied, as a prerequisite to an elucidation of the pathogenic mechanisms of PXE.
Patients, Material, and Methods
Patients
DNA samples from patients examined in the present study were collected in the United Kingdom by F.M.P., in the United States and South Africa by S.T. and L.B. (on behalf of PXE International, Inc.), in Italy and Germany by I.P.R., and in Belgium by A.D.P. The diagnosis of PXE in all unrelated patients was consistent with consensus criteria reported elsewhere (Christiano et al. 1992). The affected status of a patient was determined on the basis of examinations of dermal lesions and the presence of ocular and/or cardiovascular findings, and the status was confirmed by the observation of calcified elastic fibers during histologic examination of a skin biopsy specimen treated with von Kossa stain. Genomic DNA obtained from a panel of 100 unrelated and unaffected individuals was used for control purposes. The panel comprised white individuals with origins in Europe or the United States. All blood samples were obtained from unaffected individuals and from patients who gave informed consent according to protocols approved by the appropriate institutional review board (or equivalent committee for human subjects in countries other than the United States). Genomic DNA was isolated from EDTA-anti-coagulated whole blood and was purified with a QIAamp blood kit (Qiagen), according to the manufacturer’s instructions. Purified DNA was stored at −80°C in a mixture (pH 8.0) of 1 mM Tris-HCl and 0.1 mM EDTA.
PCR and DNA Sequence Analysis
Intron-derived primers for PCR amplification of all 31 ABCC6 exons were synthesized using sequence information obtained from BAC clone CIT987SK-A-962B4 (GenBank accession number U91318). Intron/exon boundaries were deduced from information available in the Institute for Genome Research database and by comparison with the ABCC6 cDNA (GenBank accession number NM_001171). Typical PCR reactions, which have been described elsewhere (Le Saux et al. 1999), were performed in the presence of [32P]-labeled primers in a 9700 thermocycler (PerkinElmer). PCR products were 150–550 bp and included complete intron/exon boundaries. Radioactive PCR products were analyzed either by single-stranded conformation polymorphism (SSCP) analysis (Orita et al. 1989) using MDE-PAGE (BioWhittaker Molecular Applications), according to the manufacturer’s instructions, and by conformation-sensitive gel electrophoresis (CSGE) analysis, as described elsewhere (Ganguly et al. 1993). DNA conformers were eluted in water from gel slices and were reamplified and sequenced with the same primers used to generate the PCR products. Alternatively, PCR products were amplified directly from genomic DNA before sequencing. DNA sequence analysis was performed using ABI BigDye terminator cycle sequencing with an ABI 310 automated DNA sequencer. The information generated by the sequencer was analyzed using the ABI Prism genetic analyzer software, version 1.2.2. The Sequencher 3.1 program was used to identify variations between the sequences of putative mutations and control DNA.
Definition of Disease-Associated Alleles
For the purposes of this study, we have defined a sequence variant as phenotype-modifying (i.e., causative of PXE) by using criteria described elsewhere for defining a nucleotide sequence variant as disease causing (Cotton and Scriver 1998): (1) ABCC6 sequence variants predicted to result in nonsense or splice-site alterations were categorized as disease-associated alleles. These sequence variants included premature stop codons, small and large insertions/deletions resulting in a frameshift, and sequence alterations within ABCC6 that had the potential to affect constitutive intron/exon splicing. To be considered as causative alleles, these sequence variants were required to be absent in a panel of 200 alleles from unaffected and unrelated control individuals. (2) Other potentially pathogenic nucleotide variants resulting in amino acid substitution were considered as disease-causing when (a) the substitution involved a conserved amino acid, (b) the variant was absent in a panel of 200 alleles of unaffected control individuals, and (c) the variant was shown to cosegregate with the PXE phenotype in available kindreds. Conserved amino acids were identified on the basis of an alignment of nine human ABCC proteins, performed with the ClustalW program. The term “mutation,” in the present article, refers to a putative disease-causing allele defined according to these criteria. All exonic and intronic changes that did not meet these criteria were regarded as neutral or silent variants.
Haplotype Analysis
Microsatellite markers described elsewhere (Le Saux et al. 1999) were used for high-resolution mapping of chromosome 16p13.1. Each microsatellite was amplified by PCR, using primers end-labeled with γ[32P]-ATP. The radiolabeled PCR products were analyzed by autoradiography after electrophoresis on 6% polyacrylamide gels containing 6 M urea, 37.5% formamide, and a buffer containing 0.09 M Tris, 0.09 M boric acid, and 0.01 M EDTA. Electrophoresis was conducted at 80 W for 4–6 h, and gels were then dried and exposed to Fuji X-ray film at −80°C.
PCR Characterization of Alu-Mediated Deletions
The characterization of large deletions involving exon 15 and exons 23–29 of ABCC6 was conducted using PCR amplification of the regions of interest, with primers positioned within flanking introns. The initial characterization of the deletions was performed with a combination of existing primers used to amplify the coding regions corresponding to exons 14, 16, 22, and 30. These primers were ABCC6Ex14m, 5′-AGG AAG CTG TTG CCA CAC at-3′; ABCC6Ex16a, 5′-GCG AGG AAG TGG GAC TTT CA-3′; ABCC6Ex21m, 5′-GCC TGA GGG TTA GGC ACA TA-3′; and ABCC6Ex30a, 5′-CAG GAC CCC TCC AGC TCT AA-3′. The primers used to determine the precise boundaries of these deletions were located 5′ and 3′ of Alu repeats in introns 14, 15, 22, and 29. The sequences of these primers were ABCC6IVS15a, 5′-TGC TGA GGT TGA GAA ACC CA–3′; ABCC6IVS14m, 5′-GGC TAA GCC ATG AGA GCT TT-3′; ABCC6IVS29a, 5′-CTG TAG GCA GGT CAT TCA AA-3′; and ABCC6IVS22m, 5′-TCC CCT AAA GAT GGA GAG AT-3′.
Results
Mutational Screening and Analysis
ABCC6 was screened in a cohort of 122 apparently unrelated patients with PXE from families with an autosomal recessive mode of inheritance of the PXE phenotype. Affected individuals originated from several European countries (Belgium, Germany, Holland, Italy, and the United Kingdom), South Africa, and the United States. All exons of ABCC6 from patients and available unaffected relatives were amplified by PCR. The PCR products were analyzed by a combination of SSCP, heteroduplex scanning, and direct sequencing to detect single-base substitutions and small insertions/deletions. In this cohort of patients with PXE, we found a total of 36 different mutations according to our strict criteria (see Definition of Disease-Associated Alleles section). Among these, 8 mutations had been described elsewhere, and 28 mutations were novel. Mutations identified in the present study, together with previously published mutations, bring the total number of known PXE mutations to 43. This information is summarized in table 1.
Table 1.
ABCC6 Mutations in a Cohort of Patients with PXE
|
Change in |
|||||
| Amino Acid | Nucleotide | Statusa | Origin(s)b | Exon(s)c | Reference(s) |
| … | 179–195del | ht | Belgium | 2 | Present study |
| … | 938–939insT | ch, ht | SA, UK | 8 | Present study |
| N411K | 1233T→G | ht | US | 10 | Present study |
| A455P | 1363G→C | Nd | Nd | 11 | Uitto et al. (2001) |
| R518Q | 1553G→A | ch, ht | Belgium | 12 | Present study, Uitto et al. (2001) |
| F568S | 1703T→C | ch | US | 13 | Present study |
| … | ABCC6del15 | hm | SA | 15 | Present study |
| … | 1944del22 | ht | Holland | 16 | Bergen et al. (2000) |
| … | 1995delG | ht | Germany | 16 | Present study |
| L673P | 2018T→C | ch | SA | 16 | Present study |
| R765Q | 2294G→A | ht | Germany | 18 | Present study |
| Y768X | 2304C→A | ch, ht | SA | 18 | Present study |
| … | 2322delC | ht | US | 18 | Present study |
| … | 2542delG | Nd | Nd | 19 | Uitto et al. (2001) |
| … | IVS21+1G→T | ch | US, Germany | i-21 | Present study, Uitto et al. (2001) |
| R1030X | 3088C→T | ht | SA, UK | 23 | Present study |
| R1114P | 3341G→C | hm | UK | 24 | Present study |
| S1121W | 3362C→G | ch | Germany | 24 | Present study |
| R1138W | 3412C→T | hm | Nd | 24 | Ringpfeil et al. (2000) |
| R1138P | 3413G→C | ch | Germany | 24 | Present study |
| R1138Q | 3413G→A | ch | UK, US | 24 | Present study, Ringpfeil et al. (2000) |
| R1141X | 3421C→T | All | All | 24 | Present study and othersd |
| R1164X | 3490C→T | ch | Germany, UK | 24 | Ringpfeil et al. (2001) |
| G1203D | 3608G→A | ch | Germany | 25 | Present study |
| … | IVS26-1G→A | ch | Belgium | i-26 | Present study, Ringpfeil et al. (2000, 2001) |
| Q1237X | 3709C→T | ch | Belgium | 26 | Present study |
| … | 3775delT | ht, hm | SA, US, Holland | 27 | Present study, Bergen et al. (2000) |
| V1298F | 3892G→T | ht | US | 28 | Present study |
| T1301I | 3902C→T | ch | Belgium | 28 | Present study |
| G1302R | 3904G→A | hm | US | 28 | Present study |
| A1303P | 3907G→C | ch | Belgium | 28 | Present study |
| R1314W | 3940C→T | hm | US | 28 | Present study |
| R1314Q | 3941G→A | ch | Germany | 28 | Present study |
| G1321S | 3961G→A | ht | US | 28 | Present study |
| R1339C | 4015C→T | All | SA, US | 28 | Present study, Struk et al. (2000) |
| Q1347H | 4041G→C | hm | US | 28 | Present study |
| D1361N | 4081G→A | ch | Germany | 29 | Present study |
| … | 4104delC | ch | Belgium | 29 | Present study |
| R1398X | 4192C→T | ch | Belgium | 29 | Present study |
| … | ABCC6del23–29 | ch | US | 23–29 | Present study, Ringpfeil et al. (2001) |
| … | 4220insAGAA | ht | Holland | 30 | Bergen et al. (2000) |
| I1424T | 4271T→C | ht | US | 30 | Present study |
| … | ABCC6del | ht | Holland | all | Bergen et al. (2000) |
Nd = not determined; hm = homozygote; ht = heterozygote; ch = compound heterozygote.
UK = United Kingdom; US = United States; SA = South Africa; Nd = not determined.
Exon indicates which exon(s) contains the mutation or is deleted by the mutation; i- = the intron that contains the mutation.
Mutation-Detection Rate
The overall detection rate of disease-causing sequence variants within the 244 chromosomes that we analyzed was 63.9%. The detection rate was slightly lower (58.9%) when patients from known consanguineous and Afrikaner families, for which a founder effect is suspected (Torrington and Viljoen 1991), were not included. The mutation-detection rate was 54.3% for the European cohort, 64.9% for the United States cohort (table 2), and 85.3% in patients of Afrikaner origin. Within the Afrikaner population, the mutation-detection rate was higher, because most alleles appear to be identical by descent (O.L.S., K.B., C.S., C.T., E.W.J., L.B., S.T., D.L.V., and C.D.B., unpublished data). In our cohort of 122 patients, 104 (85.2%) individuals were found to have at least one disease-causing allele. Of these, two ABCC6 mutant alleles were detected in 52 patients, one allele was found in 52 patients, and no disease-associated allele could be identified in the 18 remaining subjects with PXE.
Table 2.
Frequencies of Mutant Alleles Found in a Cohort of 101 Unrelated Patients with PXE[Note]
|
Overall |
Europe |
United States |
||||
| Mutationa | No. of Alleles | Frequency(%) | No. of Alleles | Frequency(%) | No. of Alleles | Frequency(%) |
| R1141X | 38 | 18.8 | 33 | 28.4 | 3 | 4.1 |
| ABCC6del23–29 | 26 | 12.9 | 5 | 4.3 | 21 | 28.4 |
| IVS21+1G→T | 7 | 3.5 | 4 | 3.4 | 3 | 4.1 |
| G1302R | 4 | 2.0 | 0 | .0 | 4 | 5.4 |
| A1303P | 4 | 2.0 | 3 | 2.6 | 1 | 1.4 |
| R1314W | 3 | 1.5 | 0 | .0 | 3 | 4.1 |
| R518Q* | 3 | 1.5 | 1 | .9 | 1 | 1.4 |
| 3775delT* | 3 | 1.5 | 2 | 1.7 | 0 | .0 |
| R1138Q | 2 | 1.0 | 1 | .9 | 1 | 1.4 |
| V1298F | 2 | 1.0 | 0 | .0 | 2 | 2.7 |
| R1339C | 2 | 1.0 | 0 | .0 | 2 | 2.7 |
| Q1347H | 2 | 1.0 | 0 | .0 | 2 | 2.7 |
| 4104delC* | 2 | 1.0 | 1 | .9 | 0 | .0 |
| 179–195del | 1 | .5 | 1 | .9 | 0 | .0 |
| 938–939insT* | 1 | .5 | 0 | .0 | 0 | .0 |
| N411K | 1 | .5 | 0 | .0 | 1 | 1.4 |
| F568S | 1 | .5 | 0 | .0 | 1 | 1.4 |
| 1995delG | 1 | .5 | 1 | .9 | 0 | .0 |
| R765Q | 1 | .5 | 1 | .9 | 0 | .0 |
| 2322delC | 1 | .5 | 0 | .0 | 1 | 1.4 |
| R1030X* | 1 | .5 | 0 | .0 | 0 | .0 |
| R1114P | 1 | .5 | 1 | .9 | 0 | .0 |
| S1121W | 1 | .5 | 1 | .9 | 0 | .0 |
| R1138P | 1 | .5 | 1 | .9 | 0 | .0 |
| G1203D | 1 | .5 | 1 | .9 | 0 | .0 |
| IVS26−1G→A | 1 | .5 | 1 | .9 | 0 | .0 |
| Q1237X | 1 | .5 | 1 | .9 | 0 | .0 |
| W1241C | 1 | .5 | 1 | .9 | 0 | .0 |
| T1301I | 1 | .5 | 1 | .9 | 0 | .0 |
| R1314Q | 1 | .5 | 1 | .9 | 0 | .0 |
| D1361N | 1 | .5 | 1 | .9 | 0 | .0 |
| R1398X | 1 | .5 | 1 | .9 | 0 | .0 |
| G1321S | 1 | .5 | 0 | .0 | 1 | 1.4 |
| I1424T | 1 | .5 | 0 | .0 | 1 | 1.4 |
| ? | 83 | 41.1 | 52 | 44.8 | 26 | 35.1 |
| MDR | … | 58.9 | … | 55.2 | … | 64.9 |
Note.— Forty-two alleles from founder-derived and consanguineous families are not included in this table. The total number of alleles represented is 202; 116 alleles were of European origin; 74 were of US origin; and 12 originated elsewhere.
* = Allele also found in a group of 12 non-European/non–United States alleles; ? = unknown allele; MDR = mutation-detection rate.
PXE Mutations
The most-prevalent mutations detected in the ABCC6 gene were missense substitutions (21 [58.3%] mutations, see table 1). Other ABCC6 mutant alleles included five (13.9%) nonsense and six (16.7%) frameshift mutations. Among these, four mutations involved the deletion of a single nucleotide, one mutation was an insertion of thymidine, and one mutation corresponded to the deletion of 17 bp. Two other mutations were predicted to affect constitutive exon/intron splicing of ABCC6 pre-mRNA, and two large out-of-frame deletions of 1.6 kb and 16.4 kb were characterized (these mutations are described in detail in the sections below). Each of the two splice-site mutations and each of the two deletion mutations each accounted for 5.6% of all mutations detected in the present study. Although most of the mutations reported here appear to be unique, a few disease-causing variants have been found to occur frequently in apparently unrelated individuals; R1141X was found at a frequency of 18.8%, and ABCC6del23–29 occurred at a frequency of 12.9% (table 2) in our cohort of patients. Of a total of 244 chromosomes, 42 were not taken into account in these calculations, because they were derived from consanguineous families or from the founder-derived Afrikaner population. Most individuals with two mutant alleles were compound heterozygotes. Interestingly, alleles R1141X and ABCC6del23–29 were found to occur at different frequencies when patient origin was considered. R1141X was far more frequently represented in the European cohort (33 [28.4%] of 116 alleles) than in the U.S. cohort of patients with PXE (3 [4.1%] of 74). Also, this nonsense mutation was unequally distributed among the European countries considered. Indeed, 15 of 26 Belgians, 8 of 15 Germans, 3 of 12 British, 2 of 6 Dutch, and 1 of 1 Italian carried an R1141X allele in a homozygous, compound heterozygous, or heterozygous state. In the U.S. cohorts, three patients carried a single R1141X allele. Among the South African patients, 2 of the 5 who were of British ancestry had the nonsense mutation in the heterozygous or compound heterozygous state, and 2 of the 17 of the Afrikaner patients carried a heterozygous or compound heterozygous R1141X. In contrast to the latter mutation, ABCC6del23–29 was found to represent nearly one-third (21 [28.4%] of 74 alleles) of the United States alleles, whereas only 5 (4.3%) of 116 alleles analyzed among European patients with PXE were identified as ABCC6del23–29. The difference in frequency is statistically significant (Fisher’s exact test; P=.0004 and .0014, respectively). The frequency of individuals homozygous for these two frequently occurring disease-causing alleles (R1141X and ABCC6del23–29) was observed to be in Hardy-Weinberg equilibrium in the European and United States cohorts, respectively.
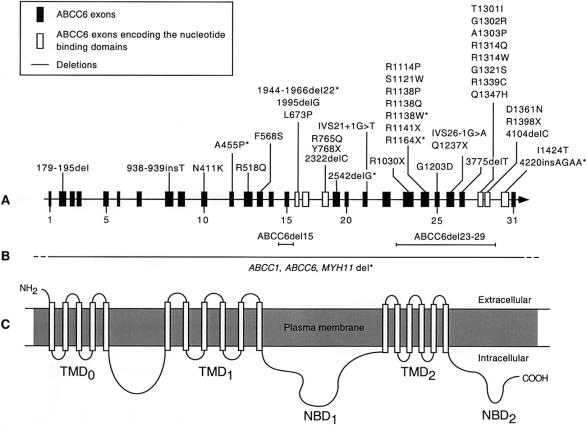
When previously published mutations and those reported in the present study are taken together (a total of 43 mutations), 36 substitutions and deletions (85.7%) were predicted to affect the C-terminal half of the protein. Fifteen mutations (35.7%) were notably clustered within the C-terminal nucleotide-binding domain (NBD2) (fig. 1), which is encoded by exons 28, 29, and 30. It is noteworthy that seven mutations (16.7%) were located in exon 24, and three of these mutations were predicted to influence the codon for a single residue, R1138. Remarkably, approximately one-third (12) of all mutations affected arginyl residues. Also, it is interesting to note that all but two mutations (F568S and G1203D) located in the last transmembrane domain were predicted to reside in cytoplasmic regions of ABCC6. Most individuals in whom two mutant alleles were identified were compound heterozygotes.
Figure 1.
Localization of mutations causing PXE in ABCC6. A, The positions of mutations presented as either nucleotide changes or amino acid substitutions are indicated with respect to the exon/intron structure of ABCC6. The asterisk indicates mutations that have been described only by others. Boxes represent exons drawn approximately to scale. The number of every fifth exon is indicated. Unshaded boxes show exons encoding elements of NBD1 and NBD2. B, Horizontal lines indicate three large deletions involving some or all ABCC6 exons. The horizontal line with dashes depicts a large deletion involving all of ABCC6, with deletion breakpoints both upstream and downstream of ABCC6. C, The predicted topology of ABCC6 contains three transmembrane domains: TMD0, TMD1, and TMD2, with five, six, and six membrane-spanning segments, respectively. NBD1 and NBD2 are also indicated. The various domains of ABCC6 are approximately aligned with the various exons encoding these regions.
Small and large deletions, insertions, and nonsense mutations (which accounted for 13 of the 36 mutations characterized in the present study) were all predicted to result in premature termination of translation and would therefore be pathogenic. Premature termination mutations frequently result in the nonsense-mediated decay (NMD) of mutant mRNA products and significantly reduce mutant transcript levels (Maquat 1996; Nagy and Maquat 1998; Frischmeyer and Dietz 1999). In some cases, however, premature termination mutations can escape NMD and result in a substantial amount of truncated proteins. These mutations are often located close to the end of the open reading frame (Nijbroek et al. 1995; Mickle et al. 1998; Nagy and Maquat 1998). Alternatively, premature termination mutations could result in exon skipping. Such consequences have been described in several inherited disorders, notably for cystic fibrosis, which is caused by mutations in the ABCC7 gene (Dietz et al. 1993; Hull et al. 1994; Messiaen et al. 1997). Because several premature termination mutations in ABCC6 are located near both the 5′ and 3′ ends of the gene, the consequences of premature termination mutations should therefore be experimentally assessed case by case. One nonsense mutation (R1141X) that we have previously characterized in a homozygous state was indeed associated with no detectable ABCC6 messenger in RNA samples obtained from skin fibroblasts (Le Saux et al. 2000), and the absence of any ABCC6 mRNA is probably caused by NMD and would result in a null allele. The consequence of the out-of-frame deletion ABCC6del23–29 was recently investigated (Ringpfeil et al. 2001). This mutation was shown to result in the deletion of 1,213 nucleotides of the mRNA, associated with a premature termination codon in exon 30 that probably results in a truncated protein. The splice-site mutations that we have identified were also likely to cause disease, given that they would probably result in either exon skipping or activation of a cryptic splice site that would lead to either a truncated protein or an aberrant one. The putative splice-site mutation 4041G→C is of particular interest, because it may result in aberrant mRNA splicing in addition to an amino acid substitution, (Q1347H). Indeed, the mutation occurring at the last nucleotide of exon 28 at the donor splice site lowered the splice-potential score (Shapiro and Senapathy 1987) from 92.1 to 78.6. Moreover, an identical mutation in ABCC7 (CFTR [GenBank accession number NM_000492]), which is responsible for cystic fibrosis, involved the same glutaminyl residue, Q1291H (3873G→C), at the last nucleotide of exon 20 of the ABCC7 gene and resulted in a glutaminyl-to-histidinyl residue substitution. RNA-based PCR analysis revealed that Q1291H in the ABCC7 gene was also a splice mutation. Both correctly and aberrantly spliced mRNAs were produced from the Q1291H allele. The incorrectly spliced product resulted from the use of a nearby cryptic splice site (Jones et al. 1992). We predict that a similar aberrant splice product could occur as a consequence of the Q1347H mutation in ABCC6.
All missense mutations presented in tables 1 and 2 were considered pathogenic, because they were not found in our control panel of 200 alleles and because they involved residues conserved among all nine human ABCC proteins.
Neutral Variants
Nucleotide changes in the ABCC6 gene that are predicted to result in amino acid substitutions and are categorized as neutral (non–disease-causing) variants are reported in table 3. These neutral variants did not reveal any clustering. Three of the neutral variants (K281E, I319V, and H632Q) could not be detected by SSCP or CSGE. These changes were identified only by nucleotide sequencing of PCR conformers that contain other silent changes (data not shown) or mutations. The frequencies of K281E, I319V, and H632Q in our PXE cohort and control panel were, consequently, not assessed. Most of the neutral variants were unique; however, two of these alleles (V614A and R1268Q) were found at 81.9% and 9.4%, respectively, in the cohort of patients with PXE.
Table 3.
Missense Neutral Variants Identified in the ABCC6 Gene in a Cohort of 122 Patients
|
Change in |
||||||
| Amino Acid | Nucleotide | Statusa | Origin(s)b | Exon(s) | No. of Alleles/PXE Chromosomes | No. of Alleles/Control Chromosomesc |
| G61D | 182G→A | ht | SA | 2 | 1/244 | 0/200 |
| G207R | 619G→A | ht | Belgium | 6 | 1/244 | 0/200 |
| R265G | 793A→G | ht | Belgium | 7 | 1/244 | 0/200 |
| K281Ed | 841A→G | ht, hm | SA | 8 | 5/8d | Nd |
| I319Vd | 955A→G | ht, hm | SA | 8 | 5/8d | Nd |
| N497K | 1489C→A | ht | Belgium | 12 | 1/244 | 0/200 |
| V614A | 1841T→C | ht, hm | All | 14 | 200/244 | 163/200 |
| H632Qd | 1896C→A | ht, hm | SA, Belgium | 15 | 17/24d | Nd |
| L953H | 2858T→A | ht | US | 22 | 1/244 | 0/200 |
| W1241C | 3723G→C | ht | Germany | 26 | 1/244 | 0/200 |
| R1268Q | 3803G→A | ht | All | 27 | 23/244 | 31/200 |
ht = heterozygote; hm = homozygote.
SA = South Africa.
Nd = not determined.
Neutral variant characterized by sequencing only.
Characterization of Two Alu-Mediated Deletions
ABCC6del23–29
A loss of heterozygosity of an R1141X allele was detected in three affected children of a two-generation family from the United States. These affected individuals seemed homozygous for R1141X; whereas the mother appeared to be heterozygous for this allele, and the father appeared to have two normal alleles. Haplotype analysis performed with 12 microsatellites on chromosome 16p13.1 (Le Saux et al. 1999) indicated, however, a heterozygous carrier status for the nonsense mutation of the three affected offspring. These results suggested that the affected offspring in this family had inherited a single maternal R1141X mutation and a paternally derived deletion involving exon 24, thereby reducing the nonsense mutation to hemizygosity in all three patients. Several polymorphic variants in exons 14 and 19, intron 25, and the ABCC6 3′ untranslated region (UTR) (Le Saux et al. 2000) were subsequently analyzed by SSCP to determine the extent of this deletion. The experiment revealed the deletion in this family to be identical to a recently reported Alu-mediated deletion of ABCC6 exons 23–29 in several apparently unrelated families (Ringpfeil et al. 2001). PCR analysis, using intron 21 and 30 primers flanking the deletion breakpoints, confirmed the paternal origin of this 16,415-bp deletion within this pedigree (data not shown). We have not found this deletion mutation in our control panel of 200 alleles. Moreover, we observed a frequency of this deletion of 12.9% in our cohort of patients with PXE (table 2).
ABCC6del15
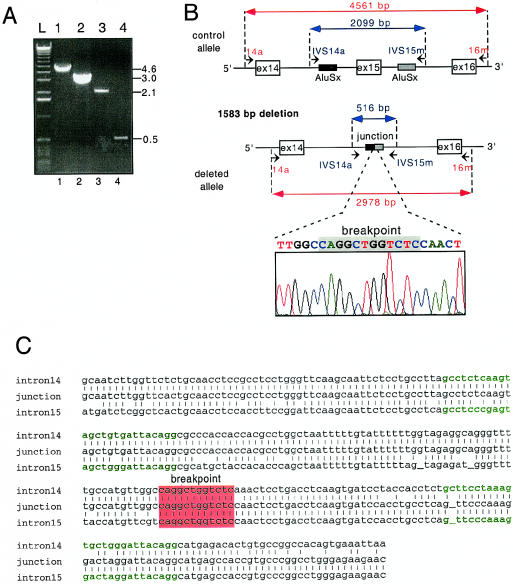
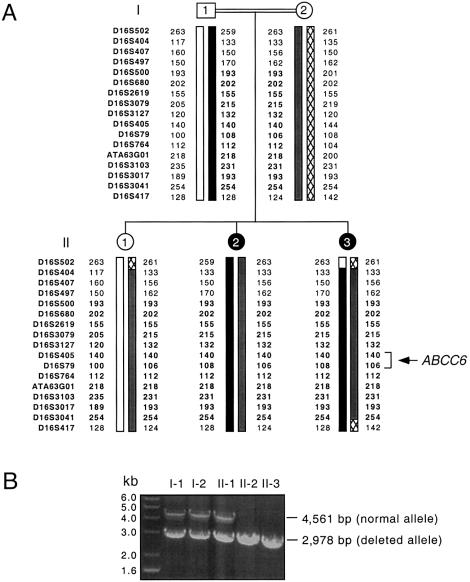
We observed a different out-of-frame deletion in ABCC6 in a single family of Indian ancestry originating from South Africa. This mutation was inferred from unsuccessful attempts to use PCR to amplify exon 15 of ABCC6 from an affected member of this family, in an analysis in which all other ABCC6 exons could successfully be amplified. Using PCR primers in introns 13 and 16, we obtained a PCR product of ∼3.0 kb, significantly shorter than the 4.6-kb PCR product obtained from a control amplification (fig. 2A and 2B). We designed a new pair of primers flanking two AluSx sequences present in introns 14 and 15. The patient-derived PCR product obtained using these new primers was ∼0.5 kb; when the same primers were used with control DNA, the PCR product was 2.1 kb. Both sets of PCR experiments indicated a 1.6-kb deletion within ABCC6 that resulted in the loss of exon 15 (fig. 2A and 2B). Sequence analysis revealed a 1,583-nucleotide deletion that included all of exon 15 (ABCC6del15). The breakpoint occurred within a 12-bp domain (fig. 2C) flanked by two 26-bp Alu core sequences (Rudiger et al. 1995), suggesting an Alu-mediated deletion. This deletion of exon 15 would result in a frameshift within the ABCC6 mRNA synthesized from the mutant allele, substituting the first codon of exon 16 for a nonsense mutation (TAA). If an ABCC6 protein were encoded from this mutant allele, the resulting polypeptide would be shortened by 880 amino acids and would lack both nucleotide-binding domains (NBDs). A haplotype analysis of this Indian family, which was performed with 17 microsatellites spanning the region p13.1 of chromosome 16, revealed a stretch of 12 consecutive homozygous markers both paternally and maternally inherited in the affected daughters (fig. 3A). The homozygosity of this region of chromosome 16 was in agreement with the consanguineous nature of this family. The unaffected sister, however, was heterozygous for this deleted allele and had inherited a maternal chromosome that conferred her carrier status, which was confirmed by PCR (fig. 3B). ABCC6del15 was not found in our control panel of 200 alleles nor was this deletion observed in any of the other 122 patients included in the present study.
Figure 2.
Characterization of a homozygous deletion of ABCC6 exon 15 in a patient with PXE who was of Indian origin. A, PCR analysis of the deletion mutation. The amplification of a region corresponding to exons 14, 15, and 16 using primers 14a and 16m with genomic DNA from a control individual (lane 1) and from the Indian patient with PXE (lane 2) resulted in PCR products of 4.6 kb and 3.0 kb, respectively. Using primers flanking Alu repeats present in intron 14 and 15 (IVS14a and IVS15m), we obtained PCR products of 2.0 kb and 0.5 kb from the same control individual (lane 3) and the Indian patient with PXE (lane 4), indicating that a homozygous deletion of about 1.6 kb occurred in the subject with PXE. The position of the primers used to PCR-amplify exon 15 are shown in panel B. B, Schematic representation of the genomic region surrounding ABCC6 exon 15 in an unaffected individual (top) and in the Indian patient with PXE (bottom). The sequence of the junction of the deleted allele is also shown. C, The highest sequence homology of the two Alu Sx repeats within intron 14 and 15 was noted in a 12-bp region that contains the 1,583-bp deletion breakpoint (red). Nucleotide sequence denoted in green represents the 26-bp core sequences flanking the breakpoint domain in intron 14 and 15.
Figure 3.
Haplotype analysis of a family with PXE of Indian ancestry originating from South Africa. A, The analysis was performed with 17 D16S polymorphic markers spanning ∼25 cM of 16p13.1. This analysis allowed the assignment of paternal and maternal chromosomes to the three offspring in this pedigree. Markers indicated in bold correspond to a stretch of 12 microsatellites found in a homozygous state and suggested consanguinity in this pedigree. B, The cosegregation of the deletion ABCC6del15 and the phenotype was verified using PCR amplification of genomic DNA from the family members. The deleted alleles generated a PCR product of 2,978 bp, whereas the PCR product derived from the normal allele was 4,561 bp. Individuals I-1, I-2, and II-1 carried both normal and mutant alleles, and both affected individuals, II-2 and II-3, displayed a homozygous deletion.
ABCC6 Pseudogenes
During the course of the present mutational analysis of the ABCC6 gene, several sequence variants that we detected seemed to be derived from possible duplicated regions of 16p13.1 that could correspond to one or more ABCC6 pseudogenes. Indeed, a nucleotide substitution in exon 9—that would result in a nonsense mutation Q378X (1132C→T)—and a single insertion in exon 2 (196–197insT) were observed in all Afrikaner and several European patients who had already been shown to be homozygous or compound heterozygous for other ABCC6 mutations. Moreover, all 26 affected and unaffected individuals tested thus far were found to be heterozygous for both these apparent mutations. Exons 2 and 9 of ABCC6 of 15 Afrikaner patients, 4 European patients, and 3 unaffected control individuals were sequenced. Mutations 196–197insT and Q378X were found to be randomly associated with all of the homozygous or compound heterozygous mutant alleles. Because these mutations would lead to premature termination of translation, both Q378X and 196–197insT were potentially disease-causing alleles. The high frequency of these variants in our control population suggested that these alleles were, perhaps, part of a pseudogene. Indeed, BLAST searches of the draft of the Human Genome sequence at Web sites of the National Center for Biotechnology Information (NCBI) and Celera revealed two pseudogenelike sequences on chromosome 16 that contained regions highly homologous to ABCC6 exons 1–4 and exons 1–9, respectively.
The sequence divergence between these putative pseudogenes and ABCC6 (absent in any published ABCC6 mRNA sequences) corresponded to several nucleotide substitutions and insertions that we have detected in our cohort of patients with PXE, including Q378X and 196–197insT (table 4). Interestingly, three of these substitutions—1171A→G (V39V), 190T→C/191G→A (W64Q) and 189G→C (L63L)—were found in a homozygous state, indicating that these alleles can be found both in the pseudogenes and the parental ABCC6. The region homologous to exons 1–9 of ABCC6 was found in the Celera database, but no location could be deduced. However, the region homologous to exons 1–4 was identified on a chromosome 16 working-draft segment in the NCBI database (GenBank accession number NT_010393) at 275 kb centromeric to the parental ABCC6 gene and 55 kb centromeric to the NPIP gene (GenBank accession number XM_017612) (Le Saux et al. 1999; Cai et al. 2000). However, this pseudogene was not contained within a contig of four overlapping BAC clones, 962B4, 13F4, 589H1, and 256A9 (GenBank accession numbers, U91318, AC002039, AC002045, and AC002492, respectively) that cover ∼415 kb of genomic DNA that extends centromeric of the ABCC6 gene (Le Saux et al. 1999; Cai et al. 2000). Moreover, the NCBI map of chromosome 16 currently reports at least nine gaps of unknown length between ABCC6 and the putative pseudogene containing exons 1–4. It is therefore likely that the ABCC6 pseudogene containing exons 1–4 is located in a proximal region of chromosome 16 at a distance >415 kb. The sequence surrounding this pseudogene diverged from ABCC6 at breakpoints located 3.4 kb 5′ of the exon 1 homologous sequence and 4.4 kb 3′ of the exon 4 homologous sequence. The domain within the pseudogene homologous to the 5′ UTR of ABCC6 contained two deleted regions of 142 and 315 bp, located at 2.8 kb and 0.9 kb, respectively, from the exon 1 homologous sequence.
Table 4.
Nucleotide and Apparent Amino Acid Changes Found in Two ABCC6 Pseudogenes
|
Change in |
||
| Amino Acid | Nucleotide | Exon |
| V39Va | 117A→G | 2 |
| L63La | 189G→C | 2 |
| W64Qa | 190T→C/191G→A | 2 |
| … | 196–197insT | 2 |
| A78T | 232G→A | 3 |
| A158V | 473C→T | 4 |
| S359S | 1077A→G | 9 |
| Q378X | 1132C→T | 9 |
| L381L | 1141T→C | 9 |
Variants found in homozygous states.
Genotype-Phenotype Correlation
The clinical information of 11 patients with PXE from Belgium and 15 from South Africa was available at the time of the present study. Among the four Belgian patients with homozygous R1141X alleles, three had typical skin lesions, and all patients displayed moderate-to-severe ocular involvement. In one patient the skin lesions were very mild. Five South African patients carried homozygous R1339C alleles. They also presented typical light-to-moderate skin lesions and moderate-to-severe ocular symptoms. However, three of these patients with PXE had light-to-severe cardiovascular abnormalities. In the group of 17 patients who carried compound heterozygous mutations, the skin lesions appeared typical in most, with a significant variability in ocular and cardiovascular findings. Although the severity of dermal and ocular lesions seems to correlate moderately with the age of the patients (as has been observed elsewhere [Neldner 1988]), no specific correlation appears between the phenotype and the type or position of the mutations.
Discussion
The present study describes the identification of a spectrum of mutations in PXE. Thirty-six mutations were characterized, and, among these, 28 were novel variants. Most mutations were missense variants. Indeed, 21 nucleotide substitutions were predicted to result in an amino acid change. In addition, we have detected six small insertions or deletions, five nonsense mutations, two nucleotide substitutions likely to involve aberrant mRNA splicing, and two large deletions. Therefore, the total number of PXE mutations known to date is 43. Putative disease-causing mutations were identified in ∼64% of the 244 chromosomes studied in affected individuals, and at least one disease allele was found in 85.2% of affected individuals.
PXE Mutations
The high degree of allelic heterogeneity observed in PXE was comparable to that of other autosomal recessive diseases. Although most mutations observed in ABCC6 were unique, two variants (R1141X and ABCC6del23–29) occurred at a high frequency (table 2). Overall, both mutations were found to occur at similar frequencies, 18.8% and 12.9%, respectively. However, when patient ancestries were considered, R1141X was found to occur at a greater frequency in Europe, whereas ABCC6del23–29 was preponderant in the United States (table 2). Such high frequencies suggest that these alleles are likely to be transmitted identical by descent from common ancestral alleles. Seventy percent of the individuals from the United States are of European ancestry, with some patients and families originating in the United Kingdom. Four of the five ABCC6del23–29 alleles found in Europe were identified in British patients. It is therefore possible that the ABCC6del23–29 alleles are copies of a founder mutation that originated in the British Isles before the first wave of immigrants to the New World, in spite of divergent haplotypes encountered in several families with this Alu-mediated deletion (Ringpfeil et al. 2001). It is indeed reasonable to expect a rapid haplotype divergence in chromosomal regions extending ∼10 cM on each side of the ABCC6 gene over a period of >200 years. A similar hypothesis could explain the high frequency of R1141X alleles in the European sample. However, to demonstrate that the various R1141X and ABCC6del23–29 alleles are identical by descent, the analysis of additional microsatellite markers and single-nucleotide polymorphisms within and in close proximity to ABCC6 will be required.
The mutation-detection rate obtained in the present study indicated that a significant number of mutations escaped detection by SSCP and CSGE. However, the percentage of detected mutations is comparable to those in similar PCR-based studies that have defined a spectrum of mutations in ABCA4 (Lewis et al. 1999; Maugeri et al. 1999; Rivera et al. 2000). Mutations in ABCA4, which also encodes an ABC transporter, are responsible for Stargardt disease (Allikmets et al. 1997b) and retinitis pigmentosa (Cremers et al. 1998; Martinez-Mir et al. 1998), and they could also be a factor in age-related macular degeneration (Allikmets et al. 1997a). To determine whether our screening methods were a limitation in the mutation-detection rate, all 31 exons of ABCC6 were sequenced from seven patients who had one or no mutant alleles identified. No additional pathogenic sequence alteration could be detected, suggesting that other sequence variants could be involved in PXE. Such sequence variants may include large deletions, duplications, and alterations of sequence within introns, within the promoter domain, and within the 3′UTR of ABCC6. All of these rearrangements would be undetected in our PCR-based analysis of exonic regions. It is also possible that the mutation-detection rate could have been affected by the presence of patients who have mutated alleles in a heterozygous state and who have developed a partial phenotype (van Soest et al. 1997; Bacchelli et al. 1999; Rubegni et al. 2000; Sherer et al. 2001). In addition, our criteria for defining a disease allele included the prerequisite that such an allele not be present in 200 control chromosomes derived from unaffected individuals. Because it is not unreasonable to expect the occasional presence of disease alleles in a heterozygous state among unaffected individuals, particularly for a disease of predominantly recessive inheritance, this rule probably led us to overlook true alleles predisposing to the disease.
It is remarkable that the two large deletions found in this study, ABCC6del23–29 and ABCC6del15, were mediated by the same type of Alu repeats (AluSX), both containing a core sequence of 26 bp (Rudiger et al. 1995). The analysis of the noncoding regions of ABCC6 revealed a high proportion of repetitive elements (Ringpfeil et al. 2001), suggesting that this gene might be susceptible to a significant level of genomic rearrangements. Indeed, ABCC6 intronic sequences contained 38 AluSX repeats, of which 19 presented a 26-bp core sequence that was ⩾95% conserved. The presence of these numerous repetitive sequences supports our hypothesis that a significant fraction of undetected mutations (table 2) could derive from DNA rearrangements that have yet to be characterized.
Mutation Clusters
The distribution pattern of ABCC6 mutations showed a cluster of disease-causing variants in a region of the gene that encodes the C-terminal end of the protein (fig. 1), in contrast to the neutral variants, which appeared to be evenly distributed (table 3). Indeed, most (36 of 43) mutations, including deletions, were located in the second half of the gene, and, remarkably, 24 (56%) mutations were located in exon 24 and NBD2, which is encoded by exons 28–30 (fig. 1). The significance of this apparent clustering of mutations in the NBD2 domain is not known. However, 70% of all missense mutations occur within two domains of ABCC6: a large cytoplasmic loop of 70 amino acids encoded by exon 24 and NBD2 and only two missense mutations (L673P and R765Q; see fig. 1) were located in NBD1. The overwhelming number of substitutions detected in NBD2 in patients with PXE suggests that the two NBDs of ABCC6 are functionally nonequivalent. Moreover, the clustering of missense mutations in NBD2 suggests that altered function of this ATP-binding domain is critical to the development of the PXE phenotype.
The distinct role of NBDs of several other ABC transporters—such as ABCC7 (CFTR), ABCC8 (SUR1 [GenBank accession number NM_000352]), and, notably, ABCC1 (MRP1 [GenBank accession number XM_017599])—has been demonstrated (Szabo et al. 1999; Ueda et al. 1999; Gao et al. 2000; Nagata et al. 2000; Aleksandrov et al. 2001). However, there are some distinctive differences in the proposed models of channel and transport activities for these transmembrane proteins. The binding and hydrolysis of ATP at NBD1 in ABCC7 and ABCC8 initiate channel activity. In contrast, the functional model of ABCB1 (P-glycoprotein [GenBank accession number NM_000927]) involves the alternate use of NBD1 and NBD2, because the two NBDs appear to be functionally equivalent (Urbatsch et al. 1995; Senior and Gadsby 1997). The suggested transport mechanism of ABCC1, which presents a high degree of identity with ABCC6 (44.6%), is initiated with the binding and hydrolysis of ATP at NBD2, followed by the trapping of ADP (Gao et al. 2000; Hou et al. 2000; Nagata et al. 2000). The energy released from the hydrolysis of the nucleotide at NBD2 is used for the active transport of a substrate. The transport would result in a conformational change favorable to ATP binding at NBD1. The hydrolysis of this ATP would then be used to return the pump to its initial conformation (Hou et al. 2000). Mutational analysis of both ABCC1 NBDs showed that mutations at NBD2 abolished transport, whereas mutations in NBD1 only resulted in the decrease of transport activity by ∼70% (Gao et al. 2000; Hou et al. 2000). Because of the high similarity with ABCC1, it would be reasonable to infer a similar structure/function relationship for ABCC6. Hence, mutations within or in close proximity to the NBD2 Walker motifs of ABCC6 could abolish all transport activity. The presence of 50% of all missense mutations in NBD2 (exons 28, 29, and 30) is in good agreement with this hypothesis. Furthermore, homozygous or compound heterozygous mutations in NBD1 may not contribute significantly to the development of the PXE phenotype, because an intact NBD2 could sustain a residual transport activity (30%), which could be comparable to the overall transport capacity of an asymptomatic individual carrying a single disease-causing allele (50%).
Of the five mutations in exon 24, four amino acid substitutions involve a change in charged residues. On the basis of observations that substitutions of charged residues in the CL4 domain of ABCC7 (CFTR) resulted in protein misprocessing, it is possible to infer that mutations in exon 24 of ABCC6, which encodes an equivalent domain, could interfere with the proper folding of ABCC6 (Seibert et al. 1996). Missense mutations in other ABCC6 domains, such as transmembrane regions (A455P, F568S, and G1203D), could also affect the protein processing or its integration into the cytoplasmic membrane. Moreover, the introduction of a prolyl residue at position L673, which is one of the two substitutions in NBD1, could, similarly, result in an incorrect folding. Indeed, it has been suggested that the proper folding of ABCC7 (CFTR) depends on the integrity of NBD1 and on complex interactions between both NBDs and their surrounding environment (Dalemans et al. 1991; Pollet et al. 2000).
Phenotype-Genotype Correlation
No significant correlation could be established between the clinical manifestations and disease-causing alleles of the 26 patients with PXE for whom this information was available. The small size of this cohort may be a factor in the lack of obvious relation between the phenotype and genotype. Although the severity of the phenotype seemed to be related to the age of the patients, it appeared not to be mutation dependent. Indeed, the mutations identified in these patients (premature termination mutations and missense mutations located in NBD2) are all predicted to result in the complete loss of ABCC6 function. It is therefore reasonable to assume that PXE mutations predispose carriers to the typical PXE clinical lesions, but the contribution of other factors—such as trauma, in the case of ocular lesions, and, perhaps, diet or lifestyle—may determine the age at onset and the severity of the clinical manifestations.
Mutations in genes encoding many of the ABC transporters produce phenotypes consistent with the overall functions of these different transporters (de Vree et al. 1998; Strautnieks et al. 1998; Brooks-Wilson et al. 1999; Rust et al. 1999). In contrast, it is quite unclear how mutations in ABCC6 will influence the assembly and deposition of elastic fibers and the subsequent development of the ocular, dermal, and vascular phenotype of PXE. It has been shown recently that the liver and kidney tissues express relatively abundant levels of ABCC6 mRNA (Kool et al. 1999), and very recent immunolocalization studies have identified a basolateral location for ABCC6 in both hepatocytes and renal epithelial cells (Madon et al. 2000). It is possible, therefore, that an aberrant transport function of ABCC6 in the kidney and/or the liver could precipitate secondary changes in the dermis and the arterial wall that lead to calcification of elastic fibers and the development of PXE. Indeed, for this reason, Uitto and coworkers (2001) have suggested that perhaps PXE is more a heritable metabolic disorder than a connective tissue disease. Alternatively, altered renal and hepatic ABCC6 function may be of little consequence to PXE, and subtle changes in the expression of this ABC transporter in the tissue affected in patients with PXE may be instrumental in influencing an appropriate extracellular matrix environment for the correct assembly of elastic fibers. Although the substrate transported by ABCC6 is unknown, a comparison with other ABCC transporters suggests that it is unlikely that calcification of elastic fibers in the tissue of patients with PXE results from direct changes in calcium transport by ABCC6. Glutathione conjugates of simple or complex anions are thought, in general, to be the substrates for ABCC6, and the continuing work to identify these substrates will certainly provide valuable insight into the functional relationships between ABCC6 and the complex PXE phenotype.
Acknowledgments
We are very grateful to all the patients and their relatives, whose cooperation made this study possible. We would like to thank Drs. András Váradi and Balázs Sarkadi for valuable comments regarding the role and functional consequences of mutations in ABCC6. We are also grateful for the various contributions of the members of the PXE International Research Consortium, particularly Patrick Terry. This work was supported by National Institutes of Health grant EY13019 (to C.B.), Research Centers in Minority Institutions grant RR03061 from National Center for Research Resources to the Pacific Biomedical Research Center of the University of Hawaii, and Telethon-Italy grant 696 (to I.P.R.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Celera, http://www.celera.com/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for BAC clone CIT987SK-A-962B4 [accession number U91318], BAC clone CIT987SK-A-13F4 [accession number AC002039], BAC clone CIT987SK-A-589H1 [accession number AC002045], BAC clone CIT987SK-A-256A9 [accession number AC002492] ABCC6 cDNA [accession number NM_001171], chromosome 16 working-draft segment [accession number NT_010393], ABCC7 cDNA [accession number NM_000492], ABCC8 cDNA [accession number NM_000352], ABCC1 cDNA [accession number XM_017599], ABCB1 cDNA [accession number NM_000927], and NPIP cDNA [accession number XM_017612])
- Institut de Biologie et Chimie des Protéines, http://www.ibcp.fr (for ClustalW program)
- Institute for Genomic Research, The, http://www.tigr.org
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PXE [MIM 177850 and MIM 264800])
References
- Aleksandrov L, Mengos A, Chang X-B, Aleksandrov A, Riordan JR (2001) Differential interactions of nucleotides at the two nucleotide binding domains of the cystic fibrosis transmembrane conductance regulator. J Biol Chem 276:12918–12923 [DOI] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, Dean M, Lupski JR, Leppert M (1997a) Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science 277:1805–1807 [DOI] [PubMed] [Google Scholar]
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR (1997b) A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet 15:236–246 [DOI] [PubMed] [Google Scholar]
- Baccarani-Contri M, Boraldi F, Taparelli F, De Paepe A, Ronchetti IP (1996) Matrix proteins with high affinity for calcium ions are associated with mineralization within the elastic fibers of pseudoxanthoma elasticum dermis. Am J Pathol 148:569–577 [PMC free article] [PubMed] [Google Scholar]
- Bacchelli B, Quaglino D, Gheduzzi D, Taparelli F, Boraldi F, Trolli B, Le Saux O, Boyd CD, Ronchetti IP (1999) Identification of heterozygote carriers in families with a recessive form of pseudoxanthoma elasticum (PXE). Mod Pathol 12:1112–1123 [PubMed] [Google Scholar]
- Bergen AA, Plomp AS, Schuurman EJ, Terry S, Breuning M, Dauwerse H, Swart J, Kool M, van Soest S, Baas F, ten Brink JB, de Jong PT (2000) Mutations in ABCC6 cause pseudoxanthoma elasticum. Nat Genet 25:228–231 [DOI] [PubMed] [Google Scholar]
- Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, et al (1999) Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet 22:336–345 [DOI] [PubMed] [Google Scholar]
- Cai L, Struk B, Adams MD, Ji W, Haaf T, Kang HL, Dho SH, et al (2000) A 500-kb region on chromosome 16p13.1 contains the pseudoxanthoma elasticum locus: high-resolution mapping and genomic structure. J Mol Med 78:36–46 [DOI] [PubMed] [Google Scholar]
- Christiano AM, Lebwohl MG, Boyd CD, Uitto J (1992) Workshop on pseudoxanthoma elasticum: molecular biology and pathology of the elastic fibers. Jefferson Medical College, Philadelphia, Pennsylvania, June 10, 1992. J Invest Dermatol 99:660–663 [DOI] [PubMed] [Google Scholar]
- Cotton RG, Scriver CR (1998) Proof of “disease causing” mutation. Hum Mutat 12:1–3 [DOI] [PubMed] [Google Scholar]
- Cremers FP, van de Pol DJ, van Driel M, den Hollander AI, van Haren FJ, Knoers NV, Tijmes N, Bergen AA, Rohrschneider K, Blankenagel A, Pinckers AJ, Deutman AF, Hoyng CB (1998) Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt's disease gene ABCR. Hum Mol Genet 7:355–362 [DOI] [PubMed] [Google Scholar]
- Dalemans W, Barbry P, Champigny G, Jallat S, Dott K, Dreyer D, Crystal RG, Pavirani A, Lecocq JP, Lazdunski M (1991) Altered chloride ion channel kinetics associated with the delta F508 cystic fibrosis mutation. Nature 354:526–528 [DOI] [PubMed] [Google Scholar]
- Darier J (1896) Pseudo-xanthome élastique. IIIème Congrès Internationale de Dermatologie de Londres 289–295 [Google Scholar]
- de Vree JM, Jacquemin E, Sturm E, Cresteil D, Bosma PJ, Aten J, Deleuze JF, Desrochers M, Burdelski M, Bernard O, Oude Elferink RP, Hadchouel M (1998) Mutations in the MDR3 gene cause progressive familial intrahepatic cholestasis. Proc Natl Acad Sci USA 95:282–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz HC, Valle D, Francomano CA, Kendzior RJ Jr, Pyeritz RE, Cutting GR (1993) The skipping of constitutive exons in vivo induced by nonsense mutations. Science 259:680–683 [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8:1893–1900 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 90:10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Cui H-R, Loe DW, Grant CE, Almquist KC, Cole SPC, Deeley RG (2000) Comparison of the functional characteristics of the nucleotide binding domains of multidrug resistance protein 1. J Biol Chem 275:13098–13108 [DOI] [PubMed] [Google Scholar]
- Germain DP, Perdu J, Remones V, Jeunemaitre X (2000) Homozygosity for the R1268Q mutation in MRP6, the pseudoxanthoma elasticum gene, is not disease-causing. Biochem Biophys Res Commun 274:297–301 [DOI] [PubMed] [Google Scholar]
- Hou Y-X, Cui L, Riordan JR, Chang X-B (2000) Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J Biol Chem 275:20280–20287 [DOI] [PubMed] [Google Scholar]
- Hull J, Shackleton S, Harris A (1994) The stop mutation R553X in the CFTR gene results in exon skipping. Genomics 19:362–364 [DOI] [PubMed] [Google Scholar]
- Jones CT, McIntosh I, Keston M, Ferguson A, Brock DJ (1992) Three novel mutations in the cystic fibrosis gene detected by chemical cleavage: analysis of variant splicing and a nonsense mutation. Hum Mol Genet 1:11–17 [DOI] [PubMed] [Google Scholar]
- Kool M, van der Linden M, de Haas M, Baas F, Borst P (1999) Expression of human MRP6, a homologue of the multidrug resistance protein gene MRP1, in tissues and cancer cells. Cancer Res 59:175–182 [PubMed] [Google Scholar]
- Le Saux O, Urban Z, Goring HH, Csiszar K, Pope FM, Richards A, Pasquali-Ronchetti I, Terry S, Bercovitch L, Lebwohl MG, Breuning M, van den Berg P, Kornet L, Doggett N, Ott J, de Jong PT, Bergen AA, Boyd CD (1999) Pseudoxanthoma elasticum maps to an 820-kb region of the p13.1 region of chromosome 16. Genomics 62:1–10 [DOI] [PubMed] [Google Scholar]
- Le Saux O, Urban Z, Tschuch C, Csiszar K, Bacchelli B, Quaglino D, Pasquali-Ronchetti I, Pope FM, Richards A, Terry S, Bercovitch L, de Paepe A, Boyd CD (2000) Mutations in a gene encoding an ABC transporter cause pseudoxanthoma elasticum. Nat Genet 25:223–227 [DOI] [PubMed] [Google Scholar]
- Lebwohl M, Schwartz E, Lemlich G, Lovelace O, Shaikh-Bahai F, Fleischmajer R (1993) Abnormalities of connective tissue components in lesional and non-lesional tissue of patients with pseudoxanthoma elasticum. Arch Dermatol Res 285:121–126 [DOI] [PubMed] [Google Scholar]
- Lewis RA, Shroyer NF, Singh N, Allikmets R, Hutchinson A, Li Y, Lupski JR, Leppert M, Dean M (1999) Genotype/phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am J Hum Genet 64:422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madon J, Hagenbuch B, Landmann L, Meier PJ, Stieger B (2000) Transport function and hepatocellular localization of mrp6 in rat liver. Mol Pharmacol 57:634–641 [DOI] [PubMed] [Google Scholar]
- Maquat LE (1996) Defects in RNA splicing and the consequence of shortened translational reading frames. Am J Hum Genet 59:279–286 [PMC free article] [PubMed] [Google Scholar]
- Martinez-Mir A, Paloma E, Allikmets R, Ayuso C, del Rio T, Dean M, Vilageliu L, Gonzalez-Duarte R, Balcells S (1998) Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet 18:11–12 [DOI] [PubMed] [Google Scholar]
- Maugeri A, van Driel MA, van de Pol DJ, Klevering BJ, van Haren FJ, Tijmes N, Bergen AA, Rohrschneider K, Blankenagel A, Pinckers AJ, Dahl N, Brunner HG, Deutman AF, Hoyng CB, Cremers FP (1999) The 2588G→C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am J Hum Genet 64:1024–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen L, Callens T, De Paepe A, Craen M, Mortier G (1997) Characterisation of two different nonsense mutations, C6792A and C6792G, causing skipping of exon 37 in the NF1 gene. Hum Genet 101:75–80 [DOI] [PubMed] [Google Scholar]
- Mickle JE, Macek M, Jr., Fulmer-Smentek SB, Egan MM, Schwiebert E, Guggino W, Moss R, Cutting GR (1998) A mutation in the cystic fibrosis transmembrane conductance regulator gene associated with elevated sweat chloride concentrations in the absence of cystic fibrosis. Hum Mol Genet 7:729–735 [DOI] [PubMed] [Google Scholar]
- Nagata K, Nishitani M, Matsuo M, Kioka N, Amachi T, Ueda K (2000) Nonequivalent nucleotide trapping in the two nucleotide binding folds of the human multidrug resistance protein MRP1. J Biol Chem 275:17626–17630 [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23:198–199 [DOI] [PubMed] [Google Scholar]
- Neldner KH (1988) Pseudoxanthoma elasticum. Int J Dermatol 27:98–100 [DOI] [PubMed] [Google Scholar]
- Nijbroek G, Sood S, McIntosh I, Francomano CA, Bull E, Pereira L, Ramirez F, Pyeritz RE, Dietz HC (1995) Fifteen novel FBN1 mutations causing Marfan syndrome detected by heteroduplex analysis of genomic amplicons. Am J Hum Genet 57:8–21 [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Endo M, Koyanagi H, Ichihara T, Takao A, Maruyama M (1990) Coronary artery bypass in a 15-year-old girl with pseudoxanthoma elasticum. Ann Thorac Surg 49:483–485 [DOI] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 86:2766–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passi A, Albertini R, Baccarani Contri M, de Luca G, de Paepe A, Pallavicini G, Pasquali Ronchetti I, Tiozzo R (1996) Proteoglycan alterations in skin fibroblast cultures from patients affected with pseudoxanthoma elasticum. Cell Biochem Funct 14:111–120 [DOI] [PubMed] [Google Scholar]
- Pollet JF, Van Geffel J, Van Stevens E, Van Geffel R, Beauwens R, Bollen A, Jacobs P (2000) Expression and intracellular processing of chimeric and mutant CFTR molecules. Biochim Biophys Acta 1500:59–69 [DOI] [PubMed] [Google Scholar]
- Rigal D (1881) Observation pour servir à l'histoire de la chéloide diffuse xanthélasmique. Ann Dermatol Syphilol 2:491–501 [Google Scholar]
- Ringpfeil F, Lebwohl MG, Christiano AM, Uitto J (2000) Pseudoxanthoma elasticum: mutations in the MRP6 gene encoding a transmembrane ATP-binding cassette (ABC) transporter. Proc Natl Acad Sci USA 97:6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringpfeil F, Nakano A, Uitto J, Pulkkinen L (2001) Compound heterozygosity for a recurrent 16.5-kb Alu-mediated deletion mutation and single-base-pair substitutions in the ABCC6 gene results in pseudoxanthoma elasticum. Am J Hum Genet 68:642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A, White K, Stohr H, Steiner K, Hemmrich N, Grimm T, Jurklies B, Lorenz B, Scholl HP, Apfelstedt-Sylla E, Weber BH (2000) A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am J Hum Genet 67:800–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubegni P, Mondillo S, De Aloe G, Agricola E, Bardelli AM, Fimiani M (2000) Mitral valve prolapse in healthy relatives of patients with familial pseudoxanthoma elasticum. Am J Cardiol 85:1268–1271 [DOI] [PubMed] [Google Scholar]
- Rudiger NS, Gregersen N, Kielland-Brandt MC (1995) One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res 23:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denefle P, Assmann G (1999) Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat Genet 22:352–355 [DOI] [PubMed] [Google Scholar]
- Seibert FS, Linsdell P, Loo TW, Hanrahan JW, Clarke DM, Riordan JR (1996) Disease-associated mutations in the fourth cytoplasmic loop of cystic fibrosis transmembrane conductance regulator compromise biosynthetic processing and chloride channel activity. J Biol Chem 271:15139–15145 [DOI] [PubMed] [Google Scholar]
- Senior AE, Gadsby DC (1997) ATP hydrolysis cycles and mechanism in P-glycoprotein and CFTR. Semin Cancer Biol 8:143–150 [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer DW, Bercovitch L, Lebwohl M (2001) Pseudoxanthoma elasticum: significance of limited phenotypic expression in parents of affected offspring. J Am Acad Dermatol 44:534–537 [DOI] [PubMed] [Google Scholar]
- Strautnieks SS, Bull LN, Knisely AS, Kocoshis SA, Dahl N, Arnell H, Sokal E, Dahan K, Childs S, Ling V, Tanner MS, Kagalwalla AF, Nemeth A, Pawlowska J, Baker A, Mieli-Vergani G, Freimer NB, Gardiner RM, Thompson RJ (1998) A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet 20:233–238 [DOI] [PubMed] [Google Scholar]
- Struk B, Cai L, Zach S, Ji W, Chung J, Lumsden A, Stumm M, Huber M, Schaen L, Kim CA, Goldsmith LA, Viljoen D, Figuera LE, Fuchs W, Munier F, Ramesar R, Hohl D, Richards R, Neldner KH, Lindpaintner K (2000) Mutations of the gene encoding the transmembrane transporter protein ABC-C6 cause pseudoxanthoma elasticum. J Mol Med 78:282–286 [DOI] [PubMed] [Google Scholar]
- Struk B, Neldner KH, Rao VS, St Jean P, Lindpaintner K (1997) Mapping of both autosomal recessive and dominant variants of pseudoxanthoma elasticum to chromosome 16p13.1. Hum Mol Genet 6:1823–1828 [DOI] [PubMed] [Google Scholar]
- Szabo K, Szakacs G, Hegedus T, Sarkadi B (1999) Nucleotide occlusion in the human cystic fibrosis transmembrane conductance regulator: different patterns in the two nucleotide binding domains. J Biol Chem 274:12209–12212 [DOI] [PubMed] [Google Scholar]
- Torrington M, Viljoen DL (1991) Founder effect in 20 Afrikaner kindreds with pseudoxanthoma elasticum. S Afr Med J 79:7–11 [PubMed] [Google Scholar]
- Ueda K, Komine J, Matsuo M, Seino S, Amachi T (1999) Cooperative binding of ATP and MgADP in the sulfonylurea receptor is modulated by glibenclamide. Proc Natl Acad Sci USA 96:1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, Boyd CD, Lebwohl MG, Moshell AN, Rosenbloom J, Terry S (1998) International Centennial Meeting on Pseudoxanthoma Elasticum: progress in PXE research. J Invest Dermatol 110:840–842 [DOI] [PubMed] [Google Scholar]
- Uitto J, Pulkkinen L, Ringpfeil F (2001) Molecular genetics of pseudoxanthoma elasticum: a metabolic disorder at the environment-genome interface? Mol Med Today 7:13–17 [DOI] [PubMed] [Google Scholar]
- Uitto J, Shamban A (1987) Heritable skin diseases with molecular defects in collagen or elastin. Dermatol Clin 5:63–84 [PubMed] [Google Scholar]
- Urbatsch IL, Sankaran B, Bhagat S, Senior AE (1995) Both P-glycoprotein nucleotide-binding sites are catalytically active. J Biol Chem 270:26956–26961 [DOI] [PubMed] [Google Scholar]
- van Soest S, Swart J, Tijmes N, Sandkuijl LA, Rommers J, Bergen AA (1997) A locus for autosomal recessive pseudoxanthoma elasticum, with penetrance of vascular symptoms in carriers, maps to chromosome 16p13.1. Genome Res 7:830–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weenink AC, Dijkman G, de Meijer PH (1996) Pseudoxanthoma elasticum and its complications: two case reports. Neth J Med 49:24–29 [DOI] [PubMed] [Google Scholar]