Abstract
A comprehensive analysis of somatic and germline mutations related to DNA mismatch–repair (MMR) genes can clarify the prevalence and mechanism of inactivation in colorectal carcinoma (CRC). In the present study, 257 unselected patients referred for CRC resection were examined for evidence of defective DNA MMR. In particular, we sought to determine the frequency of hereditary defects in DNA MMR in this cohort of patients. MMR status was assessed by testing of tumors for the presence or absence of hMLH1, hMSH2, and hMSH6 protein expression and for microsatellite instability (MSI). Of the 257 patients, 51 (20%) had evidence of defective MMR, demonstrating high levels of MSI (MSI-H) and an absence of either hMLH1 (n=48) or hMSH2 (n=3). All three patients lacking hMSH2, as well as one patient lacking hMLH1, also demonstrated an absence of hMSH6. DNA sequence analysis of the 51 patients with defective MMR revealed seven germline mutations—four in hMLH1 (two truncating and two missense) and three in hMSH2 (all truncating). A detailed family history was available for 225 of the 257 patients. Of the seven patients with germline mutations, only three had family histories consistent with hereditary nonpolyposis colorectal cancer. Of the remaining patients who had tumors with defective MMR, eight had somatic mutations in hMLH1. In addition, hypermethylation of the hMLH1 gene promoter was present in 37 (88%) of the 42 hMLH1-negative cases available for study and in all MSI-H tumors that showed loss of hMLH1 expression but no detectable hMLH1 mutations. Our results suggest that, although defective DNA MMR occurs in ∼20% of unselected patients presenting for CRC resection, hereditary CRC due to mutations in the MMR pathway account for only a small proportion of patients. Of the 257 patients, only 5 (1.9%) appear to have unequivocal evidence of hereditary defects in MMR. The epigenetic (nonhereditary) mechanism of hMLH1 promoter hypermethylation appears to be responsible for the majority of the remaining patients whose tumors are characterized by defective DNA MMR.
Introduction
Familial polyposis (FAP [MIM 175100]) and hereditary nonpolyposis colorectal cancer (HNPCC [MIM 114500]) are two common autosomal dominant disorders predisposing to the development of colorectal cancer (CRC) (Bellacosa et al. 1996; Khine et al. 1996; Fante et al. 1997; Lynch et al. 1997; Soravia et al. 1997). FAP is characterized by the presence of hundreds to thousands of adenomatous polyps. HNPCC, on the other hand, is characterized by the occurrence of colorectal cancer at an early age (fourth to sixth decade), a tendency to develop multiple primary cancers, and an increased risk for the development of certain other cancers, particularly endometrial and stomach cancer (Lynch and Lynch 1995). It is generally easier to ascertain a diagnosis of FAP than HNPCC, because most patients with FAP eventually develop profuse polyposis and, sometimes, other tumor types (e.g., osteomas and periampullary carcinomas) that can aid in the diagnosis. Patients with HNPCC, on the other hand, lack distinctive phenotypic features. Consequently, the diagnosis of HNPCC has historically been based on family history. As a result, the true incidence of this disease and the underlying molecular defects have been difficult to establish. The estimated percentage of CRC cases caused by HNPCC has varied from ∼0.5% to 10% (Aaltonen et al. 1994b; Bellacosa et al. 1996; Brassett et al. 1996; Evans et al. 1997; Peel et al. 2000).
The Amsterdam criteria (AC) were established in the early 1990s to define the clinical criteria used to identify patients with HNPCC (Vasen et al. 1991). These criteria call for the occurrence of verifiable colorectal cancer in three individuals (one of whom is the first-degree relative of the other two), the presence of cancer in at least two successive generations, and an age at onset of CRC of <50 years in at least one case. These criteria were created to help identify families that have a high probability of having an hereditary form of colorectal cancer not accompanied by polyposis (i.e., HNPCC). It was understood, however, that families classified, on the basis of the AC, as having HNPCC might represent a heterogeneous group with respect to the underlying basis of disease. Nonetheless, it was an important step in attempting to determine the genetic basis of colorectal cancer.
For clinical purposes, however, the AC are now recognized to be too restrictive, since other malignancies (e.g., gastric and endometrial) frequently occur in patients with HNPCC and may be the presenting cancers in such families (Beck et al. 1997b; Bapat et al. 1999). As a result, several modifications that include consideration of extracolonic malignancies in the diagnosis have been suggested (Beck et al. 1997b; Bapat et al. 1999; Vasen et al. 1999). In addition to family history, however, there are now laboratory approaches that may aid in establishing this diagnosis in a subset of patients with HNPCC. These have arisen from studies that revealed the underlying genetic defect in many—but not all—families diagnosed with HNPCC; namely, defective DNA-mismatch repair (MMR).
Of the seven human DNA MMR genes that have been identified (hMLH1, hMSH2, hMSH3, hMSH6, hPMS1, hMLH3, and hPMS2), germline mutations have been identified in all but two (hMSH3 and hMLH3) in families with HNPCC (Fishel et al. 1993; Lindblom et al. 1993; Papadopoulos et al. 1994, 1995; Lipkin et al. 2001). Germline mutations are detected most frequently in patients with HNPCC who satisfy the AC, although some individuals who do not fulfill these criteria also carry germline mutations (Beck et al. 1997a). The majority of these HNPCC patients have mutations in either hMLH1 or hMSH2, with less frequent involvement of other genes. However, there are also some families with HNPCC in which mutations in MMR genes have not been identified and whose tumors do not demonstrate phenotypic evidence of defective DNA MMR—that is, they do not demonstrate either the presence of tumor microsatellite instability (MSI) or the absence of protein expression for one of the genes involved in DNA MMR (Aaltonen et al. 1994a; Craanen et al. 1996; Moslein et al. 1996; Lamberti et al. 1999; Peel et al. 2000).
MSI is a phenotypic indicator of defective DNA MMR and is detected in tumors from HNPCC patients, as well as in a low percentage of various sporadic cancers, such as colon, endometrial, and stomach cancer (Aaltonen et al. 1993; Han et al. 1993; Ionov et al. 1993; Thibodeau et al. 1993; Burks et al. 1994; Duggan et al. 1994; Keller et al. 1995; Akiyama et al. 1996; Caduff et al. 1996; Battista et al. 1997; Gurin et al. 1999). In CRC, several phenotypes of MSI have been defined: MSI-H (MSI in >30% of loci examined), MSI-L (MSI in <30% loci examined), and MSS (microsatellite stable, showing no instability at any site) (Boland et al. 1998). Sporadic CRC with the MSI-H phenotype has distinct clinicopathological features, including a tendency to occur in the proximal colon, high frequency in females, high grade, diploidy, and improved overall survival compared with those without widespread microsatellite instability (Thibodeau et al. 1993; Fujita et al. 1996; Sankila et al. 1996; Lynch et al. 1997). Almost all such cases are due to functional loss of either hMLH1 or hMSH2. The MSI-L phenotype, on the other hand, is not associated with these clinicopathological features or with altered hMLH1 or hMSH2. The majority of HNPCC-associated tumors are characterized by MSI-H, whereas MSI-H, MSI-L, and MSS are found in sporadic CRC.
In spite of numerous studies on the frequency of defective MMR in CRC, there have been few studies examining the frequency of inherited CRC due to defective MMR in a series of patients not selected for family history of colon cancer. Aaltonen et al. (1998), in a study of 509 consecutive Finnish patients with colorectal cancer, found that inherited defects in MMR occurred in ∼2% of the cases studied. In their study, all cases were examined for evidence of defective MMR, irrespective of family history; however, half (5/10) of the germline mutations identified were determined to originate from a single common founder mutation, found in >30 families in Finland and Sweden. No other population-based studies have been performed. The purpose of the present study was to determine the frequency of defective DNA MMR, as well as the underlying mechanism, in an unselected prospective series of colorectal cancers in patients referred to the Mayo Clinic. More specifically, we sought to determine the frequency of inherited defects in this series of patients. The present article details the first set of 257 unselected cancers in an ongoing prospective study. Tumors were examined for the presence or absence of: (a) tumor MSI; (b) hMLH1, hMSH2, and hMSH6 protein expression; (c) hMLH1, hMSH2, and hMSH6 gene mutations; and (d) hMLH1 and hMSH2 promoter hypermethylation. Patients’ family histories were obtained to determine the familial component, and the clinical characteristics of the patients were analyzed.
Material and Methods
Patient Population
A total of 514 patients underwent surgical resection for CRC at the Mayo Clinic during a 1.5-year period, from December, 1995, to April, 1997. Four hundred and sixty-six patients were approached about the study, and 267 (57.3%) consented to participate. Of the consenting patients, seven had inadequate cancer tissue available for study, one had only paraffin-embedded tissue available, and two had only metastatic disease. These patients were not included, leaving 257 available for further study. Of the 257 patients, 237 were white (92%), 4 (1.5%) were of African American descent, 3 (1%) were of Hispanic origin, and 13 (5%) had no designated race. Seventy-five percent (192) came from Minnesota (103), Iowa (32), Wisconsin (10), North Dakota (6), South Dakota (7), or Illinois (34). The remainder came from an additional 27 states within the continental United States.
Patient chart reviews were performed to obtain clinical characteristics of the tumor, including tumor site, stage, and age at diagnosis. For tumor site, tumors of the proximal colon were defined as those CRCs occurring in the cecum, the ascending colon, or the transverse colon. Distal tumors were defined as those occurring in the descending or sigmoid colon or in the rectum. Family histories were documented by telephone follow-up for 225 of the 257 patients, through use of a detailed questionnaire. On the basis of this information, three- to four-generation pedigrees were constructed. On the basis of the presence of family history of CRC, patients fulfilling the AC (n=7) (Vasen et al. 1991) and patients fulfilling the modified AC (n=9, including the seven AC) (Vasen et al. 1999) were identified. For the purposes of the present study, an HNPCC-related cancer included cancers of the colon, small bowel, ureter, endometrium, and renal pelvis (Vasen et al. 1999). FAP was diagnosed in three patients, and chronic ulcerative colitis (CUC) was diagnosed in another four; these patients were not excluded from the analysis.
DNA Extraction
DNA was extracted from frozen or paraffin-embedded tissues, as described elsewhere (Thibodeau et al. 1998). In brief, DNA from microdissected frozen tissue sections (10 μm) was extracted by a standard phenol/chloroform procedure. For tumor DNA, only those areas containing >70% tumor cells were used. For DNA extraction from paraffin-embedded tissues, the Qiamp Tissue kit (Qiagen, Inc.) was used, according to the manufacturer's instructions. The corresponding normal control DNA for each patient was derived from peripheral blood. For these specimens, DNA was extracted using the Puregene nucleic acid–isolation kit (Gentra).
Microsatellite Instability
Paired normal and tumor DNA were analyzed for microsatellite instability with six dinucleotide microsatellite markers (D5S346, TP53, D18S34, D18S49, D18S61, and ACTC) and one mononucleotide repeat (BAT 26). PCR and gel electrophoresis were performed as described by Thibodeau et al. (1993). Tumors were classified as MSH-H if ⩾30% markers demonstrated instability, as MSH-L if <30% demonstrated MSI, and as MSS if no marker exhibited MSI (Boland et al. 1998; Thibodeau et al. 1998). These studies were performed with DNA isolated from paraffin-embedded material.
Immunohistochemical Analysis
The expression of hMLH1, hMSH2, and hMSH6 protein was assessed as described elsewhere (Thibodeau et al. 1998). In brief, 5-μm tissue sections from formalin-fixed, paraffin-embedded tissue were stained with antibody to hMLH1 (clone G168 728; 1 mg/ml [PharMingen]), hMSH2 (clone FE11; 0.5 mg/ml [Oncogene Science]), and hMSH6 (clone 44, 0.5 μg/ml [Transduction Laboratories]). Tumor cells that showed an absence of nuclear staining in the presence of normal positive staining in surrounding cells were interpreted as having an absence of expression of these proteins.
Sequencing
Exons 1–19 of hMLH1 and/or exons 1–16 of hMSH2 were sequenced in all tumors lacking expression of hMLH1 (n=48) or hMSH2 (n=3), using the ThermoSequenase kit (Amersham), essentially as described by Moslein et al. (1996). For hMSH6, only exon 5 was sequenced (Parc et al. 2000), since this exon contains repeat sequences commonly altered in tumors with defective DNA MMR (Yin et al. 1997; Iino et al. 2000). These studies were performed with DNA isolated from both leukocytes and fresh frozen tumor (when available).
Methylation Analysis
The methylation status of the promoter regions of both hMLH1 and hMSH2 was assessed as described elsewhere (Cunningham et al. 1998), using an HpaII-based PCR assay. As a control for the PCR, a calcitonin sequence devoid of HpaII sites was used. The hMLH1 and hMSH2 primers amplified fragments containing HpaII-sensitive sites. DNA was first digested with the restriction enzyme HpaII. The digested DNA was then subjected to multiplex PCR; the products were electrophoresed on 8% polyacrylamide gels and then were analyzed for the relative amplification of hMLH1 or hMSH2 and calcitonin. The sizes of the unmethylated PCR products were 114 bp, 107 bp, and 145 bp, for hMLH1, hMSH2, and calcitonin, respectively. Ratios of either hMLH1 or hMSH2 to calcitonin of <.2 indicated no methylation (as in all the normal samples tested), whereas a ratio of >.8 was scored as positive for hypermethylation. Some samples had intermediate values and were considered to have either partial methylation or hemimethylation.
Some of the samples were also examined using the methylation-specific PCR (MSP) assay, as described by Herman et al. (1998). These samples were also subjected to DNA sequence analysis, after bisulfite treatment. For the sequence analysis, the following primers for PCR amplification and sequencing were designed to amplify both methylated and unmethylated modified DNA, corresponding to the regions assessed in the HpaII (region 3) and MSP (region 2): using the U83845 sequence, region 3, forward 5′ position 339, tagattaggtatagggtttta and reverse 5′ position 603, aaatataccaataaaaaca; region 2, forward 5′ position 158, gaggtttgtaygagtagtttt and reverse 5′ position 381, ataaacacrttatttaat (r = a or g, y = c or t). The underlined nucleotides represent those cytosine residues that are converted to uracil/thymine (if unmethylated) or are retained (if methylated) in the modification. The PCRs were performed in 25-μl reactions containing PCR buffer 1 (Perkin Elmer) with 1.5 mM (region 3) or 2 mM (region 2) MgCl2, 10 pmol primer, 1.25 units AmpliTaq Gold (Perkin Elmer), and 1 μl bisulfite-modified DNA (equivalent to 50 ng genomic DNA). After an initial preheating step of 12 min at 95°C, PCR was performed over 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 30 min. PCR products were cleaned using a HighPure PCR purification column (Boeringer Mannheim) and were sequenced with the forward primer used in the initial PCR, on an ABI 377 (ABI Biosystems). Methylation studies were performed with DNA isolated from both leukocytes and fresh frozen tumor (when available).
Results
Patient Sample
The clinical characteristics of the 257 participating patients are shown in table 1. Patient ages at diagnosis were 29–91 years (median 69 years), and the male-to-female ratio was 1.47 (153 male, 104 female). For the nonparticipants (n=199), the male-to-female ratio was 1.1 (102 male, 97 female; P=.08), indicating that males were more likely to participate than females. The nonparticipants were also slightly older than were the participants (median age 73 vs. 69 years; P=.0001).
Table 1.
Patient Characteristics
|
No. (%) of Individuals |
|||||||||||
| Sexb |
Tumor Sitec |
Dukes Staged |
Age at Diagnosise |
||||||||
| Sample (No. and %of Total Sample) | Median Age (Range)a(years) | Male | Female | Proximal | Distal | A | B | C | D | ⩽50 years | >50 years |
| All Patients (N=257) | 69 (29–91) | 153 (59.0) | 104 (41.0) | 120 (46.7) | 137 (53.3) | 16 (6.2) | 112 (43.6) | 86 (33.5) | 42 (16.7) | 24 (9.3) | 233 (90.7) |
| MSI-H (n=51, 19.8%) | 72 (35–91) | 23 (45.1) | 28 (54.9) | 44 (86.3) | 7 (13.7) | 6 (11.8) | 31 (60.8) | 11 (21.6) | 3 (5.9) | 3 (5.9) | 48 (94.1) |
| MSI-L (n=18, 7.0%) | 65.5 (29–84) | 13 (72.2) | 5 (27.8) | 8 (44.4) | 10 (55.6) | 0 (0.0) | 9 (50.0) | 7 (38.9) | 2 (11.1) | 3 (16.7) | 15 (83.3) |
| MSS (n=188, 73.2%) | 67 (34–90) | 117 (62.2) | 71 (37.8) | 68 (36.2) | 120 (63.8) | 10 (5.3) | 72 (38.3) | 68 (36.2) | 38 (20.2) | 18 (9.6) | 170 (90.4) |
P=.02 (Kruskal-Wallis test).
P=.05 (χ2 test).
P=.001 (χ2 test).
P=.01 (χ2 test). Dukes Stage A—invasion limited to submucosa, with no nodal metastasis; B—invasion to at least the level of muscularis propria, with no nodal metastasis; C—invasion at any level, with regional nodal metastasis but no distant metastasis; D—distant metastasis present (Astler and Coller 1954).
P=.39 (χ2 test).
MSI and Immunohistochemistry
Of the 257 patients with CRC available for study, 51 (20%) were MSI-H, 18 (7%) were MSI-L, and 188 (73%) were MSS (table 1, fig. 1). All but one of the MSI-H tumors and none of the MSI-L or MSS tumors demonstrated MSI at BAT 26. Consistent with previous observations, significant associations were observed between tumors with the MSI-H phenotype and proximal tumor location, female gender, and Dukes’ stage (table 1).
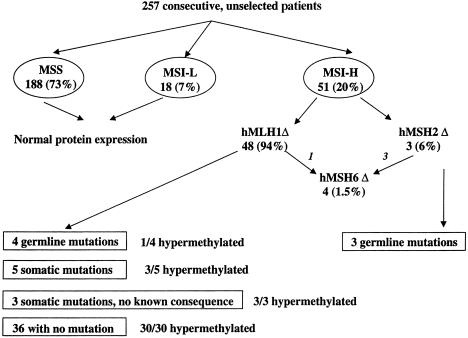
Figure 1.
Flow diagram for the results of testing of the 257 consecutive, unselected patients with CRC. Tumors were assessed for the presence or absence of tumor MSI (MSI-H, MSI-L, or MSS); hMLH1, hMSH2, and hMSH6 protein expression (Δ indicates loss of expression); hMLH1, hMSH2, and hMSH6 gene mutations; and hMLH1 and hMSH2 promoter hypermethylation.
Protein expression for hMLH1, hMSH2, and hMSH6 was then examined in paraffin-embedded tissues (immunohistochemistry not shown; results summarized in fig. 1). All 51 MSI-H tumors showed an absence of protein expression of either hMLH1 (n=48 [94%]) or hMSH2 (n=3 [6%]). None of the tumors demonstrated an absence of hMSH6 only; however, all three tumors lacking hMSH2, as well as one with an absence of hMLH1, also showed an absence of hMSH6. All of the MSI-L and MSS tumors demonstrated normal expression for all three of these proteins (two MSS tumors were not scored for hMLH1, because of technical failure, but had normal expression of hMSH2 and hMSH6). All of the patients with FAP and CUC had normal protein expression; one patient with FAP exhibited an MSI-L phenotype, and the remaining patients exhibited an MSS phenotype.
Mechanism of Gene Inactivation
DNA sequence analysis and promoter hypermethylation studies were used to explore the mechanisms underlying defective DNA MMR in the MSI-H patients (n=51). Normal leukocyte DNA was available for study from all 51 MSI-H patients; however, DNA from fresh frozen tumor was available for study from only 44 of the 51 MSI-H patients (42 of 48 lacked hMLH1, and 2 of 3 lacked hMSH2). Thus, all MSI-H patients were examined for germline mutations, but only 44 of the 51 patients were examined for somatic alterations (tumor DNA sequencing and promoter methylation). The single MSI-H patient that did not show instability at BAT-26 was not examined for somatic alterations, since no frozen tissue was available for DNA extraction (this patient did not show evidence of any germline alteration in hMLH1).
Of the 48 patients lacking hMLH1 protein expression, 12 had hMLH1 gene alterations, 4 of whom had germline alterations and 8 of whom had somatic alterations only (table 2, fig. 1). Two of the four germline hMLH1 mutations were splicing alterations, which are likely to be pathogenic, whereas the other two were missense changes, which have unknown pathogenic consequences (table 2). One of these missense mutations (in patient 209) occurred at a conserved codon and might therefore have pathological consequences, whereas the other mutation (in patient 83) occurred at a nonconserved site. Two of the patients with germline mutations (patients 274 and 209) also demonstrated the presence of a somatic mutation in the tumor. Three of the eight tumors with somatic hMLH1 alterations had mutations with uncertain pathological consequences; of these three, two were within introns (in patients 441 and 302) and one resulted in a silent change (in patient 298). These three mutations were analyzed with a splice-predictor program at the Berkeley Drosophila Genome Project Web site, and none of them were predicted to alter splicing. Of the remaining five mutations, four would be expected to result in truncated proteins (in patients 75, 53, 61, and 91), and one was a missense alteration occurring at a highly conserved codon (in patient 379).
Table 2.
The Spectrum of Germline and Somatic Alterations in hMLH1 and hMSH2
| Gene, MutationType, andPatient Number | Exon/Intron | Nucleotide Changea | Consequence |
| hMLH1: | |||
| Germline: | |||
| 274 | I6 | IVS5-1, g→a | Splice site |
| 21 | I9 | IVS9+1, g→a | Splice site |
| 83 | E12 | 1321 G→A | A441T |
| 209 | E12 | 1217 G→A | S406N |
| Somatic: | |||
| 274 | E7 | 583 del A | G201 stop |
| 274 | E19 | 2146 G→A | V716M |
| 209 | E8 | 677 G→A | R226Q |
| 75 | E6 | 469 del T | K160 stop |
| 53 | E7 | 583 del A | G201 stop |
| 61 | E17 | 1912 G→T | G638 stop |
| 91 | I16-E17 | IVS17-7, del 17 bp | Splice site |
| 379 | E18 | 2000 A→G | D667G |
| 298 | E7 | 557 C→T | H186H |
| 441 | I8 | IVS8+16, del a | Unknown |
| 302 | I11 | IVS11+9, g→a | Unknown |
| hMSH2: | |||
| Germline: | |||
| 46 | I4 | IVS4+1, g→a | Splice site |
| 90 | E8 | 1316-1317del CT | T441 stop |
| 403 | E13 | 2038 C→T | R680 stop |
| Somatic: | |||
| 46 | E7 | 1165 C→T | R389 stop |
Lowercase letters indicate nucleotides in intron; uppercase letters indicate nucleotides in exon.
All three tumors with an absence of hMSH2 expression had a germline hMSH2 mutation (table 2, fig. 1), each of which would be expected to result in a truncated product. Patient 46 also showed the presence of a somatic alteration.
In addition to those alterations described above, six germline polymorphisms—four in hMLH1 and two in hMSH2—were also noted (table 3). Three of the hMLH1 polymorphisms and both of the hMSH2 polymorphisms have been described elsewhere (for a list of published and unpublished mutations and polymorphisms, see the Web site of the International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer). The one unreported hMLH1 sequence variant was a silent change in exon 17 (leucine to leucine) and was found in only a small number of cases.
Table 3.
Spectrum of Germline Sequence Variants in hMLH1 and hMSH2
| Gene andExon/Intron | Nucleotide Changea | Consequence | Frequency |
| hMLH1: | |||
| E8 | 655 A→G | I219V | 19/48 |
| I13 | IVS13+14, g→a | Unknown | 2/48 |
| I14 | IVS15-19, a→g | Unknown | 27/48 |
| E17b | 1959 G→T | L653L | 4/48 |
| hMSH2: | |||
| I9 | IVS10-9, t→a | Unknown | 3/3 |
| I12 | IVS13-6, t→c | Unknown | 1/3 |
Lowercase letters indicate nucleotides in intron; uppercase letters indicate nucleotides in exon.
Not previously reported.
The promoter regions of both hMLH1 and hMSH2 were initially examined for hypermethylation through use of an HpaII-based PCR assay. Forty-four MSI-H tumors were examined, 42 of 48 for hMLH1 and 2 of 3 for hMSH2. Hypermethylation of the hMSH2 promoter was not detected for the MSI-H/hMSH2–negative tumors tested. Of the MSI-H/hMLH1–negative tumors, 34 demonstrated hMLH1 hypermethylation by the HpaII-based PCR assay. Three MSI-H tumors lacked expression of hMLH1 but had no detectable hMLH1 mutations and did not exhibit hMLH1 hypermethylation by the HpaII assay. We examined these 3 tumors, as well as another 17 that did show promoter hypermethylation by the HpaII assay, for methylation, using the MSP assay. The 3 tumors that did not show evidence of hypermethylation with the HpaII assay now showed evidence of methylation with the MSP assay, and the methylation status of the other 17 tumors was confirmed.
To further explore the molecular basis for the differences between the three discordant cases, the hMLH1 5′ regions examined by the two assays (HpaII site, region 3, and MSP, region 2) were sequenced using bisulfite-modified DNA. For all three of these cases, all CpG sites were methylated, with the exception of the HpaII site. Thus, 37 of 42 tumors (88%) demonstrated promoter hypermethylation for hMLH1. Four tumor samples had both mutations in hMLH1 and hypermethylation (fig. 1). Two of these mutations were missense changes, and two were frameshifts; three were somatic, and one was a germline alteration. The one germline alteration was a missense change at a nonconserved site (in patient 83). All of the tested tumors lacking a germline or somatic alteration showed evidence of promoter hypermethylation (30/30). Thus, all of the MSI-H tumors tested that lacked hMLH1 expression had either hypermethylation and/or a mutation in hMLH1.
Analysis of hMSH6
Three of four available tumors that lacked hMSH6 expression were examined for the presence of a mutation in exon 5 of hMSH6. Patient 46 had a C insertion, and case 83 had both a C insertion and a C deletion within the C(8) repeat. Patient 19 showed the presence of an ins3405T mutation. All of these alterations are somatic, and all result in a frameshift.
Defective MMR and a Family History of CRC
Forty-seven (92%) of the MSI-H patients, all 18 (100%) of the MSI-L patients, and 160 (85%) of the MSS patients had family histories available for review. Of the 225 unrelated patients, 7 (3.1%) fulfilled the AC for HNPCC. Defective DNA MMR was noted in four of these seven individuals. Of the four patients with defective MMR, three were found to have germline defects—one in hMLH1 (patient 274) and two in hMSH2 (patients 46 and 403)—whereas the other tumor (in patient 19) demonstrated only hMLH1 hypermethylation. It is possible that this tumor does indeed harbor a germline alteration not detected by our sequencing strategy. Notably, three of the seven tumors from patients with HNPCC did not exhibit evidence of defective DNA MMR. An additional two patients fulfilled the modified AC for HNPCC, only one of whom had an MSI-H tumor. No additional germline alterations were identified.
The family characteristics of those patients with germline mutations are presented in table 4. Only three (patients 46, 274, and 403) of the seven patients with germline mutations fulfilled the AC. Two of these patients were aged <50 years (patients 35 and 44) at first diagnosis; the third was aged 55 years. Patient 403, who had a truncating hMSH2 mutation, had a father who developed stomach cancer at age 38 years. Although patient 403 was not in a branch of the family that satisfied the AC, additional individuals on the paternal side of the family (aunt, uncle, cousin, and second cousin) had CRC, satisfying the AC for this family branch. We were not able to obtain family-history information for the one patient (patient 83) with a tumor that was associated with a germline missense hMLH1 mutation. This was also the only tumor from a germline mutation–bearing patient that exhibited hMLH1 hypermethylation. One additional patient (patient 21) had no documented family history, and two patients (patients 90 and 209) had no indication of a family history of either CRC or extracolonic HNPCC tumors.
Table 4.
Family Characteristics of Patients with Germline Mutations[Note]
|
No. of CRC Cases in |
|||||||
| Mutation Typeand PatientNumber | Germline Mutation | Age at Onset(years) | Multiple CRC in Proband | First-DegreeRelatives | Second-Degree andHigher Relatives | No. of OtherHNPCC Cancersin Familya | Total No. ofCRC Casesin Family |
| Pathogenic: | |||||||
| 274b | hMLH1 IVS5-1 | 35 | Yes | 4 | 3 | 0 | 8 |
| 21 | hMLH1 IVS9+1 | 68 | No | 2 | NK | NK | 3 |
| 46b | hMSH2 IVS4+1 | 44 | No | 1 | 7 | 0 | 9 |
| 90 | hMSH2 T441stop | 35 | No | 0 | 0 | 0 | 1 |
| 403b | hMSH2 R680stop | 55 | No | 0 | 4 | 0 | 5 |
| Uncertain pathogenicity: | |||||||
| 83 | hMLH1 A441T | 82 | No | NK | NK | NK | NK |
| 209 | hMLH1 S406N | 65 | No | 0 | 0 | 0 | 1 |
Note.— NK = not known. In the case of patient 83, no family history was available, and, in the case of patient 21, only limited history was available.
Includes cancer of the endometrium, ovary, stomach, renal pelvis, ureter, and small bowel, in first-degree and higher relatives.
Patient fit the AC.
Discussion
There has been considerable discussion about both the frequency of HNPCC and the means of identifying these individuals. The purpose of the present prospective study was to determine the frequency of inherited defects in DNA MMR (which we will refer to as “hereditary defective mismatch repair syndrome” or HDMMR), which is one particular cause of HNPCC. This was a prospective study in which all consenting surgical cases of CRC were examined for evidence of defective DNA MMR, without prior selection based on family history.
Overall, evidence of defective DNA MMR was found in 20% of the 257 patients with CRC. This is within the range (12%–24%) previously published for colorectal cancer (Ionov et al. 1993; Thibodeau et al. 1993; Nakashima et al. 1997). All of the tumors with evidence of MSI-H (a phenotypic indicator of defective MMR) had altered protein expression of either hMLH1 or hMSH2. Abnormal protein expression for these two genes was not found in the remaining tumors. Furthermore, in the overall group of patients, absence of hMSH6 occurred only in the context of an abnormality in hMSH2 or hMLH1. Thus, it is likely that all cases of defective MMR that involve (a) hMSH2, (b) hMLH1, (c) hMSH6, and/or (d) an MSI phenotype have been identified in this group of patients.
Unequivocal pathogenic (truncating) germline mutations were identified in five patients, and missense germline mutations of unknown significance were identified in two additional patients. Thus, in this cohort of patients, the incidence of hereditary defective MMR is ∼2%. This may be an underestimate, since we did not test for the presence of large deletions, which have been noted to occur (Wijnen et al. 1998). However, our findings suggest that the true frequency of HDMMR in this group of patients is not likely to be much higher, since somatic mutations and/or hypermethylation of the hMLH1 promoter appears to account for the MSI-H phenotype in all of the remaining tumors. Although the Finnish study by Aaltonen et al. (1998) found a somewhat lower proportion of CRC to have defective DNA MMR (12% of cases), the frequency of germline hMSH2 and hMLH1 mutations was similar to that found in the present study. In the study by Aaltonen et al. (1998), tumors were screened for MSI but mutations were tested for in hMSH2 or hMLH1 only; hMSH6 was not examined. Additionally, immunohistochemical analysis was not included in their study. Cumulatively, these two studies suggest that the frequency of inherited defects in MMR in a population of patients with CRC referred for surgery is ∼2%. The present study is the first large study in the United States that has looked at the frequency of disease in an unselected group of patients who were referred for CRC resection and in which no known founder mutations are described.
The germline mutations detected in the present study would be expected to produce a truncated or altered protein in all of the hMSH2 and in two of the four hMLH1 cases. The remaining two hMLH1 alterations were missense changes; one of these mutations occurred in a highly conserved codon (S406N). Recent evidence indicates that some missense hMLH1 mutations can affect the interaction of hMLH1 with hPMS2 (Guerrette et al. 1999); however, neither of the germline missense mutations identified in the present study occurred in this critical region. Nonetheless, the tumor with the S406N missense alteration also had a second somatic missense mutation, suggesting the possibility that this germline alteration may have functional consequences. The second missense alteration (A441T) occurred at a nonconserved codon. This tumor also exhibited hMLH1 promoter hypermethylation and, given the late age at onset for the patient (age 82 years), the A441T more likely represents a rare normal variant rather than a causative mutation. At this point, however, functional studies would be needed to distinguish between these two possibilities.
There is no clear consensus about how best to identify patients to be tested for HNPCC. Aaltonen et al. (1998) suggested that either family history, young age at diagnosis, or a history of multiple CRC or related cancers warranted testing for defective DNA MMR. This position was challenged in an editorial (Lynch and Smyrk 1998), which argued that family history alone may be the most appropriate means of identifying HNPCC. In the present study, tumors from three of the seven patients fulfilling the AC did not exhibit evidence of defective DNA MMR; germline defects were evident in only three of the patients with defective MMR. A fourth patient with defective MMR may have a mutation not detected by the methods employed in the present study. It is likely, therefore, that other genes not involving MMR are responsible for the colorectal cancer in the three patients with HNPCC who have normal MMR. Additionally, neither of the two additional patients satisfying the modified AC were found to harbor a germline alteration. Conversely, when the family characteristics of the patients with germline mutations were examined, only three of seven patients fulfilled the AC. It is becoming increasingly apparent that not all individuals fulfilling the AC or modified AC for HNPCC will have inherited defects in DNA MMR, and not all individuals with inherited defects have a strong family history of cancer. HDMMR is a genetic diagnosis based on the finding of defective DNA MMR in the tumor and of a germline mutation in one of the DNA MMR genes, whereas HNPCC is a clinical diagnosis. Not all patients with HNPCC will have HDMMR, and not all patients with HDMMR will fulfill the AC (or other clinical criteria) for HNPCC. However, HDMMR is likely to be a more homogenous entity (despite allelic heterogeneity), unlike HNPCC, which is heterogeneous and probably includes cases of HDMMR, other hereditary forms of colorectal cancer (some for which the genes may not have identified), and the chance familial clustering of patients with sporadic colorectal cancer. It may be important to think about and treat these two syndromes as different entities. The Bethesda guidelines (Rodriguez-Bigas et al. 1997) may improve the predictive potential for identification of patients with defective DNA MMR genes over that provided by family history alone (Pistorius et al. 2000), since they take both the clinicopathological and family characteristics into account. We will report on the use of these guidelines, within this data set, in a future article.
Interestingly, all of the hMSH2-deficient cases were the result of a germline mutation. Although there are only three cases in the series, the data suggest that an absence of hMSH2 protein expression in CRC, when detected, is predictive of the presence of a germline mutation. Larger studies will be required to confirm such findings. It is also interesting to note that hMSH6 protein expression was lost in all three hMSH2 cases, but in only one of the hMLH1 cases. Our data suggest that, in such cases, the loss of hMSH6 is secondary to a mutation in either hMLH1 or hMSH2, a result which supports the work of others (Wu et al. 1999; Planck et al. 2000). The complete concordance for loss of hMSH2 and hMSH6 is most likely due to the physical proximity of these two genes on chromosome 2. A germline mutation in hMSH2 could be followed by loss of the second allele, by a gross chromosomal event. Such an event would also eliminate one normal hMSH6 allele. Defective MMR resulting from loss of hMSH2 could then lead to instability of the hMSH6 poly C tract in exon 5 and subsequent inactivation of the second allele. As before, additional studies will be required to test this hypothesis.
In summary, whereas the frequency of defective DNA MMR in this referral-based study was found to be relatively common (20%), the frequency of inherited defects (HDMMR) was found to be low (∼2%). The primary mechanism of DNA MMR gene inactivation appears to be epigenetic, with hMLH1 hypermethylation occurring in 100% of hMLH1-negative MSI-H tumors without detectable mutations. Two conclusions can be drawn from the family history information. First, although a strong family history correlates with the presence of germline mutations, it did not identify all the potential at-risk individuals. Second, with only four of seven patients with HNPCC having evidence of defective MMR (and none of those having a family history of HNPCC-related tumors), it is clear that there must be other factors that predispose to cancer development in these families. Molecular techniques can help in the identification of patients with defects in DNA MMR and can be used to complement family history and other guidelines in the clinical setting. This will greatly facilitate the diagnosis of HDMMR and the identification of individuals at high risk of cancer development.
Acknowledgments
We thank Dr. Gloria Petersen, for her helpful comments on the manuscript, and Karen Erwin, for secretarial support. This work was supported by National Institutes of Health grant R01 CA68535.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Berkeley Drosophila Genome Project, http://www.fruitfly.org/ (for splice-predictor program)
- International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer, http://www.nfdht.nl/ (for listing of published and unpublished mutations and polymorphisms)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HNPCC [MIM 114500] and APC [MIM 175100])
References
- Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, Petersen G, Kinzler K, Vogelstein B, de la Chapelle A (1993) Clues to the pathogenesis of familial colorectal cancer. Science 260:812–816 [DOI] [PubMed] [Google Scholar]
- Aaltonen LA, Peltomaki P, Mecklin JP, Jarvinen H, Jass JR, Green JS, Lynch HT, Watson P, Tallqvist G, Juhola M (1994a) Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res 54:1645–1648 [PubMed] [Google Scholar]
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A (1998) Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 338:1481–1487 [DOI] [PubMed] [Google Scholar]
- Aaltonen LA, Sankila R, Mecklin JP, Jarvinen H, Pukkala E, Peltomaki P, de la Chapelle A (1994b) A novel approach to estimate the proportion of hereditary nonpolyposis colorectal cancer of total colorectal cancer burden. Cancer Detect Prev 18:57–63 [PubMed] [Google Scholar]
- Akiyama Y, Nakasaki H, Nihei Z, Iwama T, Nomizu T, Utsunomiya J, Yuasa Y (1996) Frequent microsatellite instabilities and analyses of the related genes in familial gastric cancers. Jpn J Cancer Res 87:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astler VB, Coller FA (1954) The prognostic significance of direct extension of the carcinoma of the colon and rectum. Ann Surg 139:846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat BV, Madlensky L, Temple LKF, Hiruki T, Redston M, Baron DL, Xia L, Marcus VA, Soravia C, Mitri A, Shen W, Gryfe R, Berk T, Chodirker BN, Cohen Z, Gallinger S (1999) Family history characteristics, tumor microsatellite instability and germline MSH2 and MLH1 mutations in hereditary colorectal cancer. Hum Genet 104:167–176 [DOI] [PubMed] [Google Scholar]
- Battista P, Palmirotta R, Vitullo P, Veri MC, Colalongo C, Rigoli L, Fedele F, Caruso R, Inferrera C, Romano F, Mariani-Costantini R, Frati L, Cama A (1997) Microsatellite instability in early gastric cancer. Int J Oncol 10:65–70 [PubMed] [Google Scholar]
- Beck NE, Tomlinson IP, Homfray TF, Frayling IM, Hodgson SV, Bodmer WF (1997a) Frequency of germline hereditary non-polyposis colorectal cancer gene mutations in patients with multiple or early onset colorectal adenomas. Gut 41:235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck NE, Tomlinson IP, Homfray T, Hodgson SV, Harocopos CJ, Bodmer WF (1997b) Genetic testing is important in families with a history suggestive of hereditary non-polyposis colorectal cancer even if the Amsterdam criteria are not fulfilled. Br J Surg 84:233–237 [PubMed] [Google Scholar]
- Bellacosa A, Genuardi M, Anti M, Viel A, Ponz de Leon L (1996) Hereditary nonpolyposis colorectal cancer: review of clinical, molecular genetics, and counseling aspects. Am J Med Genet 62:353–364 [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58:5248–5257 [PubMed] [Google Scholar]
- Brassett C, Joyce JA, Froggatt NJ, Williams G, Furniss D, Walsh S, Miller R, Evans DG, Maher ER (1996) Microsatellite instability in early onset and familial colorectal cancer. J Med Genet 33:981–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burks RT, Kessis TD, Cho KR, Hedrick L (1994) Microsatellite instability in endometrial carcinoma. Oncogene 9:1163–1166 [PubMed] [Google Scholar]
- Caduff RF, Johnston CM, Svoboda-Newman SM, Poy EL, Merajver SD, Frank TS (1996) Clinical and pathological significance of microsatellite instability in sporadic endometrial carcinoma. Am J Pathol 148:1671–1678 [PMC free article] [PubMed] [Google Scholar]
- Craanen ME, Blok P, Offerhaus GJ, Tytgat GN (1996) Recent developments in hereditary nonpolyposis colorectal cancer. Scand J Gastroenterol Suppl 218:92–97 [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN (1998) Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res 58:3455–3460 [PubMed] [Google Scholar]
- Duggan BD, Felix JC, Muderspach LI, Tourgeman D, Zheng J, Shibata D (1994) Microsatellite instability in sporadic endometrial carcinoma. J Natl Cancer Inst 86:1216–1221 [DOI] [PubMed] [Google Scholar]
- Evans DG, Walsh S, Jeacock J, Robinson C, Hadfield L, Davies DR, Kingston R (1997) Incidence of hereditary non-polyposis colorectal cancer in a population-based study of 1137 consecutive cases of colorectal cancer. Br J Surg 84:1281–1285 [PubMed] [Google Scholar]
- Fante R, Benatti P, di Gregorio C, De Pietri S, Pedroni M, Tamassia MG, Percesepe A, Rossi G, Losi L, Roncucci L, Ponz de Leon L (1997) Colorectal carcinoma in different age groups: a population-based investigation. Am J Gastroenterol 92:1505–1509 [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R (1993) The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer [published erratum appears in Cell 1994 Apr 8;77(1):167]. Cell 75:1027–1038 [DOI] [PubMed] [Google Scholar]
- Fujita S, Moriya Y, Sugihara K, Akasu T, Ushio K (1996) Prognosis of hereditary nonpolyposis colorectal cancer (HNPCC) and the role of Japanese criteria for HNPCC. Jpn J Clin Oncol 26:351–355 [DOI] [PubMed] [Google Scholar]
- Guerrette S, Acharya S, Fishel R (1999) The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem 274:6336–6341 [DOI] [PubMed] [Google Scholar]
- Gurin CC, Federici MG, Kang L, Boyd J (1999) Causes and consequences of microsatellite instability in endometrial carcinoma. Cancer Res 59:462–466 [PubMed] [Google Scholar]
- Han HJ, Yanagisawa A, Kato Y, Park JG, Nakamura Y (1993) Genetic instability in pancreatic cancer and poorly differentiated type of gastric cancer. Cancer Res 53:5087–5089 [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB (1998) Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 95:6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino H, Simms L, Young J, Arnold J, Winship IM, Webb SI, Furlong KL, Leggett B, Jass JR (2000) DNA microsatellite instability and mismatch repair protein loss in adenomas presenting in hereditary non-polyposis colorectal cancer. Gut 47:37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M (1993) Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363:558–561 [DOI] [PubMed] [Google Scholar]
- Keller G, Rotter M, Vogelsang H, Bischoff P, Becker KF, Mueller J, Brauch H, Siewert JR, Hofler H (1995) Microsatellite instability in adenocarcinomas of the upper gastrointestinal tract. Relation to clinicopathological data and family history. Am J Pathol 147:593–600 [PMC free article] [PubMed] [Google Scholar]
- Khine K, Smith DR, Goh HS (1996) Use of molecular methods in the early diagnosis of familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer. Ann Acad Med Singapore 25:64–70 [PubMed] [Google Scholar]
- Lamberti C, Kruse R, Ruelfs C, Caspari R, Wang Y, Jungck M, Mathiak M, Malayeri HR, Friedl W, Sauerbruch T, Propping P (1999) Microsatellite instability—a useful diagnostic tool to select patients at high risk for hereditary non-polyposis colorectal cancer: a study in different groups of patients with colorectal cancer. Gut 44:839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom A, Tannergard P, Werelius B, Nordenskjold M (1993) Genetic mapping of a second locus predisposing to hereditary non-polyposis colon cancer. Nat Genet 5:279–282 [DOI] [PubMed] [Google Scholar]
- Lipkin SM, Wang V, Stoler DL, Anderson GR, Kirsch I, Hadley D, Lynch HT, Collins FS (2001) Germline and somatic mutation analyses in the DNA mismatch repair gene MLH3: Evidence for somatic mutation in colorectal cancers. Hum Mutat 17:389–396 [DOI] [PubMed] [Google Scholar]
- Lynch HT, Lynch JF (1995) Clinical implications of advances in the molecular genetics of colorectal cancer. Tumori 81:19–29 [PubMed] [Google Scholar]
- Lynch HT, Smyrk TC (1998) Identifying hereditary nonpolyposis colorectal cancer. N Engl J Med 338:1537–1538 [DOI] [PubMed] [Google Scholar]
- Lynch HT, Smyrk T, Lynch J (1997) An update of HNPCC (Lynch syndrome). Cancer Genet Cytogenet 93:84–99 [DOI] [PubMed] [Google Scholar]
- Moslein G, Tester DJ, Lindor NM, Honchel R, Cunningham JM, French AJ, Halling KC, Schwab M, Goretzki P, Thibodeau SN (1996) Microsatellite instability and mutation analysis of hMSH2 and hMLH1 in patients with sporadic, familial and hereditary colorectal cancer. Hum Mol Genet 5:1245–1252 [DOI] [PubMed] [Google Scholar]
- Nakashima H, Mori M, Mimori K, Inoue H, Baba K, Shibuta K, Kusumoto H, Haraguchi M, Ueo H, Akiyoshi T (1997) Microsatellite instability in Japanese colorectal carcinoma. Oncol Rep 4:387–389 [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Liu B, Parsons R, Lengauer C, Palombo F, D'Arrigo A, Markowitz S, Willson JK, Kinzler KW (1995) Mutations of GTBP in genetically unstable cells. Science 268:1915–1917 [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD (1994) Mutation of a mutL homolog in hereditary colon cancer. Science 263:1625–1629 [DOI] [PubMed] [Google Scholar]
- Parc YR, Halling KC, Wang L, Christensen ER, Cunningham JM, French AJ, Burgart LJ, Price-Troska TL, Roche PC, Thibodeau SN (2000) HMSH6 alterations in patients with microsatellite instability-low colorectal cancer. Cancer Res 60:2225–2231 [PubMed] [Google Scholar]
- Peel DJ, Ziogas A, Fox EA, Gildea M, Laham B, Clements E, Kolodner RD, Anton-Culver H (2000) Characterization of hereditary nonpolyposis colorectal cancer families from a population-based series of cases. J Natl Cancer Inst 92:1517–1522 [DOI] [PubMed] [Google Scholar]
- Pistorius SR, Kruppa C, Haas S, Plaschke J, Kruger S, Bulitta CJ, Nagel M, Saeger HD, Schackert HK (2000) Clinical consequences of molecular diagnosis in families with mismatch repair gene germline mutations. Int J Colorectal Dis 15:255–263 [DOI] [PubMed] [Google Scholar]
- Planck M, Wenngren E, Borg A, Olsson H, Nilbert M (2000) Somatic frameshift alterations in mononucleotide repeat-containing genes in different tumor types from an HNPCC family with germline MSH2 mutation. Genes Chromosomes Cancer 29:33–39 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, Srivastava S (1997) A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst 89:1758–1762 [DOI] [PubMed] [Google Scholar]
- Sankila R, Aaltonen LA, Jarvinen HJ, Mecklin JP (1996) Better survival rates in patients with MLH1-associated hereditary colorectal cancer. Gastroenterology 110:682–687 [DOI] [PubMed] [Google Scholar]
- Soravia C, Bapat B, Cohen Z (1997) Familial adenomatous polyposis (FAP) and hereditary nonpolyposis colorectal cancer (HNPCC): a review of clinical, genetic and therapeutic aspects. Schweiz Med Wochenschr 127:682–690 [PubMed] [Google Scholar]
- Thibodeau SN, Bren G, Schaid D (1993) Microsatellite instability in cancer of the proximal colon. Science 260:816–819 [DOI] [PubMed] [Google Scholar]
- Thibodeau SN, French AJ, Cunningham JM, Tester D, Burgart LJ, Roche PC, McDonnell SK, Schaid DJ, Vockley CW, Michels VV, Farr GH Jr, O'Connell MJ (1998) Microsatellite instability in colorectal cancer: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res 58:1713–1718 [PubMed] [Google Scholar]
- Vasen HF, Mecklin JP, Khan PM, Lynch HT (1991) The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum 34:424–425 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Watson P, Mecklin JP, Lynch HT (1999) New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology 116:1453–1456 [DOI] [PubMed] [Google Scholar]
- Wijnen J, Vanderklift H, Vasen H, Khan PM, Menko F, Tops C, Heijboer HM, Lindhout D, Moller P, Fodde R (1998) MSH2 genomic deletions are a frequent cause of HNPCC. Nat Genet 20:326–328 [DOI] [PubMed] [Google Scholar]
- Wu Y, Berends MJW, Mensink RGJ, Kempinga C, Sijmons RH, van der Zee AGJ, Hollema H, Kleibeuker JH, Buys CHCM, Hofstra RMW (1999) Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet 65:1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Kong D, Wang S, Zou TT, Souza RF, Smolinski KN, Lynch PM, Hamilton SR, Sugimura H, Powell SM, Young J, Abraham JM, Meltzer SJ (1997) Mutation of hMSH3 and hMSH6 mismatch repair genes in genetically unstable human colorectal and gastric carcinomas. Human Mutation 10:474–478 [DOI] [PubMed] [Google Scholar]