Abstract
Type 1 diabetes (T1D) is a genetically complex disorder of glucose homeostasis that results from the autoimmune destruction of the insulin-secreting cells of the pancreas. Two previous whole-genome scans for linkage to T1D in 187 and 356 families containing affected sib pairs (ASPs) yielded apparently conflicting results, despite partial overlap in the families analyzed. However, each of these studies individually lacked power to detect loci with locus-specific disease prevalence/sib-risk ratios (λs) <1.4. In the present study, a third genome scan was performed using a new collection of 225 multiplex families with T1D, and the data from all three of these genome scans were merged and analyzed jointly. The combined sample of 831 ASPs, all with both parents genotyped, provided 90% power to detect linkage for loci with λs = 1.3 at P=7.4×10-4. Three chromosome regions were identified that showed significant evidence of linkage (P<2.2×10-5; LOD scores >4), 6p21 (IDDM1), 11p15 (IDDM2), 16q22-q24, and four more that showed suggestive evidence (P<7.4×10-4, LOD scores ⩾2.2), 10p11 (IDDM10), 2q31 (IDDM7, IDDM12, and IDDM13), 6q21 (IDDM15), and 1q42. Exploratory analyses, taking into account the presence of specific high-risk HLA genotypes or affected sibs' ages at disease onset, provided evidence of linkage at several additional sites, including the putative IDDM8 locus on chromosome 6q27. Our results indicate that much of the difficulty in mapping T1D susceptibility genes results from inadequate sample sizes, and the results point to the value of future international collaborations to assemble and analyze much larger data sets for linkage in complex diseases.
Introduction
Type 1 diabetes (T1D [MIM 222100]) is characterized by autoimmune destruction of pancreatic β cells and a subsequent complete dependence on exogenously administered insulin for the regulation of blood glucose levels. The increased concordance rates for T1D in identical twins and the tendency for the disorder to aggregate within families suggest that at least some portion of susceptibility to T1D is genetically determined. However, within families, the disorder follows no clear mode of inheritance and is generally thought to result from the combined effects of multiple genes interacting with nongenetic factors (Risch 1987; Thomson et al. 1988; Rich 1990).
T1D has a long history of studies evaluating candidate genes for allelic association with disease status in either family-based or case-control formats. Two chromosomal regions have emerged from these studies, with consistent and significant evidence of association with T1D across multiple reports. The first is the HLA region at 6p21.3 (IDDM1). HLA-DQB1 (MIM 604305) and HLA-DRB1 (MIM 142857) are the major components of IDDM1, with at least two other loci involved, including HLA-DPB1 (MIM 142858) (Todd et al. 1987; Horn et al. 1988; Easteal et al. 1990; Erlich et al. 1996; Lie et al. 1999; Noble et al. 2000; Cucca et al. 2001).
The second chromosome region that displays consistent evidence of allelic association with T1D is the insulin (INS) gene region (IDDM2 [MIM 125852]) on chromosome 11p15. Detailed association mapping using polymorphic markers spanning this region, as well as family-based haplotype association studies, indicate that susceptibility is tightly associated with alleles at a variable number of tandem repeats (VNTR) marker located 596 bp upstream of the start site of transcription for the INS gene (Bell et al. 1984; Lucassen et al. 1993; Owerbach and Gabbay 1993, 1994; Julier et al. 1994; Bennett et al. 1995). Alleles at this VNTR have been demonstrated to affect the steady-state level of insulin mRNA in both the thymus (Pugliese et al. 1997; Vafiadis et al. 1997) and the pancreatic islets (Bennett et al. 1995; Kennedy et al. 1995; Vafiadis et al. 1996), providing plausible mechanisms for the effect of this polymorphism on disease susceptibility.
Many other functional candidate genes have been tested for their association with T1D. Most such studies have been unreported, and, with the possible exception of CTLA4 on chromosome 2q33 (IDDM12 [MIM 601388]), no locus has been identified as a likely one for T1D. Genomewide scans for association are not yet feasible, owing to the cost of genotyping and the incomplete knowledge of the polymorphism and haplotype content of the 30,000–50,000 genes predicted to constitute the human genome. Therefore, most recent attempts to identify T1D genes have focused primarily on genetic linkage approaches that use affected sib pairs (ASPs). T1D was the first genetically complex disorder to be analyzed on a genomewide basis for linkage in ASPs, and several whole-genome scans have been reported (Davies et al. 1994; Hashimoto et al. 1994; Concannon et al. 1998; Mein et al. 1998). These genomewide scans have provided strong evidence of linkage between IDDM1 and T1D. The relative risk to siblings, λs, associated with allele sharing at IDDM1 has been estimated to be 2.5–3.4. Depending on the model selected for interaction between IDDM1 and other loci, inheritance at IDDM1 may account for as much as half of the familial clustering of the disease (Risch 1987). This strong effect of IDDM1 may overshadow the contributions of other loci and may raise the power requirements for studies that seek to identify them. Approximately 20 additional putative diabetes loci have been reported, although, in most cases, the aggregate supporting evidence has not attained a genomewide significance level (P⩽2.2×10-5). IDDM3–IDDM13 (MIMs 600318, 600319, 600320, 601941, 600321, 600883, 601942, 601208, 601388, and 601318), IDDM15 (MIM 601666), and IDDM18 (MIM 605598) correspond to regions displaying increased LOD scores in genomewide scans and/or evidence of association by transmission/disequilibrium testing (TDT) (Spielman et al. 1993; Reijonen and Concannon 2001). The symbols IDDM14 and IDDM16 have been reserved but not published. IDDM17 (MIM 603266) was identified in an extended Bedouin pedigree with multiple affected individuals in whom T1D was associated with HLA-DR3 (Verge et al. 1998). Several other regions that may harbor susceptibility loci, including 1q42, Xp11, and 16q22-q24, have also been implicated in genomewide scans (Concannon et al. 1998; Cucca et al. 1998; Mein et al. 1998).
The large number of T1D loci reported, although promising, also reflects an underlying problem: Studies from different laboratories have, by and large, yielded discordant findings. This problem is most clearly illustrated by a comparison of the results reported from the two largest genomewide scans completed in T1D ASPs, one that included 356 families from the United Kingdom (Mein et al. 1998) and another that analyzed 187 families from the United States, with follow-up of positive findings in 429 additional families (Concannon et al. 1998). Apart from human leukocyte antigen (HLA)/IDDM1, there was no agreement between the positive results of the two studies. The UK study obtained P<7.4×10-4 for chromosomes 11p15 (INS/IDDM2), 10p11, and 16q22-q24; whereas the US study detected 1q42 at P<7.4×10-4. Such samples of 200–400 ASPs have only limited power to detect loci with λs ⩽1.4 (Hauser et al. 1996); therefore, the lack of concordance in the two studies may have resulted from random sampling differences, given their overall lack of power. If the INS/IDDM2 odds ratio of 3 is typical of non-HLA loci (equates to a λs of 1.12), then at least 2,500 ASPs would be required to obtain convincing evidence of linkage at reasonable levels of statistical power. In addition, simulation studies suggest that substantially larger sample sizes are required to reproduce an initial finding of linkage when the same criteria for significance are applied in follow-up as in an initial study (Suarez et al. 1994).
In the current study, a genome scan was performed on 225 new multiplex families with T1D. The data from this genome scan were merged with those from our previous, independently published studies (Davies et al. 1994; Concannon et al. 1998; Mein et al. 1998) and were jointly analyzed. Evidence of interaction between loci was sought by assessing the correlation of family-specific nonparametric linkage (NPL) scores for all loci at which there was nominal evidence of linkage. For HLA, a weighting scheme based on the relative risk associated with different combinations of HLA haplotypes was used to condition the linkage data. Finally, stratified analyses of families ordered by age at onset were performed. These studies provide a consensus view of the evidence of linkage to T1D in families whose DNA samples are currently available from public repositories. Suggestive evidence of linkage (P⩽7.4×10-4) was observed at seven sites in the genome, with linkage in other potential regions conditional on stratification by HLA or age.
Subjects and Methods
Subjects
A total of 767 families were studied. The families were drawn from the Human Biological Data Interchange (HBDI) repository (N=389) (Lernmark et al. 1990), the Diabetes UK Warren I repository (N=356) (Bain et al. 1990), and the Children’s Hospital of Philadelphia (CHOP) (N=22). All families studied were of white European ancestry and resided in either the United Kingdom or the United States. Mein et al. (1998) established criteria for inclusion that required at least one affected sib with onset of disease at age <17 years and no affected sib with age at onset >29 years. In the present study, 756 of 767 families met these criteria. In the remaining 11 families, ages at onset were 18–36 years. Only four of these families were included in genome scans; the other seven were used only for follow-up studies at selected sites.
In the full collection of 767 families, there were 710 with two affected siblings, 51 with three affected siblings, 1 with four affected siblings, and 2 with five affected siblings; in addition, 3 extended pedigrees from the HBDI repository had both affected sibs and affected relative pairs with more distant relationships. In total, there were 831 pairwise independent ASPs, 8 avuncular pairs, and 4 first-cousin pairs.
Genotyping
Genotypes were derived from three sources. For the present report, 225 families from the HBDI repository were genome-scanned at 10-cM resolution by the Mammalian Genotyping Service at the Marshfield Institute. These data, which are reported here for the first time, were merged with data from our previous genome scans of 187 families (101 from Warren I, 64 from HBDI, and 22 from CHOP) (Concannon et al. 1998) and a further 356 families from Warren I (Mein et al. 1998). There was no overlap between the 225 HBDI families who underwent genome scanning for the present study and those similarly analyzed in the two previously published studies. The studies by Concannon et al. (1998) and Mein et al. (1998) had 100 Warren I families in common. When this overlap is taken into account, the total number of families in the current analysis that have undergone genomewide scanning is 667 (356 Warren I, 289 HBDI, and 22 CHOP). Data from an additional 100 HBDI families genotyped only within selected candidate regions were also incorporated. Genotype data from all sources were merged into a single database. Discrepancies in family-naming conventions were resolved. Individual marker names were modified to indicate the laboratory of origin for the genotyping; no markers were deleted. There was ∼20% overlap in marker usage among the three merged data sets. Markers that overlapped between the different data sets were given distinct designations and were treated as unique markers spaced 0.1 cM apart.
Analyses
The complete data set was screened for Mendelian inconsistencies, using PedCheck (O'Connell and Weeks 1998). Inconsistencies were eliminated by retyping or by removal from the analysis. Multipoint linkage analysis was performed using the S (pairs) option of GeneHunter-Plus, and maximized LOD scores were calculated under an exponential model with δ constrained between 0 and 2 (Kong and Cox 1997).
To search for evidence of statistical interaction between unlinked loci (Cox et al. 1999), correlations were calculated using family-specific NPL scores for loci yielding nominal evidence of linkage to T1D (LOD ⩾0.59; P<.05). This analysis was performed in two stages, focusing first on those loci where the evidence of linkage was suggestive (LOD ⩾2.2) and then on all loci with LOD ⩾0.59. P values were calculated from a t statistic with 766 df and corrected, by the Bonferroni method, for the number correlations examined. This test is slightly conservative, because the number of comparisons for different loci varied but was always less than the total number of families used to set the degrees of freedom.
In the specific case of IDDM1 (HLA), family-specific weights were assigned on the basis of the degree of risk associated with the HLA-DR and -DQ genotypes of the affected members of each pedigree. Families received weights on a scale of 1.0–5.0, with 5.0 corresponding to the highest HLA-encoded risk (families in which all affected individuals had the high-risk genotype combination DR3 and DR4) and 1.0 corresponding to the lowest risk (no affected individuals with either DR3 or DR4). Intermediate risks were assigned on the basis of the mixture of DR3, DR4, and DRX (where “X” is not 3 or 4) present in the affected members of the pedigrees. DR4 haplotypes carrying the DQB1*0301 allele were treated as lower risk than those with the DQB1*0302 allele. A total of 453 families in the data set had genotypes for DR and DQ available. To establish a baseline for conditional analyses, these families were assigned weight 1, all other families were assigned weight 0, and LOD scores were calculated. Multipoint analyses were then performed in subsets of families defined by HLA risk, by assigning weight 1 to the families in which genotypes for DR and DQ were available and assigning weight 0 to all others.
As an initial screen for significance in these conditional analyses, all sites were identified where conditioning resulted in an increase ⩾1.44 in the LOD score over that obtained in the baseline analysis. The increase over baseline LOD score obtained by family weighting multiplied by 2ln(10) is asymptotically distributed as a χ2 with 1 df, under the null hypothesis of no interaction. When this test is used, a cutoff of 1.44 corresponds to a P value of .01. Although appropriate as a screening tool, this test does not take into account the possibility of false positives resulting from multiple testing, nor does it consider that there are different proportions of families in the different weighted subsets. Therefore, the significance of any observed increase in the LOD score at specific chromosomal positions was assessed by simulations in which families were randomly assigned to have weight 1 or weight 0, with the number of families of each type determined by the actual number of families assigned weights 1 and 0 for that HLA-based analysis. All simulations included at least 1,000 replicates. For the region on chromosome 10p13, 10,000 replicates were analyzed.
For age-at-onset analyses, families were ranked on the basis of the oldest affected sibling's age at onset. Extended pedigrees with more-distant relative pairs and families with incomplete age-at-onset information were excluded by assigning them weight 0 in all analyses. This excluded group accounted for 85 of the overall pool of 767 families. Among the remaining 682 families, there was no significant difference between the mean age at onset in first, second, or third affected siblings (11.1, 11.0, and 11.2 years, respectively). Genomewide analysis was performed with only a single weighting scheme that assigned weight 1 to families in which both affected sibs had onset at age ⩽11 years and assigned weight 0 to all other families.
Power Calculations
Calculations of the power of this collection of families to detect linkage were done assuming a fully informative marker locus with no recombination to the disease-susceptibility locus, using a formula derived from Gu and Rao (1997).
Results
Multipoint Linkage Analyses
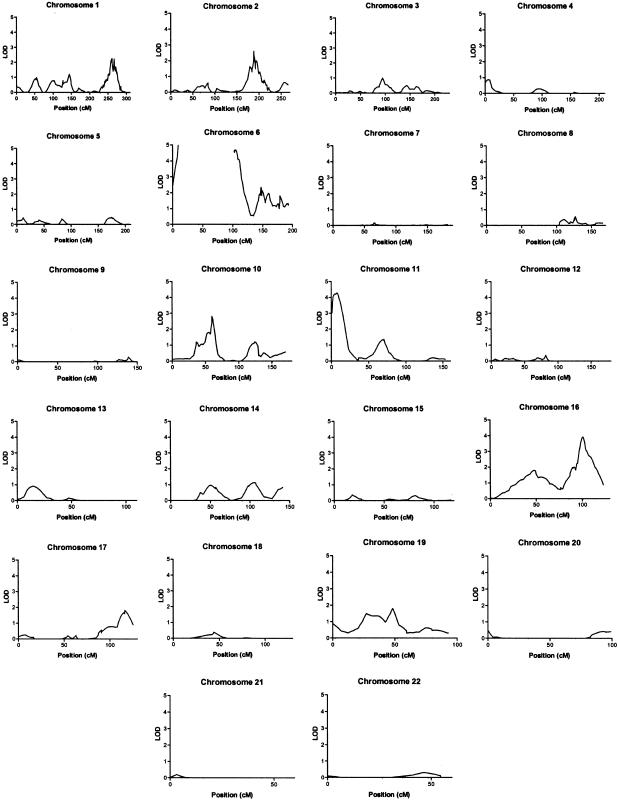
The current study incorporates genotype data from a total of 767 multiplex families with T1D: 356 UK families from the Diabetes UK Warren I repository and 411 US families from the HBDI repository and CHOP. The US families include 225 families that were newly genotyped at 10-cM resolution specifically for this report. The total of 767 families presented here (667 with genomewide-scan data) constitutes a substantial increase over the largest collection of families with T1D studied for linkage to date. Figure 1 shows the results of multipoint linkage analysis in this data set.
Figure 1.
Multipoint linkage analysis, using GeneHunter-Plus, in merged T1D data set. LOD scores were calculated using ASM under an exponential model, with δ constrained between 0 and 2. LOD scores are plotted against chromosomal position (in cM).
There were seven sites at which the multipoint evidence of linkage was either suggestive (LOD ⩾2.2; P⩽7.4×10-4) or significant (LOD ⩾3.6; P⩽2.2×10-5), according to previously proposed criteria (Lander and Kruglyak 1995). As expected, the contribution of the IDDM1 region was easily detected (LOD=65.8). The IDDM2 region also showed significant evidence of linkage (LOD=4.28). To exclude possible bias in the detection of IDDM2 resulting from the inclusion of markers selected for their reported allelic association with T1D, a second analysis of the data for chromosome 11 was performed with these markers excluded. When data for only the framework markers D11S922 and D11S2362 flanking the region were included, the maximum multipoint LOD score in the IDDM2 region was still suggestive of linkage (LOD=2.53).
One additional region on 16q, near the marker D16S3098, also displayed significant evidence of linkage (LOD=3.93). Mein et al. (1998) have reported suggestive evidence of linkage to this same region, and Cordell et al. (1995) suggested, on the basis of a two-locus conditional analysis, the possibility of interaction between this locus and IDDM1. Additional genotyping of the marker D16S3098 was performed in families collected by HBDI, and this resulted in a further increase in the LOD score to 4.13.
Suggestive evidence of linkage was observed at four additional sites. On chromosome 10p11, near the marker D10S565, a LOD score of 2.80 was obtained. The position of this site corresponds approximately to that of the putative IDDM10 locus (Reed et al. 1997; Mein et al. 1998). On chromosome 2q, three putative loci, IDDM7 (2q31, D2S152 associated), IDDM12 (2q33, CTLA4 associated) and IDDM13 (2q34-q34, NRAMP1 associated), have been described elsewhere (Copeman et al. 1995; Morahan et al. 1996; Nistico et al. 1996; Esposito et al. 1998). A single peak was apparent on chromosome 2q31, near the marker D2S1391, with a maximum LOD of 2.62. On chromosome 6q, a LOD score of 2.36 was observed, corresponding to the putative locus IDDM15 (Delepine et al. 1997). Caution is required in the interpretation of these results for IDDM15, because of the potential confounding effects of the strong sharing at IDDM1. Finally, on chromosome 1q, there were two closely spaced peaks (LOD=2.2) that corresponded to the previously reported linkage at 1q42 (Concannon et al. 1998).
There were five other chromosome regions where the multipoint LOD was >1.5. In addition to the peaks at IDDM1 and IDDM15, there were two peaks on 6q, with LOD scores of 1.96 and 1.81, that may correspond to the loci reported elsewhere: IDDM5 (Davies et al. 1994) and IDDM8 (Luo et al. 1995). Other peaks occurred on chromosomes 16p11-p13 (LOD=1.74), 17q25 (LOD=1.81), and 19q11 (LOD=1.80).
Testing for Interaction between Unlinked Loci
Analytic methods that take into account the joint effects that multiple genetic loci have on risk may strengthen the evidence of their existence and contribute to their localization. Evidence of statistical interactions between unlinked loci in the present study was evaluated by calculating correlations between family-specific NPL scores for pairs of loci in a two-step process (Cox et al. 1999). Initially, calculations were performed only for pairs of loci, excluding IDDM1, that displayed evidence of linkage (P⩽7.4×10-4) in the overall genome scan. A total of 15 correlations were calculated using NPL scores at the regional multipoint maximums corresponding to IDDM2, IDDM15, 16q22-q24, IDDM10, IDDM7, and 1q42. A single position at 1q42 was chosen, because the two peaks had equivalent LOD scores, and the NPL scores at the two sites were highly correlated (r=.92; P<.0001). The number of families considered in each calculation was 727–747. Only one correlation (1q42 with IDDM2) was nominally significant (P<.05), but not after correction for the number of tests performed.
The testing of loci that individually display substantial evidence of linkage to T1D is useful as a means of modeling interactions between established susceptibility loci. However, there may be additional loci that do not individually provide significant evidence of linkage but that may be revealed on the basis of evidence of their interaction. Accordingly, the screen for correlation between family-specific NPL scores was broadened, in a second stage, to include all possible pairs of unlinked loci that displayed nominal evidence of linkage in the overall genome scan (P<.05; LOD ⩾ 0.59), again with the exception of IDDM1. A total of 422 pairs of loci were examined, 23 of which yielded results that were nominally significant (P<.05) (table 1), but again, none remained so after correction for multiple testing.
Table 1.
Interaction between NPL Scores in Unlinked Regions
|
First Locus in Pair |
Second Locus in Pair |
||||
| Chromosome | Position | Chromosome | Position | Correlation | P |
| 13 | 14.60 | 19 | .10 | .10129 | .005 |
| 2 | 84.40 | 11 | 7.90 | .09978 | .006 |
| 6 | 160.90 | 10 | 124.20 | .09756 | .007 |
| 1 | 54.70 | 2 | 84.40 | .09663 | .007 |
| 1 | 144.50 | 19 | .10 | .09436 | .009 |
| 2 | 84.40 | 14 | 50.10 | .09016 | .012 |
| 4 | 5.60 | 6 | 160.90 | .09012 | .012 |
| 1 | 260.60 | 11 | 7.90 | .08653 | .016 |
| 1 | 266.70 | 11 | 7.90 | .08186 | .023 |
| 3 | 94.80 | 6 | 160.90 | .08141 | .024 |
| 1 | 54.70 | 6 | 160.90 | .07792 | .031 |
| 6 | 179.10 | 10 | 124.20 | .07554 | .036 |
| 1 | 127.30 | 6 | 193.00 | .07540 | .037 |
| 2 | 256.80 | 11 | 70.50 | .07440 | .039 |
| 4 | 5.60 | 6 | 147.60 | .07342 | .042 |
| 6 | 147.60 | 14 | 141.30 | .07318 | .043 |
| 11 | 70.50 | 19 | .10 | .07275 | .044 |
| 3 | 94.80 | 6 | 147.60 | .07149 | .048 |
| 1 | 54.70 | 1 | 144.50 | .07110 | .049 |
| 2 | 256.80 | 16 | 101.10 | .07096 | .049 |
| 6 | 179.10 | 11 | 7.90 | −.07259 | .044 |
| 2 | 256.80 | 16 | 48.00 | −.07456 | .039 |
| 6 | 193.00 | 11 | 7.90 | −.07553 | .036 |
Conditioning on HLA Genotypes
To perform subgroup analyses based on HLA genotypes, the linkage data from individual families were weighted on the basis of the risk associated with the HLA-DR and -DQ genotypes present in the individual affected members. By applying weight 1 or weight 0 to specific categories of families, it was then possible to recalculate LOD scores, after the basic GeneHunter analysis was completed, to model various types of interactions. Initially, a baseline analysis was performed in which weight 1 was assigned to all families (N=453) for which complete HLA-DR and -DQ genotyping information was available, and weight 0 was assigned to all other families. Three different weighting schemes based on the possible contributions of HLA to T1D susceptibility were then applied. To model positive interactions, weight 1 was assigned to families in which every affected individual carried the DR3/DR4 genotype that confers the highest HLA-encoded risk (N=159), and weight 0 was assigned to all other families. As a model for heterogeneity, conditional analyses were performed in which weight 1 was assigned to families in which no affected member had DR3 or DR4 haplotype (N=19), and weight 0 was assigned to all other families. Finally, we focused on families with DR3-encoded risk of diabetes only as suggested by previous studies (Cucca et al. 1998; Verge et al. 1998). Families were assigned weight 1 if all affected members had at least one DR3 haplotype and no affected member had a DR4 haplotype (N=62), with all other families receiving weight 0.
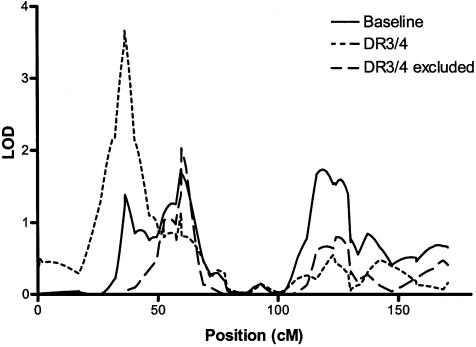
There were two sites where these conditional analyses resulted in an increase above the baseline LOD score that was significant at P⩽.01. On chromosome 3 at 94.8 cM (3p12-q13), the baseline LOD was 0.41, and the LOD score at this position in families in which the affected members had DR3, but not DR4, was 1.95 (P=.005). The second site was located on chromosome 10p13, where an increase in LOD score was observed in the subset of families in which all affected individuals had the high-risk DR3/DR4 genotype. This increase in LOD score, from a baseline value of 1.38 to 3.66, was the most significant pointwise increase observed in the conditional analyses overall (P=.0007) (fig. 2). However, it occurs within 23 cM of the peak (described above) that we observed in the overall linkage analysis at 10p11 (LOD=2.8), raising the possibility that these two findings represent a single linked region. An additional analysis, in which families with DR3/DR4 haplotype were given weight 0, suggests that the evidence of linkage at 10p13 and 10p11 derives from largely nonoverlapping sets of families (fig. 2). No significant increase in LOD score was observed at this site in a specific follow-up analysis in which families were assigned weights according to the presence of individual DR3 or DR4 haplotypes.
Figure 2.
Multipoint analysis of linkage to T1D on chromosome 10, conditioning on HLA. LOD scores are plotted against chromosomal position in centimorgans. Solid line indicates the baseline LOD score in all families for which HLA-typing data were available (N=453). Dotted line indicates LOD score in families in which all ASPs were HLA-DR3/DR4 heterozygotes (N=159). Dashed line indicates LOD score in families for which HLA-typing data were available, excluding families in which all ASPs were HLA-DR3/DR4 heterozygotes (N=294).
The effects of these conditioning schemes were examined specifically at previously reported T1D susceptibility loci. In only one case was an increase above the baseline LOD score observed that was nominally significant. This occurred in the region of the IDDM17 locus, where the baseline LOD was 1.56 but increased to 2.44 in the subset of 62 families in which all affected individuals had a DR3 haplotype and none had DR4 (P=.027).
The subset in which no affected individual had either of the T1D-associated HLA haplotypes (DR3 or DR4) contained only 19 families. This sample is so small that it is not clear there is merit in comparing results from this subset with results from the baseline analyses. Overall, no chromosome region displayed a LOD score >2.0 in this subset.
Age-at-Onset Subgroup Analysis
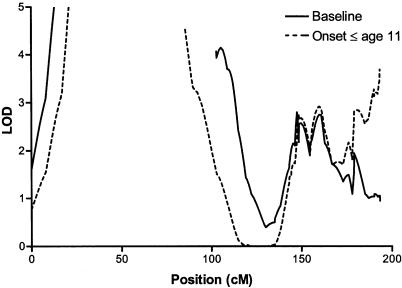
As in the case of HLA, testing for an effect of age at onset on the evidence of linkage in families is complicated by the fact that, although age at onset is a characteristic of a single individual, linkage is inherently a characteristic of groups. To take into account both individual age at onset and ASP concordance for this phenotype, families were classified according to the latest age at onset in any affected sibling. A genomewide search for linkage to T1D, conditioning on early age at onset, was performed by assigning weight 1 to families in which all affected siblings had onset of disease at an age less than or equal to the mean age at onset for the entire collection and by assigning weight 0 to all other families. Only one site in the genome displayed a significant increase in LOD score in this subset of 287 families, compared with the baseline LOD determined for 682 families. On chromosome 6q27, the baseline LOD score was 0.94, and the conditional LOD was 3.69 (P=.003) (fig. 3). This peak in the conditional analysis may correspond to the IDDM8 locus reported elsewhere (Davies et al. 1994; Luo et al. 1995; Owerbach 2000). Increases above the baseline LOD score at this point were also observed when families were selected for increasingly early ages at onset in ASPs, for example, age 8 years (N=154; LOD=2.56) or age 5 years (N=79; LOD=1.42).
Figure 3.
Multipoint analysis of linkage to T1D on chromosome 6, conditioning on age at onset. LOD scores are plotted against chromosomal position in centimorgans. Solid line indicates the baseline LOD score in all families for which age-at-onset data were available (N=682). Dashed line indicates LOD score in families in which all affected siblings had onset of disease at age ⩽11 years (N=287).
Discussion
Extensive study of the genetics of T1D, during the last three decades, has resulted in the mapping of numerous putative susceptibility loci (Reijonen and Concannon 2001). Despite these efforts, and the considerable enthusiasm with which genome-scanning approaches in ASPs have been applied since 1994, progress toward identification of the underlying susceptibility genes for T1D has been slow. The most likely explanation for this limited progress is that no locus, except HLA/IDDM1, appears to have a λs >1.4, at least in the general populations of the United Kingdom and the United States. Two collections of DNA samples from families with T1D are publicly available for genetic linkage studies, the Diabetes UK Warren 1 repository of samples from the United Kingdom and the HBDI collection of samples largely from the United States. Each collection contains <500 ASPs and is independently capable of providing power to detect loci with a λs >1.5; however, each has only modest power to detect effects in the λs range of 1.1–1.4. Nonetheless, putative diabetes loci IDDM3-13, IDDM15, IDDM17, and IDDM18, as well as unnamed regions on chromosomes 1q42, 16q22-24, and Xp11, have been reported. The loci have been identified on the basis of linkage data and, in some cases, on evidence of association (e.g., CTLA4/IDDM12 and IL12B/IDDM18). None of these regions has been independently confirmed in a fully convincing way, mainly owing to the lack of additional, independent families.
In this report we have combined the UK and US data sets and incorporated new genomewide-scan data from 225 additional multiplex families, yielding a total of 767 families and 831 pairwise ASPs. The resulting merged data set is significantly larger than any collection previously analyzed for T1D linkage and could be expected to provide at least 80% power to detect suggestive evidence of linkage (P=7.4×10-4) for loci with λs ⩾1.3 and to provide significant evidence of linkage (P=2.2×10-5) for loci with λs ⩾1.4. Linkage at IDDM1 was easily detected in this data set, as it was in smaller data sets (Davies et al. 1994). Our detection of linkage at six non-HLA regions at a level of significance (P<7.4×10-4) for which approximately one observation would be expected by chance alone indicates the presence of additional T1D loci. Overall, the probability of obtaining five loci, each with P<7.4×10-4, is .004. Confirmation of these and other, as yet undetected, loci will require a much larger number of families, at least 2,500 ASPs, to detect loci with odds ratios of 2.5–3.0, as is the case for INS/IDDM2. The odds ratio of 2.5–3.0 for the INS VNTR class I allele homozygous genotype corresponds to λs of 1.12. Although the present study analyzed data from <2,500 ASPs, a LOD score of 4.28 was obtained in the IDDM2 region. There are several possible explanations for this result. It may be a chance event, given that our study sample contained only 831 ASPs. It may also indicate that the IDDM2 region on chromosome 11p15 contains additional T1D loci. A third possibility is that the significance of the evidence of linkage in this region reflects, in part, the presence of linkage disequilibrium with several of the markers that were genotyped because of their proximity to INS. Consistent with this possibility, a substantial decrease in LOD score (from 4.28 to 2.53) in this region was observed when only framework microsatellite markers were used in the analysis, even though the INS-specific markers that were excluded were of only modest information content.
Previous studies have stratified on allele sharing at HLA-DRB1 or -DQB1 as a means of revealing additional loci whose effects might be more easily discerned on the basis of their interaction with IDDM1. However, allele sharing at DRB1 or DQB1 may not be a very powerful parameter to condition on in T1D, given (a) the high percentage of ASPs that share alleles at IDDM1 and (b) the fact that many parents in multiplex families carry two susceptible HLA class II haplotypes. A previous study of the HBDI collection of multiplex families with T1D revealed that the majority of ASPs that did not share parental alleles at HLA nevertheless had high-risk genotypes (Noble et al. 1996). Therefore, to test for possible interactions between specific HLA genotypes and allele sharing at non-HLA loci, we constructed a weighting scheme based on the risks associated with specific genotypes at DRB1 and DQB1. Given the considerations with regard to power that are discussed and cited above, these analyses, which involve limited subsets of our collection of families, have only modest power and should be considered exploratory.
Two results obtained by conditioning on HLA, both on chromosome 10, are noteworthy. On chromosome 10p13, a LOD of 3.66 was obtained in the subset of 159 families in which all affected individuals had the high-risk DR3/DR4 genotype. The increase, relative to baseline, in the LOD score at this position was significant and was specific to the DR3/DR4 combination (P=.0007, when tested by simulation in a pointwise test). Evidence of linkage at 10p13 (LOD=3.66 in the conditional analysis) and the adjacent 10p11 interval (LOD=2.80 in the overall analysis) came from largely nonoverlapping sets of families. We recognize that, given the overall evidence of linkage at 10p11, there is considerable likelihood that the localization and magnitude of that signal may drift in randomly chosen subsets of the data. Further studies in larger collections of families will be necessary to clarify whether there is a single locus or multiple loci in this region.
At chromosome 10q25, increased LOD scores were observed near the reported location of the putative IDDM17 locus. Although the increase over baseline LOD at this site was only nominally significant, the location of the increase—and its occurrence in the subset of families in which all affected individuals have DR3 and none has DR4—is consistent with the original description of IDDM17. The result raises the possibility that this locus, originally identified in a single large pedigree, may have a measurable effect in the general populations of the United Kingdom and the United States, which might help in the fine-mapping of the locus.
Lernmark and Ott (1998) suggested that differences in the findings in the genome scans of Concannon et al. (1998) and Mein et al. (1998) might have resulted from the application of specific age-at-onset criteria in the latter study. However, in merging the data from these two previous studies, we encountered only 11 (of 767) families that did not meet these age-at-onset criteria. Nevertheless, conditioning on age at onset may identify more genetically homogeneous subsets of families or one in which the role of environmental factors in susceptibility is reduced. We explored this possibility by focusing on the evidence of linkage in the subset of families in which both affected siblings had early onset of disease, as suggested by Li and Hsu (2000). Significantly increased LOD scores were observed at only one site in this subset, at chromosome 6q27. Linkage and association studies have mapped the IDDM8 locus to this same general region (Davies et al. 1994; Luo et al. 1995; Owerbach 2000), but it is not clear whether the increased LOD scores that we observed in the present conditional analysis correspond exactly to the IDDM8 locus defined in previous studies. Subsets of families with incrementally lower ages at onset for ASPs, also showed increased LOD scores over baseline at this site. A similar approach, focusing on transmissions to affected offspring with earlier ages at onset in a TDT analysis (Li and Hsu 2000), might aid in linkage disequilibrium mapping in the region.
In the present study, we used several analytic approaches to try to identify and/or clarify the contributions of loci to T1D susceptibility. However, the most effective strategy employed was also the simplest and most obvious: increasing the sample size. This was accomplished, in part, by performing a new genome scan, but the majority of the data analyzed came from the merging of information obtained from previous studies. Given the number of families that have already been studied worldwide for linkage to T1D—either in focused, site-specific studies or in genomewide scans—the most expedient approach to further progress in the identification of genes for T1D would be through the efforts of a consortium to merge and jointly analyze all extant data sets for linkage. In the meantime, the present study provides the best evidence yet for a role of multiple non-HLA genes in the familial clustering of T1D in humans, as has been previously demonstrated in spontaneous rodent models of T1D.
Acknowledgments
The authors wish to thank Patsy Byers and Mary Jones, for technical assistance, and Heather Cordell, Gillian Johnson, and Iain Eaves, for advice. This work was supported by National Institutes of Health (NIH) grants DK55889 and DK58026 (to N.J.C.), a joint grant from the Juvenile Diabetes Research Foundation (JDRF) and the Wellcome Trust (to J.A.T.), and NIH grants DK46635 and DK55970 and an independent JDRF grant (to P.C.). We thank Diabetes UK and HBDI, for making available their collections of family DNA samples, and the many patients with T1D and their family members who contributed to these collections.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Institute, http://research.marshfieldclinic.org/genetics (for genotyping of families from the HBDI repository)
- Online Mendelian Inheritance of Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for T1D [MIM 222100], HLA-DQB1 [MIM 604305], HLA-DRB1 [MIM 142857], HLA-DPB1 [MIM 142858], IDDM2 [MIM 125852], IDDM3 [MIM 600318] IDDM4 [MIM 600319], IDDM5 [MIM 600320], IDDM6 [MIM 601941], IDDM7 [MIM 600321], IDDM8 [MIM 600883], IDDM10 [MIM 601942], IDDM11 [MIM 601208], IDDM12 [MIM 601388], IDDM13 [MIM 601318], IDDM 15 [MIM 601666], and IDDM18 [MIM 605598])
References
- Bain SC, Todd JA, Barnett AH (1990) The British Diabetic Association—Warren repository. Autoimmunity 7:83–85 [DOI] [PubMed] [Google Scholar]
- Bell GI, Horita S, Karam JH (1984) A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 33:176–183 [DOI] [PubMed] [Google Scholar]
- Bennett ST, Lucassen AM, Gough SC, Powell EE, Undlien DE, Pritchard LE, Merriman ME, Kawaguchi Y, Dronsfield MJ, Pociot F, Nerup J, Bouzekri N, Cambon-Thomsen A, Ronningen KS, Barnett AH, Bain SC, Todd JA (1995) Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet 9:284–292 [DOI] [PubMed] [Google Scholar]
- Concannon P, Gogolin-Ewens KJ, Hinds D, Wapelhorst B, Morrison VA, Stirling B, Mitra M, Farmer J, Williams SR, Cox NJ, Bell GI, Risch N, Spielman RS (1998) A second-generation screen of the human genome for susceptibility to insulin-dependent diabetes mellitus (IDDM). Nat Genet 19:292–296 [DOI] [PubMed] [Google Scholar]
- Copeman JB, Cucca F, Hearne CM, Cornall RJ, Reed PW, Ronningen KS, Undlien DE, Nistico L, Buzzetti R, Tosi R, Pociot F, Nerup J, Cornelis F, Barnett AH, Bain SC, Todd JA (1995) Linkage disequilibrium mapping of a type 1 diabetes susceptibility gene (IDDM7) to chromosome 2q31-q33. Nat Genet 9:80–85 [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Todd JA, Bennett ST, Kawaguchi Y, Farrall M (1995) Two-locus maximum lod score analysis of a multifactorial trait: joint consideration of IDDM2 and IDDM4 with IDDM1 in type 1 diabetes. Am J Hum Genet 57:920–934 [PMC free article] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- Cucca F, Dudbridge F, Loddo M, Mulargia AP, Lampis R, Angius E, De Virgiliis S, Koeleman BP, Bain SC, Barnett AH, Gilchrist F, Cordell H, Welsh K, Todd JA (2001) The HLA-DPB1–associated component of the IDDM1 and its relationship to the major loci HLA-DQB1, -DQA1, and -DRB1. Diabetes 50:1200–1205 [DOI] [PubMed] [Google Scholar]
- Cucca F, Goy JV, Kawaguchi Y, Esposito L, Merriman ME, Wilson AJ, Cordell HJ, Bain SC, Todd JA (1998) A male-female bias in type 1 diabetes and linkage to chromosome Xp in MHC HLA-DR3-positive patients. Nat Genet 19:301–302 [DOI] [PubMed] [Google Scholar]
- Davies JL, Kawaguchi Y, Bennett ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, Gough SC, Jenkins SC, Palmer SM, Balfour KM, Rowq BR, Farrall M, Barnett AH, Bain SC, Todd JA (1994) A genome-wide search for human type 1 diabetes susceptibility genes. Nature 371:130–136 [DOI] [PubMed] [Google Scholar]
- Delepine M, Pociot F, Habita C, Hashimoto L, Froguel P, Rotter J, Cambon-Thomsen A, Deschamps I, Djoulah S, Weissenbach J, Nerup J, Lathrop M, and Julier C (1997) Evidence of a non-MHC susceptibility locus in type I diabetes linked to HLA on chromosome 6. Am J Hum Genet 60:174–187 [PMC free article] [PubMed] [Google Scholar]
- Easteal S, Kohonen-Corish MR, Zimmet P, and Serjeantson SW (1990) HLA-DP variation as additional risk factor in IDDM. Diabetes 39:855–857 [DOI] [PubMed] [Google Scholar]
- Erlich HA, Rotter JI, Chang JD, Shaw SJ, Raffel LJ, Klitz W, Bugawan TL, Zeidler A (1996) Association of HLA-DPB1*0301 with IDDM in Mexican-Americans. Diabetes 45:610–614 [DOI] [PubMed] [Google Scholar]
- Esposito L, Hill NJ, Pritchard LE, Cucca F, Muxworthy C, Merriman ME, Wilson A, Julier C, Delepine M, Tuomilehto J, Tuomilehto-Wolf E, Ionesco-Tirgoviste C, Nistico' L, Buzzetti R, Pozzilli P, Ferrari M, Bosi E, Pociot F, Nerup J, Bain SC, Todd JA (1998) Genetic analysis of chromosome 2 in type 1 diabetes: analysis of putative loci IDDM7, IDDM12, and IDDM13 and candidate genes NRAMP1 and IA-2 and the interleukin-1 gene cluster—IMDIAB Group. Diabetes 47:1797–1799 [DOI] [PubMed] [Google Scholar]
- Gu C and Rao DC (1997) A linkage strategy for detection of human quantitative-trait loci. I. Generalized relative risk ratios and power of sib pairs with extreme trait values. Am J Hum Genet 61:200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto L, Habita C, Beressi JP, Delepine M, Besse C, Cambon-Thomsen A, Deschamps I, Rotter JI, Djoulah S, James MR, Froguel P, Weissenbach J, Lathrop GM, Julier C (1994) Genetic mapping of a susceptibility locus for insulin-dependent diabetes mellitus on chromosome 11q. Nature 371:161–164 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M, Guo SW, Risch N (1996) Affected-sib-pair interval mapping and exclusion for complex genetic traits: sampling considerations. Genet Epidemiol 13:117–137 [DOI] [PubMed] [Google Scholar]
- Horn GT, Bugawan TL, Long CM, Erlich HA (1988) Allelic sequence variation of the HLA-DQ loci: relationship to serology and to insulin-dependent diabetes susceptibility. Proc Natl Acad Sci USA 85:6012–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier C, Lucassen A, Villedieu P, Delepine M, Levy-Marchal C, Danze PM, Bianchi F, Boitard C, Froguel P, Bell J, Lathrop GM (1994) Multiple DNA variant assocation analysis: application to the insulin gene region in type 1 diabetes. Am J Hum Genet 55:1247–1254 [PMC free article] [PubMed] [Google Scholar]
- Kennedy GC, German MS, Rutter WJ (1995) The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat Genet 9:293–298 [DOI] [PubMed] [Google Scholar]
- Kong A and Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E and Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lernmark A, Ducat L, Eisenbarth G, Ott J, Permutt MA, Rubenstein P, Spielman R (1990) Family cell lines available for research. Am J Hum Genet 47:1028–1030 [PMC free article] [PubMed] [Google Scholar]
- Lernmark A and Ott J (1998) Sometimes it's hot, sometimes it's not. Nat Genet 19:213–214 [DOI] [PubMed] [Google Scholar]
- Li H and Hsu L (2000) Effects of age at onset on the power of the affected sib pair and transmission/disequilibrium tests. Ann Hum Genet 64:239–254 [DOI] [PubMed] [Google Scholar]
- Lie BA, Todd JA, Pociot F, Nerup J, Akselsen HE, Joner G, Dahl-Jorgensen K, Ronningen KS, Thorsby E, Undlien DE (1999) The predisposition to type 1 diabetes linked to the human leukocyte antigen complex includes at least one non-class II gene. Am J Hum Genet 64:793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucassen AM, Julier C, Beressi JP, Boitard C, Froguel P, Lathrop M, Bell JI (1993) Susceptibility to insulin dependent diabetes mellitus maps to a 4.1 kb segment of DNA spanning the insulin gene and associated VNTR. Nat Genet 4:305–310 [DOI] [PubMed] [Google Scholar]
- Luo DF, Bui MM, Muir A, Maclaren NK, Thomson G, She JX (1995) Affected-sib-pair mapping of a novel susceptibility gene to insulin- dependent diabetes mellitus (IDDM8) on chromosome 6q25-q27. Am J Hum Genet 57:911–919 [PMC free article] [PubMed] [Google Scholar]
- Mein CA, Esposito L, Dunn MG, Johnson GC, Timms AE, Goy JV, Smith AN, Sebag-Montefiore L, Merriman ME, Wilson AJ, Pritchard LE, Cucca F, Barnett AH, Bain SC, Todd JA (1998) A search for type 1 diabetes susceptibility genes in families from the United Kingdom. Nat Genet 19:297–300 [DOI] [PubMed] [Google Scholar]
- Morahan G, Huang D, Tait BD, Colman PG, Harrison LC (1996) Markers on distal chromosome 2q linked to insulin-dependent diabetes mellitus. Science 272:1811–1813 [DOI] [PubMed] [Google Scholar]
- Nistico L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, Bosi E, Larrad MT, Rios MS, Chow CC, Cockram CS, Jacobs K, Mijovic C, Bain SC, Barnett AH, Vandewalle CL, Schuit F, Gorus FK, Belgian Diabetes Registry, Tosi R, Pozzilli P, Todd JA (1996) The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Hum Mol Genet 5:1075–1080 [DOI] [PubMed] [Google Scholar]
- Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, and Erlich HA (1996) The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet 59:1134–1148 [PMC free article] [PubMed] [Google Scholar]
- Noble JA, Valdes AM, Thomson G, Erlich HA (2000) The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes 49:121–125 [DOI] [PubMed] [Google Scholar]
- O'Connell JR and Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owerbach D (2000) Physical and genetic mapping of IDDM8 on chromosome 6q27. Diabetes 49:508–512 [DOI] [PubMed] [Google Scholar]
- Owerbach D and Gabbay KH (1993) Localization of a type I diabetes susceptibility locus to the variable tandem repeat region flanking the insulin gene. Diabetes 42:1708–1714 [DOI] [PubMed] [Google Scholar]
- ——— (1994) Linkage of the VNTR/insulin-gene and type I diabetes mellitus: increased gene sharing in affected sibling pairs. Am J Hum Genet 54:909–912 [PMC free article] [PubMed] [Google Scholar]
- Pugliese A, Zeller M, Fernandez A, Jr., Zalcberg LJ, Bartlett RJ, Ricordi C, Pietropaolo M, Eisenbarth GS, Bennett ST, Patel DD (1997) The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet 15:293–297 [DOI] [PubMed] [Google Scholar]
- Reed P, Cucca F, Jenkins S, Merriman M, Wilson A, McKinney P, Bosi E, Joner G, Ronningen K, Thorsby E, Undlien D, Merriman T, Barnett A, Bain S, Todd J (1997) Evidence for a type 1 diabetes susceptibility locus (IDDM10) on human chromosome 10p11-q11. Hum Mol Genet 6:1011–1016 [DOI] [PubMed] [Google Scholar]
- Reijonen H and Concannon P (2001) Genetics of type 1 diabetes. In: Krall LP, Beaser RS (eds) Joslin diabetes mellitus, 14th ed. Lea & Febiger, Philadelphia (in press) [Google Scholar]
- Rich SS (1990) Mapping genes in diabetes: genetic epidemiological perspective. Diabetes 39:1315–1319 [DOI] [PubMed] [Google Scholar]
- Risch N (1987) Assessing the role of HLA-linked and unlinked determinants of disease. Am J Hum Genet 40:1–14 [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus. Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Suarez BK, Hampe CL, Van Eerdewegh P (1994) Problems of replicating linkage claims in psychiatry. In: Gershon ES, Cloninger CR (eds) Genetic approaches to mental disorders. American Psychiatric Press, Washington, DC, pp 23–46 [Google Scholar]
- Thomson G, Robinson WP, Kuhner MK, Joe S, MacDonald MJ, Gottschall JL, Barbosa J, Rich SS, Bertrams J, Baur MP (1988) Genetic heterogeneity, modes of inheritance, and risk estimates for a joint study of Caucasians with insulin-dependent diabetes mellitus. Am J Hum Genet 43:799–816 [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Bell JI, McDevitt HO (1987) HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329:599–604 [DOI] [PubMed] [Google Scholar]
- Vafiadis P, Bennett ST, Colle E, Grabs R, Goodyer CG, Polychronakos C (1996) Imprinted and genotype-specific expression of genes at the IDDM2 locus in pancreas and leucocytes. J Autoimmun 9:397–403 [DOI] [PubMed] [Google Scholar]
- Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, Colle E, Polychronakos C (1997) Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet 15:289–292 [DOI] [PubMed] [Google Scholar]
- Verge CF, Vardi P, Babu S, Bao F, Erlich HA, Bugawan T, Tiosano D, Yu L, Eisenbarth GS, Fain PR (1998) Evidence for oligogenic inheritance of type 1 diabetes in a large Bedouin Arab family. J Clin Invest 102:1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]