Abstract
Maple syrup urine disease (MSUD) is a rare, autosomal recessive disorder of branched-chain amino acid metabolism. We noted that a large proportion (10 of 34) of families with MSUD that were followed in our clinic were of Ashkenazi Jewish (AJ) descent, leading us to search for a common mutation within this group. On the basis of genotyping data suggestive of a conserved haplotype at tightly linked markers on chromosome 6q14, the BCKDHB gene encoding the E1β subunit was sequenced. Three novel mutations were identified in seven unrelated AJ patients with MSUD. The locations of the affected residues in the crystal structure of the E1β subunit suggested possible mechanisms for the deleterious effects of these mutations. Large-scale population screening of AJ individuals for R183P, the mutation present in six of seven patients, revealed that the carrier frequency of the mutant allele was ∼1/113; the patient not carrying R183P had a previously described homozygous mutation in the gene encoding the E2 subunit. These findings suggested that a limited number of mutations might underlie MSUD in the AJ population, potentially facilitating prenatal diagnosis and carrier detection of MSUD in this group.
Originally described by Menkes in 1954, as a rapidly fatal neurodegenerative disease (Menkes et al. 1954), maple syrup urine disease (MSUD) (types Ia [MIM 248600], Ib [MIM 248611]), and II [MIM 248610]), is an inborn error of metabolism, resulting from the defective activity of branched-chain α-ketoacid dehydrogenase (BCKAD) (Chuang and Shih 2000). The enzymatic defect, transmitted in an autosomal recessive manner, results in an inability to catabolize leucine, isoleucine, and valine. BCKAD exists as a multienzyme complex composed of a multimeric core of dihydrolipoyl acyltransferase (E2), to which are bound multiple BCKAD decarboxylase (E1) and dihydrolipoamide dehydrogenase (E3) subunits (Pettit et al. 1978). Two regulatory subunits, BCKAD kinase and BCKAD phosphatase, are also associated with the E2 core (Damuni et al. 1984; Reed et al. 1985). The E1 subunit exists as a heterotetramer composed of two E1α and two E1β subunits. Because the E3 subunit is shared with the pyruvate and α-ketoglutarate dehydrogenase complexes, decreased activity of this component results in deficiencies of all three enzymes.
Five clinical MSUD phenotypes have been described, which differ in their presentation and severity. Classic MSUD, the most common clinical presentation, manifests during the newborn period and can result in severe neurological complications and death. The less-common forms are the later-onset intermediate type, an episodic intermittent type, an intermediate-like thiamin-responsive type, and E3 deficiency with lactic acidosis. Mutations in any of the E1 or E2 components can result in classic, intermediate, or intermittent MSUD, although they are more frequent in E1α and E1β than in E2 (Nellis and Danner 2001). Patients with MSUD can be effectively managed with dietary restriction of the branched-chain amino acids and maintenance of strict biochemical control (Snyderman et al. 1964). However, a significant proportion of treated individuals have impaired intellectual development or neurological complications, particularly as a result of delayed diagnosis (Kaplan et al. 1991), emphasizing the importance of newborn-screening programs. Despite early diagnosis and adequate care, episodes of life-threatening metabolic decompensation occur during periods of stress or infection and are associated with significant morbidity and mortality (Riviello et al. 1991; Levin et al. 1993; Chuang and Shih 2000).
MSUD has been described in all ethnic groups and has an estimated worldwide frequency of 1/185,000 (Naylor 1980). However, a founder effect in the Pennsylvania Mennonite population has led to an increased incidence (1/176) in this population (Marshall and DiGeorge 1981). In our clinic, we noted that a disproportionate percentage (∼30%) of families with MSUD were of Ashkenazi Jewish (AJ) heritage. Here we report the carrier frequency of a common AJ mutation, R183P, and the identification of two novel mutations.
Six unrelated AJ patients with classic MSUD (individuals 2–6 and 9), one patient with intermediate MSUD (individual 1), and the parents of one classic patient (individuals 7 and 8) participated in this study. The protocol was approved by the Mount Sinai School of Medicine Institutional Review Board, and voluntary informed consent was obtained from all participants. DNA was extracted either from peripheral blood (PureGene; Gentra) or from buccal-swab samples (MasterAmp Buccal Kit; Epicentre Technologies).
Our initial focus was on E2ΔAT, a 2-bp deletion in the second exon of the E2 gene, described by Fisher et al. (1993), in individual 9. This mutation destroys an AflIII site in the exon, allowing rapid screening of our AJ patients by amplification of the exon from genomic DNA (table 1), followed by restriction-enzyme digestion. The results of the assay demonstrated that the mutation was found only in individual 9 (not shown). We therefore identified polymorphic short tandem-repeat (STR) markers flanking the genetic loci encoding E1α, E1β, and E2, to determine whether a conserved haplotype consistent with a common mutation was present in our other patients. It is noteworthy that, in order to assess the BCKDHB locus, we first identified candidate STR markers that were linkage-mapped to 6p21-p22, the published cytogenetic localization given in the OMIM Gene Map (Zneimer et al. 1991). Alignments of gene (see the GenBank Overview web site ) and marker sequences with the genomic sequence data, by use of the Human Genome Project Working Draft at UCSC, indicated that BCKDHB maps to cytogenetic location 6q14. Radiation-hybrid mapping data from GeneMap'99 supported the localization of BCKDHB to 6q14, so markers from this region were used in the analysis.
Table 1.
Primers for Exon-Specific PCR amplifications.
|
Primer(5′→3′) |
|||
| Gene and Exon | Forward | Reverse | Length(bp) |
| E1β: | |||
| 1 | TCCAGTTCCGATTGGTCTGT | AGCTGGGATGCAAGGATTG | 434 |
| 2 | CCACAATTCAGGCACATATCAG | GCATAATATCTTTCTTTGTACCTTGA | 222 |
| 3 | TCCATACAATGGGGATATGACA | TGATCGAGATCTATGGTCCATTT | 304 |
| 4 | TCTCATTTGCCACATTAACCTTT | GGGTAGCGGCAATACTTGAA | 239 |
| 5 | CCCCGTCTTTCTTTCTGACC | TGAATCAAAATGGAAACATCAAA | 237 |
| 6 | AGCCCTTCTTAGCAGCGAGT | TTCTACAAGGAAAATATAAATCAACCA | 295 |
| 7 | TGCACAAGTGTCACCTCAGA | GCTTCCAAGCACAACGTAGG | 205 |
| 8 | TTTTTGAGGCTAGAACCTTTGT | GCCAAAGGTTTCAGGGAAAT | 214 |
| 9 | CATCAGGTTTTTCTTTTCTCTTTCA | TCTTCTGGAATTGGCATGTG | 175 |
| 10 | TTGAAGTTAAGCATCCTGACTCTG | CGTTAATGTCAGGGGCACAT | 400 |
| E2: | |||
| 2 | TTGTGTTCGCTATTTTC | CAGCAGTTGTTTTCAGGA | 122 |
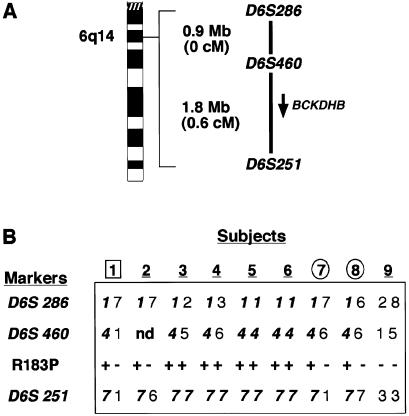
STRs tightly linked to the relevant genes were obtained from ResGen. Radiolabeling, amplification, and electrophoresis were performed by standard methods. Allele assignments were standardized across gels by use of CEPH reference individual 1331-2. The genotyping results were consistent with a disproportionately represented allele at each of the markers flanking the E1β locus (fig. 1). In particular, the 7 allele of marker D6S251 was highly overrepresented, being present in all individuals except for individual 9 and accounting for 10 of 12 total alleles. Typing of 50 control AJ chromosomes gave an estimated 7 allele frequency of ∼8%, confirming the striking allele-frequency distortion in the patients with MSUD. Markers flanking the genes encoding E1α and E2 did not display a pattern consistent with segregation of common alleles (not shown). On the basis of these suggestive results, mutation analysis of the E1β exons was performed in individual 4, a homozygote for the putative three-marker haplotype 1-4-7 at markers D6S286, D6S460, and D6S251, respectively.
Figure 1.
Genomic organization of polymorphic markers tightly linked to the genes encoding E1α, E1β, and E2 and genotype data for markers flanking E1β. A, Organization of STR markers used in analysis of the BCKDHB locus, and their relative spacing, in both physical and genetic distance. BCKDHB spans ∼240 kb, and the 3′ end lies ∼1 Mb upstream of D6S251. B, Results of genotyping analysis of patients with intermediate (boxed) or classic (underlined) MSUD. The parents of individual 5 are denoted by circles. Individual 9 is a patient with a known mutation (i.e., E2ΔAT).
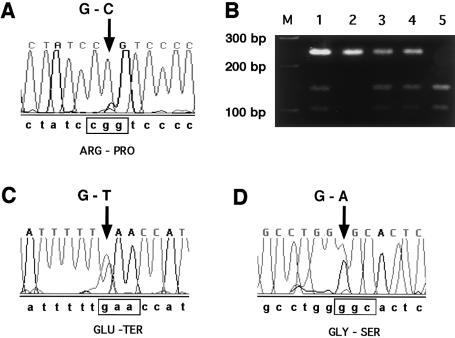
PCR primers were designed to amplify all exons and proximal intronic sequences (table 1). Amplicons were column purified by use of the Qiagen PCR Purification kit and were sequenced bidirectionally on an ABI PRISM 3700 automated sequencer (Applied Biosystems). Traces were processed by the Factura and Autoassembler software packages (Applied Biosystems). A homozygous missense mutation corresponding to 538G→C in the cDNA sequence was detected in exon 5 (fig. 2A). Overlapping MspI and NlaIV sites in the amplicon were destroyed by the base change, allowing the remaining patients to be screened with the same primer set as was used for sequencing (fig. 2B). The mutation was detected in 10 of 12 chromosomes screened and segregated with the 7 allele of marker D6S251, unambiguously in eight instances and presumptively in two instances (fig. 1B).
Figure 2.
Results of mutation analysis of AJ patients with MSUD. A, C, and D, Changes (arrows) from expected wild-type sequence, which is shown below the traces. The affected codon is boxed, and the predicted amino acid change is indicated below the codon. The different sequence traces correspond to exon 5 in individual 4 (A), to exon 10 in individual 2 (C), and to exon 7 in individual 1 (D). B, Representative digestion of exon 5 amplimers with NlaIV (New England Biolabs). The sizes of 100-bp-ladder marker fragments (lane M) are indicated to the left of the gel. The samples shown were amplified from the genomic DNA of individuals 1 (lane 1), 5 (lane 2), 6 (lane 3), 7 (lane 4), and 9 (lane 5). Undigested amplimers are 237 bp in length, whereas digested products are predicted to be 99 bp and 128 bp in length.
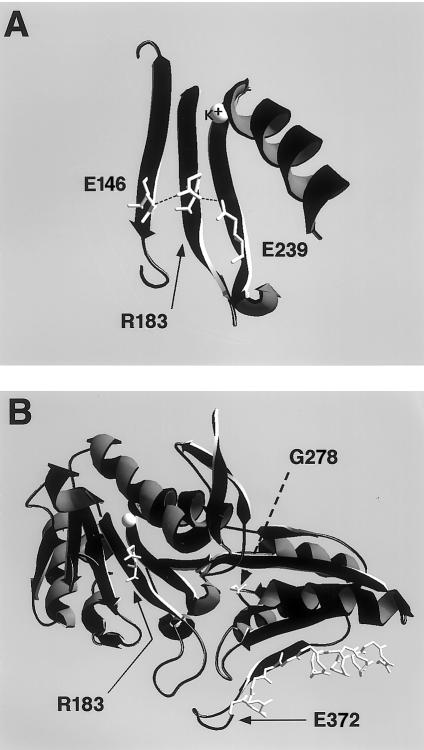
The 538G→C mutation causes an Arg→Pro substitution in E1β, at residue 183 in the mature peptide (R183P). R183 is conserved between E1β and the homologous Pseudomonas putida bacterial enzyme and is located in a β-sheet that contributes to a K+ ion-binding pocket close to the interface of the E1α and E1β subunits (Ævarsson et al. 2000). Hydrogen-bond interactions made by this residue in the E1 crystal structure were visualized by use of a SwissPdb-Viewer (fig. 3A) (Guex and Peitsch 1997). Three parallel β-sheets (d, e, and g) form extensive interactions in this region of the protein. Sheet e is central to sheets d and g, and R183 makes main-chain and side-chain hydrogen-bond interactions with residues on both adjacent sheets, notably E96 and E189, residues that were also conserved between mammalian and bacterial subunits (fig. 3A). Structural changes introduced by a Pro substitution would thus be expected to interfere with a conserved structural motif, by altering the conformation of the central β-strand and by disrupting the hydrogen-bond interactions apparently essential for proper folding of the ion-binding pocket and/or for subunit interaction. These predictions are supported by functional data recently described by Wynn et al. (2001), which demonstrate that the R183P mutation renders the E1β subunit thermolabile at physiological temperatures. Their studies indicate that the mutation confers a temperature-dependent folding defect on the E1β subunit, disrupting its ability to complex with the E1α subunit.
Figure 3.
Ribbon representations of portions of E1β crystal structure, showing positions of mutated residues. The E1 crystal-structure data were obtained from the NCBI Structure (Molecular Modeling Database). Swissprot-Pdb Viewer was used to generate models and image files. A, Ribbon diagram of β-sheets d, e, and g, with the side chains of E146, R183, and E239, respectively, which are shown in white. The mutated residue, R183, is indicated by the arrow, side-chain hydrogen-bond interactions are indicated by dashed lines, and the coordinated K+ ion is labeled directly. For clarity of presentation, main-chain hydrogen interactions have been omitted. B, Overview of entire molecule, except for structures overlying the sheet containing R183. The residues altered by missense mutations, R183 and G228, are indicated by bent and dashed arrows, respectively, and their side chains are shown in white. The α-carbon backbone of residues truncated by the nonsense mutation E372X are shown in white.
Mutation analysis of compound-heterozygous individuals 1 and 2 revealed that they contained two additional novel mutations, 833G→A and 1114G→T, respectively (fig. 2C and D). The latter mutation, found in exon 10, introduced a nonsense codon (E422X) truncating the terminal 20 amino acids. The importance that the C-terminal has for E1-E2–subunit association was pointed out by Ævarsson et al. (2000), who reported that addition of a His6 tail onto E1β did not affect E1 heterotetramer assembly but did interfere with the subsequent E2-association step. This observation raises the possibility that the truncation mutant might interfere with subunit association, although deletion of such a large region might interfere more generally with protein folding. The missense 833G→A mutation in exon 7 resulted in the substitution G278S. This residue was also conserved between mammalian and bacterial E1 homologues, and it is situated at a hairpin turn between β-sheet j and α-helix 9 (fig. 3B). The adjacent residue on helix 9, W277, forms a hydrogen-bond interaction with E231 in a hydrophobic pocket important in E1β-E1β′ interactions (Ævarsson et al. 2000). The G278S mutant might therefore destabilize E1β-subunit interactions during heterotetramer assembly. The severity of these mutations corresponds well with the observed clinical phenotypes, since G278S is associated with the intermediate phenotype and E372X is associated with the classic phenotype.
On the basis of the mutation-analysis results, we screened for the R183P mutation in DNA samples from 1,014 AJ individuals from the New York metropolitan area that were obtained with informed consent. All personal identifiers were removed, and the samples were tested anonymously. PCR amplification was performed with genomic DNA in a volume of 50 μl containing 10 pmol of exon 5–specific primers (table 1), with 100 μM of each dNTP (Roche Molecular Biochemicals), 5 U Taq DNA Polymerase (Roche), 10 mM Tris-HCl, 50 mM KCl, 0.1% Triton X-100, and 1.5 mM MgCl2. Aliquots (4 μl) of the PCR products were blotted onto replicate 8×12-cm Hybond-N+ membranes (Amersham Pharmacia Biotech), by a Biomek 2000 automated pipetting workstation (Beckman Coulter), for hybridization with allele-specific oligonucleotides (ASOs) for the R183P mutation (5′-TCACTATCCCGTCCCCTT-3′) and the wild-type sequence (5′-TCACTATCCGGTCCCCTT-3′). Mutation detection was performed by radioactive methods as described elsewhere (Fodor et al. 1998). For radioactive detection, the membranes were hybridized for 2 h at 42°C, with ∼106 cpm [32P]-ATP end-labeled probe/ml and 10-fold molar excess of the mutant or wild-type competitor oligonucleotide. The membranes were then washed sequentially in 5 × SSC for 5 min at room temperature and in 5 × SSC for 5 min at 45°C, followed by a wash in 0.1 × SSC/0.1% SDS for 5 min at 45°C, and then were exposed to autoradiography. Nine carriers were identified, which corresponded to a carrier frequency of ∼1/113. Samples giving a positive signal with the mutant ASO were reamplified, and carrier status confirmed by the NlaIV PCR-RFLP assay.
The AJ R183P allele, which accounts for 10 of 12 mutant E1β alleles in our limited series, was present at a frequency significantly higher than the estimated MSUD-carrier frequency (∼1/215) in the general population (Peinemann et al. 1993; Danner and Doering 1998; Chuang and Shih 2000) This finding is consistent with our observation of a disproportionate number of AJ cases of MSUD. The expected disease incidence for R183P homozygotes is 1/51,000. Additional compound-heterozygous cases are expected, but the degree of allelic heterogeneity in the population remains to be elucidated. A rough estimate of the number of AJ individuals born after the initiation (in 1968) of New York State newborn screening for MSUD was derived on the basis of information from the most recent (i.e., 1991) available United Jewish Appeal Federation Population Estimates and the 2000 U.S. census (U.S. Census Bureau, American Fact Finder). Assuming that the proportion of AJ individuals <30 years of age is the same as that in the general population and that the percentage of AJ families has remained constant, we estimated that ∼480,000 AJ individuals were born after the initiation of screening. The expected total of approximately nine homozygous cases is somewhat greater than our clinical observations but within reason, considering the crudeness of the estimate.
The high prevalence of R183P in our patients suggests that the identification of this mutation will facilitate prenatal diagnosis of affected families and may permit carrier screening after the identification of additional common mutations in the AJ population. Population screening is currently being offered for other diseases with similar allele frequencies as R183P, including Bloom syndrome (1/107) (Li et al. 1998), mucolipidosis type IV (1/100) (Bargal et al. 2001), Niemann-Pick type A (∼1/90) (Schuchman and Miranda 1997), and Fanconi anemia group C (1/89) (Verlander et al. 1995; Auerbach 1997). Effective carrier detection may decrease the incidence and/or the morbidity associated with delayed diagnosis of this disorder in the AJ population.
Acknowledgments
The authors would like to express their appreciation to all of the families who agreed to participate in the study and to Shihong Zhang for excellent technical assistance. G.A.D. was supported in part by Mount Sinai Child Health Research Center grant 5 P30 HD 28822.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for human branched-chain α-keto dehydrogenase E1β-subunit mRNA [accession U50708])
- GeneMap'99, http://www.ncbi.nlm.nih.gov/genemap99/
- Human Genome Project Working Draft at UCSC, http://genome.ucsc.edu
- NCBI Structure (Molecular Modeling Database), http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.shtml (for human BCKAD [accession number 1DTW])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for MSUD types Ia [MIM 248600], Ib [MIM 248611]), and II [MIM 248610])
- U.S. Census Bureau, American Fact Finder, http://factfinder.census.gov/servlet/BasicFactsServlet
References
- Ævarsson A, Chuang JL, Wynn RM, Turley S, Chuang DT, Hol WG (2000) Crystal structure of human branched-chain α-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure Fold Des 8:277–291 [DOI] [PubMed] [Google Scholar]
- Auerbach AD (1997) Fanconi anemia: genetic testing in Ashkenazi Jews. Genet Test 1:27–33 [DOI] [PubMed] [Google Scholar]
- Bargal R, Avidan N, Olender T, Ben Asher E, Zeigler M, Raas-Rothschild A, Frumkin A, Ben-Yoseph O, Friedlender Y, Lancet D, Bach G (2001) Mucolipidosis type IV: novel MCOLN1 mutations in Jewish and non-Jewish patients and the frequency of the disease in the Ashkenazi Jewish population. Hum Mutat 17:397–402 [DOI] [PubMed] [Google Scholar]
- Chuang DT, Shih VE (2000) Disorders of branched chain amino acid and keto acid metabolism. In: Scriver CR, Beaudet A, Sly WL, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 1971–2006 [Google Scholar]
- Damuni Z, Merryfield ML, Humphreys JS, Reed LJ (1984) Purification and properties of branched-chain alpha-keto acid dehydrogenase phosphatase from bovine kidney. Proc Natl Acad Sci USA 81:4335–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner DJ, Doering CB (1998) Human mutations affecting branched chain alpha-ketoacid dehydrogenase. Front Biosci 3:d517–d524 [DOI] [PubMed] [Google Scholar]
- Fisher CW, Fisher CR, Chuang JL, Lau KS, Chuang DT, Cox RP (1993) Occurrence of a 2-bp (AT) deletion allele and nonsense (G-to-T) mutant allele at the E2 (DBT) locus of six patients with maple syrup urine disease: multiple-exon skipping as a secondary effect of the mutations. Am J Hum Genet 52:414–424 [PMC free article] [PubMed] [Google Scholar]
- Fodor FH, Weston A, Bleiweiss IJ, McCurdy LD, Walsh MM, Tartter PI, Brower ST, Eng CM (1998) Frequency and carrier risk associated with common BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer patients. Am J Hum Genet 63:45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- Kaplan P, Mazur A, Field M, Berlin JA, Berry GT, Heidenreich R, Yudkoff M, Segal S (1991) Intellectual outcome in children with maple syrup urine disease. J Pediatr 119:46–50 [DOI] [PubMed] [Google Scholar]
- Levin ML, Scheimann A, Lewis RA, Beaudet AL (1993) Cerebral edema in maple syrup urine disease. J Pediatr 122:167–168 [DOI] [PubMed] [Google Scholar]
- Li L, Eng C, Desnick RJ, German J, Ellis NA (1998) Carrier frequency of the Bloom syndrome blmAsh mutation in the Ashkenazi Jewish population. Mol Genet Metab 64:286–290 [DOI] [PubMed] [Google Scholar]
- Marshall L, DiGeorge A (1981) Maple syrup urine disease in the Old Order Mennonites. Am J Hum Genet Suppl 33:139A [Google Scholar]
- Menkes J, Hurst P, Craig JM (1954) A new syndrome: progressive familial infantile cerebral dysfunction associated with an unusual urinary substance. Pediatrics 14:462–466 [PubMed] [Google Scholar]
- Naylor EW (1980) Newborn screening for maple syrup urine disease (branched-chain ketoaciduria) In: Bickel H, Guthrie R, Hammersen G (eds) Neonatal screening for inborn errors of metabolism. Springer-Verlag, Berlin, pp 19–28 [Google Scholar]
- Nellis MM, Danner DJ (2001) Gene preference in maple syrup urine disease. Am J Hum Genet 68:232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinemann F, Wendel U, Danner DJ (1993) Molecular genetic characterization of maple syrup urine disease in European families. Biochem Med Metab Biol 50:338–345 [DOI] [PubMed] [Google Scholar]
- Pettit FH, Yeaman SJ, Reed LJ (1978) Purification and characterization of branched chain α-keto acid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci USA 75:4881–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Damuni Z, Merryfield ML (1985) Regulation of mammalian pyruvate and branched-chain α-keto acid dehydrogenase complexes by phosphorylation-dephosphorylation. Curr Top Cell Regul 27:41–49 [DOI] [PubMed] [Google Scholar]
- Riviello JJ Jr, Rezvani I, DiGeorge AM, Foley CM (1991) Cerebral edema causing death in children with maple syrup urine disease. J Pediatr 119:42–45 [DOI] [PubMed] [Google Scholar]
- Schuchman EH, Miranda SR (1997) Niemann-Pick disease: mutation update, genotype/phenotype correlations and prospects for genetic testing. Genet Test 1:13–19 [DOI] [PubMed] [Google Scholar]
- Snyderman SE, Norton PM, Roitman E, Holt LE Jr (1964) Maple syrup urine disease with particular reference to dietotherapy. Pediatrics 34:454 [PubMed] [Google Scholar]
- Verlander PC, Kaporis A, Liu Q, Zhang Q, Seligsohn U, Auerbach AD (1995) Carrier frequency of the IVS4 + 4 A→T mutation of the Fanconi anemia gene FAC in the Ashkenazi Jewish population. Blood 86:4034–4038 [PubMed] [Google Scholar]
- Wynn RM, Chuang JL, Sansaricq C, Mandel H, Chuang DT (2001) Biochemical basis of type 1B (E1β) mutations in maple syrup urine disease: a prevalent allele in patients from the Druze kindred in Israel. J Biol Chem (in press) [DOI] [PubMed] [Google Scholar]
- Zneimer SM, Lau KS, Eddy RL, Shows TB, Chuang JL, Chuang DT, Cox RP (1991) Regional assignment of two genes of the human branched-chain α-keto acid dehydrogenase complex: the E1 β gene (BCKDHB) to chromosome 6p21-22 and the E2 gene (DBT) to chromosome 1p31. Genomics 10:740–747 [DOI] [PubMed] [Google Scholar]