Abstract
The hereditary disorders of peripheral nerve form one of the most common groups of human genetic diseases, collectively called Charcot-Marie-Tooth (CMT) neuropathy. Using linkage analysis we have identified a new locus for a form of CMT that we have called “dominant intermediate CMT” (DI-CMT). A genomewide screen using 383 microsatellite markers showed strong linkage to the short arm of chromosome 19 (maximum LOD score 4.3, with a recombination fraction (θ) of 0, at D19S221 and maximum LOD score 5.28, θ=0, at D19S226). Haplotype analysis performed with 14 additional markers placed the DI-CMT locus within a 16.8-cM region flanked by the markers D19S586 and D19S546. Multipoint linkage analysis suggested the most likely location at D19S226 (maximum multipoint LOD score 6.77), within a 10-cM confidence interval. This study establishes the presence of a locus for DI-CMT on chromosome 19p12-p13.2.
Charcot-Marie-Tooth (CMT) neuropathy, otherwise known as “hereditary motor and sensory neuropathy” (HMSN), is one of the most common groups of human hereditary disorders. The CMT syndrome includes many hereditary disorders of peripheral nerve affecting both motor and sensory neurons. The CMT phenotype is characterized by progressive weakness and atrophy of distal muscles, high arched feet (pes cavus), and loss of deep-tendon reflexes. CMT can be divided into two subgroups: CMT I, disorders of Schwann cells with nerve-conduction slowing, and CMT II, disorders of the distal portions of neurons, also known as the “axonal neuropathies” (Dyck and Lambert 1968). The known causes of CMT I are a 1.5-mb DNA duplication on chromosome 17p11.2-p12 (CMT1A [MIM 118220]) (Lupski et al. 1991; Raeymaekers et al. 1991) causing trisomy of the peripheral myelin protein 22 gene (PMP22 [GenBank accession number L03203]) (Matsunami et al. 1992; Patel et al. 1992; Timmerman et al. 1992; Valentijn et al. 1992b), point mutations in connexin 32 (Cx32/GJB1 [GenBank accession number XM_047682] and CMT1X [MIM 302800]; Bergoffen et al. 1993), point mutations of two myelin genes, peripheral myelin protein 22 (PMP22 in CMT1A; Valentijn et al. 1992a), myelin protein zero (MPZ/Po [GenBank accession numbers D10537 and D90501] in CMT1B [MIM 118200]; Hayasaka et al. 1993), and mutations in the transcription factor (EGR2 [GenBank accession number AF139463]; Warner et al. 1998). Motor-nerve conduction in the most common form of CMT type I (CMT1A) is typically ∼20 m/s and is usually <40 m/s (Nicholson 1991). Males with the next-most-common form (CMT1X) have median conduction velocities <45 m/s (Nicholson and Nash 1993).
In CMT II, median nerve-conduction velocity is usually >38 m/s (Harding and Thomas 1980) or >45 m/s (Dyck and Lambert 1968). Nerve conduction is not slowed until axonal degeneration occurs; then, as large fibers are lost, mild slowing of conduction occurs. Eventually, when all fibers are lost, no responses are obtained. We have used the value of 45 m/s to define the cutoff between CMT I and CMT II, since this value is in agreement with both Dyck and Lambert’s definition and with our results for CMT1A and CMT1X males.
The only known mutations causing CMT II are mutations in the neurofilament light gene (NEFL [GenBank accession number X05608]) in CMT2E (MIM 162280; Mersiyanova et al. 2000), the MPZ/Po (Marrosu et al. 1998; Senderek et al. 2000), and the microtubule motor KIF1Bβ gene (KIF1Bβ [GenBank accession number AB023656]) in CMT2A (MIM 118210; Zhao et al. 2001).
We have used the term “intermediate conduction velocity” to describe CMT families with nerve-conduction velocities, in different affected individuals, that overlap the division between CMT I and CMT II (45 m/s). A functional definition is to use the term for families with a range of nerve-conduction velocities, including both the CMT I and CMT II ranges. There has been controversy about whether an intermediate form of CMT exists; however, families of this type have been reported (Salisachs 1974; Davis et al. 1978). An Italian family has been described with a dominantly inherited form of CMT with intermediate median motor-nerve–conduction velocities (Rossi et al. 1985; Villanova et al. 1998). Two MPZ/Po mutations are associated with intermediate conduction velocities (De Jonghe et al. 1999; Mastaglia et al. 1999). We have identified a large family (DI-CMT310) with conduction velocities ranging from 24 to 54 m/s. The sural-nerve biopsy in this family shows axonal degeneration, loss of large-diameter fibers, rare segmental demyelination, and remyelination with onion bulb formation, as in the family reported by Villanova et al. (1998).
Genomic DNA was extracted from peripheral blood leukocytes, using standard techniques. A genomewide linkage screen was performed at the Australian Genome Research Facility, using 383 microsatellite markers from the ABI Prism Linkage Mapping Set Version 2 (PE Applied Biosystems). DI-CMT was assumed to be an autosomal dominant trait with a penetrance of 90% by age 20 years. The disease allele had a frequency of .0001, with a phenocopy rate of 0. Male and female recombination rates were considered to be equal. Marker allele frequencies were set at 1/n, where n is the number of alleles observed. When marker allele frequencies were varied using available population data, this had little or no effect on the LOD score results. Information on additional markers to confirm and refine the interval were accessed through the Genome Database, and samples were PCR amplified as described elsewhere (Nicholson et al. 1996). Linkage analysis was performed using the MLINK and Linkmap programs of the Linkage package (V5.1) (Lathrop et al. 1984) in the Fastlink implementation (version 4.1p; Cottingham et al. 1993). Both two-point and multipoint analyses were performed. For multipoint analysis, the genetic distances between loci were obtained from the Marshfield sex-averaged linkage map and were converted to recombination fractions using Haldane’s mapping function. The Zmax-1 method was used to determine the 95% confidence limits and support interval for the DI-CMT locus (Conneally et al. 1985). Haplotypes were assigned on the basis of the minimization of intermarker recombination.
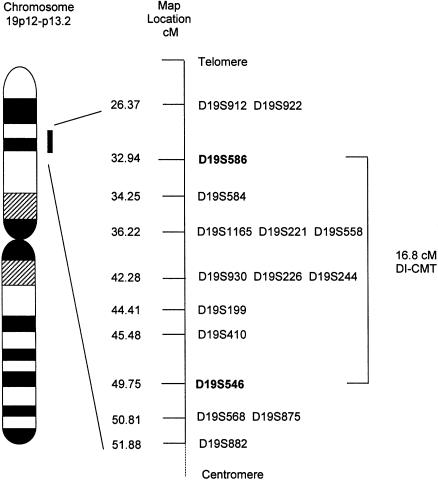
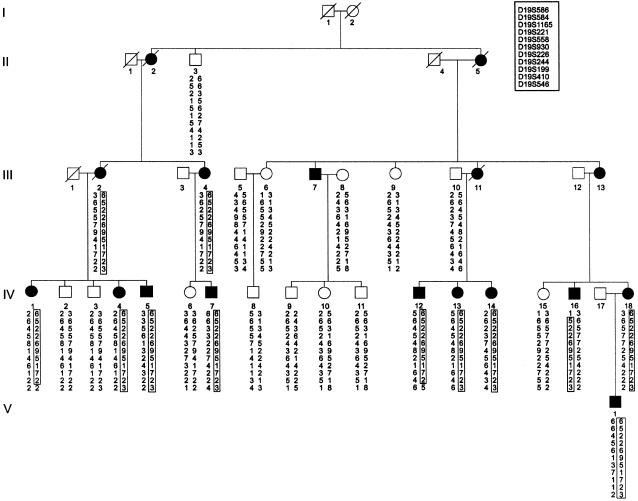
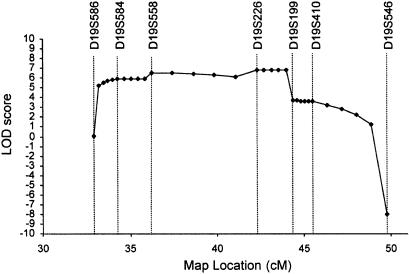
Prior to performance of a genome screen on family DI-CMT310, all known CMT I (CMT1A, CMT1B) and CMT II (CMT2A, CMT2B [MIM 600882], CMT2C [MIM 606071], CMT2D [MIM 601472], CMT2E) loci were excluded by either linkage analysis or mutation screening (data not shown). Simulation studies indicated that the family could independently demonstrate linkage (LOD score > 3.0) and was capable of excluding 11.6 cM on either side of an unlinked marker (data not shown). A genome screen was subsequently undertaken using markers spaced, on average, at 10-cM intervals. Linkage was established to chromosome 19p12-p13.2 when significant LOD scores were obtained with the consecutive markers D19S221 and D19S226 (table 1). No other markers analyzed from the genome screen gave a LOD score ⩾3.0. Fourteen additional markers were then tested in the family. The results of the two-point analysis between the disease phenotype and the additional marker loci are shown (table 1). Extended haplotypes of individuals were constructed according to the order of both the Marshfield genetic map and the chromosome 19 p-arm metric physical map (fig. 1). Haplotype analysis detected no recombination between DI-CMT and seven closely linked markers (D19S584, D19S1165, D19S221, D19S558, D19S226, D19S244, and D19S410). The markers D19S930 and D19S199 did not generate a LOD score of 3.0, but the alleles from these markers formed part of the disease haplotype segregating in this family (fig. 2). On the basis of the analysis, the proximal recombination site is between D19S546 and D19S410, as observed in two affected individuals (IV-1 and IV-12), and the distal recombination site is between D19S586 and D19S584 as observed in a single affected family member (IV-16). These results suggest the localization of DI-CMT to be within a 16.8-cM interval between D19S586 and D19S546. Multipoint linkage analysis was performed using the markers D19S586, D19S584, D19S558, D19S226, D19S199, D19S410, and D19S546. The most likely location for DI-CMT is D19S226, with a maximum multipoint LOD score of Z=6.77. By use of the Zmax-1 method, the inferred location of the DI-CMT locus has been narrowed to within a 10-cM confidence interval (8.2 cM telomeric and 1.8 cM centromeric to D19S226; see fig. 3).
Table 1.
Two-Point LOD Scores between the Dominant Intermediate CMT Locus and Microsatellite Markers on Chromosome 19p
| LOD at θ = |
|||||||
| Marker | .00 | .01 | .05 | .10 | .20 | .30 | .40 |
| D19S912 | −3.46 | −1.59 | −.40 | −.05 | .06 | −.02 | −.08 |
| D19S922 | −7.50 | −3.35 | −1.41 | −.74 | −.39 | −.40 | −.27 |
| D19S586 | −.23 | 3.19 | 3.54 | 3.38 | 2.69 | 1.75 | .71 |
| D19S584 | 4.40 | 4.32 | 3.97 | 3.51 | 2.53 | 1.48 | .47 |
| D19S1165 | 4.56 | 4.48 | 4.18 | 3.78 | 2.89 | 1.88 | .81 |
| D19S221 | 4.30 | 4.20 | 3.83 | 3.34 | 2.29 | 1.20 | .27 |
| D19S558 | 5.59 | 5.51 | 5.18 | 4.74 | 3.75 | 2.60 | 1.29 |
| D19S930 | 2.83 | 2.77 | 2.54 | 2.24 | 1.60 | .94 | .34 |
| D19S226 | 5.28 | 5.20 | 4.86 | 4.41 | 3.44 | 2.34 | 1.13 |
| D19S244 | 3.59 | 3.52 | 3.20 | 2.81 | 2.00 | 1.20 | .47 |
| D19S199 | 2.80 | 2.75 | 2.52 | 2.21 | 1.58 | .92 | .33 |
| D19S410 | 3.81 | 3.74 | 3.44 | 3.05 | 2.21 | 1.32 | .46 |
| D19S546 | −8.59 | −1.92 | −.17 | .47 | .81 | .70 | .40 |
| D19S568 | −5.61 | −1.67 | −.85 | −.51 | −.25 | −.13 | −.04 |
| D19S875 | −3.74 | −.54 | .54 | .87 | .78 | .32 | −.11 |
| D19S882 | −7.94 | −1.33 | .06 | .56 | .81 | .67 | .35 |
Figure 1.
Genetic map (sex averaged) of chromosome 19 markers used in this study. The genetic distances were obtained from the Marshfield map. Loci that appear on the same line map to the same genetic location. The order of these markers was obtained from the chromosome 19 p-arm metric physical map. Markers defining the DI-CMT genetic interval are shown in boldface.
Figure 2.
Haplotype analysis of markers from chromosome 19p12-p13.2 in family (DI-CMT310) with autosomal dominant intermediate CMT. The haplotype segregating with the disease is boxed. The markers are presented in order from telomere (top) to centromere (bottom). Blackened symbols denote affected individuals, and unblackened symbols denote unaffected individuals. Individuals are numbered consecutively in each generation, from left to right. Individuals IV-1 and IV-12 define the centromeric boundary of the disease at D19S546, whereas individual IV-16 defines the telomeric boundary at D19S586.
Figure 3.
Multipoint localization of dominant intermediate CMT. Genetic location (distance, in cM, from the telomere of chromosome 19p) is plotted against the multipoint LOD score. Markers used in the multipoint analysis are shown. A maximum multipoint LOD score of 6.77 was obtained for D19S226 at 42.28 cM. The markers D19S586 and D19S199 flank the 10-cM confidence interval.
Although the 16.8-cM interval containing the DI-CMT locus is too large for practical positional cloning, the region has been almost completely sequenced. Refinement of the critical interval, however, will be required before mutation screening of expressed sequences within the region is undertaken. The chromosome 19 p-arm metric physical map (Lawrence Livermore Laboratory) has listed 53 genes that have been mapped on a cosmid contig in the interval between D19S586 and D19S546. Several other diseases and associated genes have been mapped to the same region as DI-CMT. They include cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (NOTCH3; Joutel et al. 1996), episodic ataxia and familial hemiplegic migraine (CACNLA1A4; Jodice et al. 1997), and, more recently, a locus for autosomal dominant polycystic liver disease was mapped to this region (Reynolds et al. 2000).
DI-CMT shares clinical features with CMT I and CMT II. Genes identified for CMT type I and II could give clues to positional candidate genes for DI-CMT. Currently, 30 genetic loci have been identified for distinct forms of CMT; however, only 10 genes have been identified (Meuleman et al. 2000). Genes associated with recessive peripheral neuropathies include the gene encoding the phosphatase myotubularin-related protein 2 (MTMR2 [GenBank accession number NM_003912]; Bolino et al. 2000), a protein involved with cell-cycle regulation and signal transduction (NDRG1 [GenBank accession number XM_051273]; Kalaydjieva et al. 2001), a receptor for a neurotrophic factor (NTRK1/TrkA [GenBank accession number XM_043533]; Bodzioch et al. 2001; Houlden et al. 2001), and a protein (periaxin) associated with myelinating Schwann cells (PRX [GenBank accession number XM_047407]; Guilbot et al. 2001). The genes involved with dominant CMT I and CMT II represent a spectrum of proteins with diverse functions ranging from a structural protein (PMP22), an adhesion molecule (MPZ/Po), a transcription factor (EGR2), a mitochondrial transport protein (KIF1Bβ), and a cytoskeletal protein (NEFL). All these genes are expressed in peripheral nerve and play a role in Schwann cell biology and myelination (PMP22 and MPZ/Po), axonal structure (NEFL), axonal transport (KIF1Bβ), and differentiation of the myelinating Schwann cell (EGR2). Possible candidates for DI-CMT, on the basis of their expression profiles in peripheral nerve and spinal cord, respectively, include the growth differentiation factor 1 gene (GDF-1; Lee 1991) and the brain-specific membrane anchored protein gene (BSMAP; Elson et al. 1999). BSMAP was localized to the DI-CMT interval, according to the Unified Database for Human Genome Mapping (Weizmann Institute). In addition to these two genes, the protein kinase C substrate 80K-H (PRKCSH) could also be a positional candidate, because of its potential role in neuronal signal transduction (Ophoff et al. 1996). Screening of these genes is currently being performed. The remaining genes in the region have reported tissue expression or possible functions that eliminate their role as strong positional candidates for this disease.
Another possible clue to mechanisms involved in dominant intermediate CMT is given by the Cx32/GJB1 gene mutated in CMT1X. CMT1X has a mixed axonal and demyelinating appearance on biopsy, and affected males have slowing of nerve-conduction velocities due to demyelination. Connexin 32 is a protein that forms channels between myelin lamellae and the myelin/axonal interface. Mutations in connexin 32 disrupt the connexin channel between myelin lamellae and axons, resulting in degeneration of both myelin and axons. Dominant intermediate CMT could be caused by a gene with a similar function.
Linkage studies localizing DI-CMT to 19p12-13.2 have shown that dominant intermediate CMT exists as a separate genetic entity. Because the disease shows both axonal and demyelinating features, identification of the gene involved with this disorder may elucidate axonal and myelin interactions. Such interactions are relevant to understanding secondary axonal degeneration, a direct cause of disability in both axonal and demyelinating neuropathies.
Acknowledgments
We thank the members of this family who participated in this study. We thank Mrs. Margaret Ross, for assistance in organizing the family for this study; Dr. Lynette Kiers, for providing clinical and electrophysiological data on family members; and Professor Edward Byrne, for referring one branch of the family to the neurogenetics clinic at St. Vincent's Hospital, Melbourne. This work is supported by grants from the National Health & Medical Research Council of Australia (grant no. 153895) and the Rebecca L. Cooper Medical Research Foundation.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames.html
- Chromosome 19 p Arm Metric Physical Map, Lawrence Livermore Laboratory, http://greengenes.llnl.gov/genome-bin/loadmap?region=mp
- GenBank, http://www.ncbi.nlm.nih.gov/ (for PMP22 [accession number L03203], Cx32/GJB1 [accession number XM_047682], MPZ/Po [accession number D10537 D90501], EGR2 [accession number AF139463], MTMR2 [accession number NM_003912], NDRG1 [accession number XM_051273], NTRK1/TrkA [accession number XM_043533], PRX [accession number XM_047407], NEFL [accession number X05608], KIF1Bβ [accession number AB023656])
- Genome Database, http://www.gdb.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CMT1A [MIM 118220], CMT1B [MIM 118200], CMT1X [MIM 302800], CMT2A [MIM 118210], CMT2B [ MIM 600882], CMT2C [MIM 606071], CMT2D [MIM 601472], and CMT2E [162280])
- Unified Database for Human Genome Mapping, Weizmann Institute, http://bioinformatics.weizmann.ac.il/udb/
References
- Bergoffen J, Scherer SS, Wang S, Scott MO, Bone LJ, Paul DL, Chen K, Lensch MW, Chance PF, Fischbeck KH (1993) Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science 262:2039–2042 [DOI] [PubMed] [Google Scholar]
- Bodzioch M, Lapicka K, Aslanidis C, Kacinski M, Schmitz G (2001) Two novel mutant alleles of the gene encoding neurotrophic tyrosine kinase receptor type 1 (ntrk1) in a patient with congenital insensitivity to pain with anhidrosis: a splice junction mutation in intron 5 and cluster of four mutations in exon 15. Hum Mutat 17:72 [DOI] [PubMed] [Google Scholar]
- Bolino A, Muglia M, Conforti FL, LeGuern E, Salih MA, Georgiou DM, Christodoulou K, Hausmanowa-Petrusewicz I, Mandich P, Schenone A, Gambardella A, Bono F, Quattrone A, Devoto M, Monaco AP (2000) Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet 25:17–19 [DOI] [PubMed] [Google Scholar]
- Conneally PM, Edwards JH, Kidd KK, Lalouel JM, Morton NE, Ott J, White R (1985) Report of the committee on methods of linkage analysis and reporting. Cytogenet Cell Genet 40:356–359 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Davis CJ, Bradley WG, Madrid R (1978) The peroneal muscular atrophy syndrome: clinical, genetic, electrophysiological and nerve biopsy studies. I. Clinical, genetic and electrophysiological findings and classification. J Genet Hum 26:311–349 [PubMed] [Google Scholar]
- De Jonghe P, Timmerman V, Ceuterick C, Nelis E, De Vriendt E, Logfren A, Vercruyssen A, Verellen C, Van Maldergem L, Martin JJ, Van Broeckhoven C (1999) The Thr124Met mutation in the peripheral myelin protein zero (MPZ) gene is associated with a clinically distinct Charcot-Marie-Tooth phenotype. Brain 122:281–290 [DOI] [PubMed] [Google Scholar]
- Dyck PJ, Lambert EH (1968) Lower motor and primary sensory neuron diseases with peroneal muscular atrophy. II. Neurologic, genetic, and electrophysiologic findings in various neuronal degenerations. Arch Neurol 18:619–625 [DOI] [PubMed] [Google Scholar]
- Elson GC, de Coignac AB, Aubry JP, Delneste Y, Magistrelli G, Holzwarth J, Bonnefoy JY, Gauchat JF (1999) BSMAP, a novel protein expressed specifically in the brain whose gene is localized on chromosome 19p12. Biochem Biophys Res Commun 264:55–62 [DOI] [PubMed] [Google Scholar]
- Guilbot A, Williams A, Ravise N, Verny C, Brice A, Sherman DL, Brophy PJ, LeGuern E, Delague V, Bareil C, Megarbane A, Claustres M (2001) A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum Mol Genet 10:415–421 [DOI] [PubMed] [Google Scholar]
- Harding AE, Thomas PK (1980) The clinical features of hereditary motor and sensory neuropathy types I and II. Brain 103:259–280 [DOI] [PubMed] [Google Scholar]
- Hayasaka K, Himoro M, Wang Y, Takata M, Minoshima S, Shimizu N, Miura M, Uyemura K, Takada G (1993) Structure and chromosomal localization of the gene encoding the human myelin protein zero (MPZ). Genomics 17:755–758 [DOI] [PubMed] [Google Scholar]
- Houlden H, King RH, Hashemi-Nejad A, Wood NW, Mathias CJ, Reilly M, Thomas PK (2001) A novel trk a (ntrk1) mutation associated with hereditary sensory and autonomic neuropathy type V. Ann Neurol 49:521–525 [PubMed] [Google Scholar]
- Jodice C, Mantuano E, Veneziano L, Trettel F, Sabbadini G, Calandriello L, Francia A, Spadaro M, Pierelli F, Salvi F, Ophoff RA, Frants RR, Frontali M (1997) Episodic ataxia type 2 (ea2) and spinocerebellar ataxia type 6 (sca6) due to cag repeat expansion in the CACNA1A gene on chromosome 19p. Hum Mol Genet 6:1973–1978 [DOI] [PubMed] [Google Scholar]
- Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis EA, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E (1996) Notch3 mutations in cadasil, a hereditary adult-onset condition causing stroke and dementia. Nature 383:707–710 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, Rosenthal A, King RH, Thomas PK (2000) N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet 67:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ (1991) Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure. Proc Natl Acad Sci USA 88:4250–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, Chakravarti A, Patel PI (1991) DNA duplication associated with Charcot-Marie-Tooth disease type 1a. Cell 66:219–232 [DOI] [PubMed] [Google Scholar]
- Marrosu MG, Vaccargiu S, Marrosu G, Vannelli A, Cianchetti C, Muntoni F (1998) Charcot-Marie-Tooth disease type 2 associated with mutation of the myelin protein zero gene. Neurology 50:1397–1401 [DOI] [PubMed] [Google Scholar]
- Mastaglia FL, Nowak KJ, Stell R, Phillips BA, Edmondston JE, Dorosz SM, Wilton SD, Hallmayer J, Kakulas BA, Laing NG (1999) Novel mutation in the myelin protein zero gene in a family with intermediate hereditary motor and sensory neuropathy. J Neurol Neurosurg Psychiatry 67:174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami N, Smith B, Ballard L, Lensch MW, Robertson M, Albertsen H, Hanemann CO, Muller HW, Bird TD, White R, Chance PF (1992) Peripheral myelin protein-22 gene maps in the duplication in chromosome 17p11.2 associated with Charcot-Marie-Tooth 1A. Nat Genet 1:176–179 [DOI] [PubMed] [Google Scholar]
- Mersiyanova IV, Perepelov AV, Polyakov AV, Sitnikov VF, Dadali EL, Oparin RB, Petrin AN, Evgrafov OV (2000) A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am J Hum Genet 67:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman J, Timmerman V, Nelis E, De Jonghe P (2000) Molecular genetics of inherited peripheral neuropathies: who are the actors? Acta Neurol Belg 100:171–180 [PubMed] [Google Scholar]
- Nicholson GA (1991) Penetrance of the hereditary motor and sensory neuropathy Ia mutation: assessment by nerve conduction studies. Neurology 41:547–552 [DOI] [PubMed] [Google Scholar]
- Nicholson GA, Dawkins JL, Blair IP, Kennerson ML, Gordon MJ, Cherryson AK, Nash J, Bananis T (1996) The gene for hereditary sensory neuropathy type I (HSN-I) maps to chromosome 9q22.1-q22.3. Nat Genet 13:101–104 [DOI] [PubMed] [Google Scholar]
- Nicholson GA, Nash J (1993) Intermediate nerve conduction velocities define X-linked Charcot-Marie-Tooth neuropathy families. Neurology 43:2558–2564 [DOI] [PubMed] [Google Scholar]
- Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Mohrenweiser H, Litt M, Hofker MH, Haan J, Ferrari MD, Frants RR (1996) A 3-mb region for the familial hemiplegic migraine locus on 19p13.1-p13.2: exclusion of PRKCSH as a candidate gene. Eur J Hum Genet 4:321–328 [PubMed] [Google Scholar]
- Patel PI, Roa BB, Welcher AA, Schoener-Scott R, Trask BJ, Pentao L, Snipes GJ, Garcia CA, Francke U, Shooter EM, Lupski JR, Suter U (1992) The gene for the peripheral myelin protein pmp-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat Genet 1:159–165 [DOI] [PubMed] [Google Scholar]
- Raeymaekers P, Timmerman V, Nelis E, De Jonghe P, Hoogendijk JE, Baas F, Barker DF, Martin JJ, De V, M, Bolhuis PA, Van Broeckhoven C, HMSN Collaborative Research Group (1991) Duplication in chromosome 17p11.2 in Charcot-Marie-Tooth neuropathy type 1a (CMT1a). Neuromuscul Disord 1:93–97 [DOI] [PubMed] [Google Scholar]
- Reynolds DM, Falk CT, Li A, King BF, Kamath PS, Huston J 3d, Shub C, Iglesias DM, Martin RS, Pirson Y, Torres VE, Somlo S (2000) Identification of a locus for autosomal dominant polycystic liver disease, on chromosome 19p13.2-13.1. Am J Hum Genet 67:1598–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Paradiso C, Cioni R, Rizzuto N, Guazzi G (1985) Charcot-Marie-Tooth disease: study of a large kinship with an intermediate form. J Neurol 232:91–98 [DOI] [PubMed] [Google Scholar]
- Salisachs P (1974) Wide spectrum of motor conduction velocity in Charcot-Marie-Tooth disease. An anatomico-physiological interpretation. J Neurol Sci 23:25–31 [DOI] [PubMed] [Google Scholar]
- Senderek J, Hermanns B, Lehmann U, Bergmann C, Marx G, Kabus C, Timmerman V, Stoltenburg-Didinger G, Schroder JM (2000) Charcot-Marie-Tooth neuropathy type 2 and P0 point mutations: two novel amino acid substitutions (asp61gly; tyr119cys) and a possible “hotspot” on thr124met. Brain Pathol 10:235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman V, Nelis E, Van Hul W, Nieuwenhuijsen BW, Chen KL, Wang S, Ben O, K, Cullen B, Leach RJ, Hanemann CO, De Jonghe P, Raeymaekers P, van Ommen G-JB, Martin J-J, Muller HW, Vance JM, Fischbeck KH, Van Broeckhoven C (1992) The peripheral myelin protein gene pmp-22 is contained within the Charcot-Marie-Tooth disease type 1A duplication. Nat Genet 1:171–175 [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Baas F, Wolterman RA, Hoogendijk JE, van den Bosch NHA, Zorn I, Gabreels-Festen AAWM, de Visser M, Bolhuis PA (1992a) Identical point mutations of PMP22 in Trembler-J mouse and Charcot-Marie-Tooth disease type 1A. Nat Genet 2:288–291 [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Bolhuis PA, Zorn I, Hoogendijk JE, van den Bosch N, Hensels GW, Stanton VP Jr, Housman DE, Fischbeck KH, Ross DA, Nicholson GA, Meershoek EJ, Dauwerse HG, van-Ommen G-JB, Baas F (1992b) The peripheral myelin gene pmp-22/gas-3 is duplicated in Charcot-Marie-Tooth disease type 1A. Nat Genet 1:166–170 [DOI] [PubMed] [Google Scholar]
- Villanova M, Timmerman V, De Jonghe P, Malandrini A, Rizzuto N, Van Broeckhoven C, Guazzi G, Rossi A (1998) Charcot-Marie-Tooth disease: an intermediate form. Neuromuscul Disord 8:392–393 [DOI] [PubMed] [Google Scholar]
- Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR (1998) Mutations in the early growth response 2 (egr2) gene are associated with hereditary myelinopathies. Nat Genet 18:382–384 [DOI] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, Saito M, Tsuji S, Hirokawa N (2001) Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell 105:587–597 [DOI] [PubMed] [Google Scholar]