Abstract
Myc is a transcriptional regulator of the basic helix–loop–helix leucine zipper protein family. It has strong oncogenic potential, mutated or virally transduced forms of Myc induce lymphoid tumors in animals, and deregulated expression of Myc is associated with numerous types of human cancers. For its oncogenic activity, Myc must dimerize with the ubiquitously expressed basic helix–loop–helix leucine zipper protein Max. This requirement for dimerization may allow control of Myc activity with small molecules that interfere with Myc/Max dimerization. We have measured Myc/Max dimerization with fluorescence resonance energy transfer and have screened combinatorial chemical libraries for inhibitors of dimerization. Candidate inhibitors were isolated from a peptidomimetics library. Inhibition of Myc/Max interaction was validated by ELISA and electrophoretic mobility-shift assay. Two of the candidate inhibitors also interfere with Myc-induced oncogenic transformation in chicken embryo fibroblast cultures. Our work provides proof of principle for the identification of small molecule inhibitors of protein–protein interactions by using high-throughput screens of combinatorial chemical libraries.
Myc is a basic helix–loop–helix leucine zipper (bHLHZip) transcription factor that was first identified as the oncogenic effector of avian retroviruses inducing lymphoid tumors (1–3). The common denominator of these tumors is constitutive activation of Myc. Gain of Myc function is also seen in human tumors (4). In Burkitt's lymphoma and other lymphoid malignancies, the myc gene is translocated into the vicinity of an Ig enhancer, resulting in constitutive overexpression (5). The myc gene is amplified in lung and breast carcinomas (6–9). Elevated expression of the Myc protein is found in the majority of colon carcinomas (10). Colorectal cancers commonly show increased activity of the lymphocyte enhancer factor proteins that direct overexpression of Myc (11).
The role of Myc in tumorigenesis is linked to its activating effect on transcription and cell growth and its repressing effect on differentiation. Myc promotes oncogenic transformation and tumorigenesis by regulating target genes that drive cell proliferation and stimulate angiogenesis (12–15). Myc expression is necessary for entry of cells into S phase, and inhibition of Myc leads to withdrawal from the cell cycle and terminal differentiation (16, 17). The expression of Myc is cell context-specific and tightly depends on mitogens (18). The Myc protein has a short half-life of 20–30 min (19, 20); it is rapidly degraded by the ubiquitin-linked proteasome machinery (21). All known oncogenic functions of Myc require dimerization with Max, another bHLHZip protein (22, 23). Myc and Max dimerize through their HLHZip domains and bind to their DNA recognition site, the E-box element CACGTG, through their basic domains. Binding of Myc/Max dimers to DNA activates transcription of Myc target genes (24). Inhibitors of Myc/Max dimers could therefore regulate Myc activity and may be of pharmacological value in cancers that depend on sustained activation of Myc.
Here we describe nonpeptidic inhibitors of Myc/Max dimerization. The candidate compounds were initially identified by fluorescence resonance energy transfer (FRET) in high-throughput screens of peptidomimetic libraries. Inhibition of Myc/Max interaction was confirmed in independent in vitro assays. Two of the compounds interfered with Myc-induced oncogenic transformation of chicken embryo fibroblasts (CEF) in cell culture.
Materials and Methods
Chemical Libraries.
The synthesis of the chemical libraries from which the inhibitors emerged has been described (25). Screening hits were resynthesized for confirmation of structure and purity.
Recombinant Proteins.
The bHLHZip domain of human c-Myc (amino acids 354–434) was PCR-amplified and cloned into the BamHI/AgeI site of pECFP and pEGFP (CLONTECH). pECFP and pEGFP encode variants of cyan and green fluorescent protein (CFP and GFP, respectively) and show enhanced expression efficiency and fluorescence intensity over wild-type GFP. The insertion of the bHLHZip domain into these vectors fused the Myc sequences to the N termini of GFP or CFP, respectively. The bHLHZip domain of rat Max (amino acids 13–93 of p21 Max, 100% identical to human Max at the protein level) was PCR-amplified and cloned into the BglII/AgeI site of pEYFP-N1, a vector encoding yellow fluorescent protein (YFP) with enhanced stability and fluorescence (CLONTECH). The fusions of the bHLHZip domain of Myc or Max with the fluorescent proteins were cloned into NheI/NotI sites of the histidine-tag expression vector pET 28a (Novagen). The bHLHZip domain (amino acids 13–93) of rat Max with a C-terminal AU1 tag was also cloned into pET28a. Proteins were expressed in Escherichia coli BL21DE3 cells, purified by affinity chromatography on nickel columns, and dialyzed against buffer containing 200 mM Hepes (pH 7.0), 500 mM KCl, 30 mM MgCl2, 2 mM DTT, and 10 mM EDTA (referred to as 1× buffer).
FRET.
The protein consisting of the bHLHZip domain of Myc fused to the N terminus of CFP (MycCFP) and the analogous fusion protein between the bHLHZip domain of Max and YFP (MaxYFP) were allowed to heterodimerize at 37°C for 1 h (at 85-nM monomer concentration). Screening compounds were added to a final concentration of 25 μM and 8% DMSO, and the mixtures were incubated for 1 h at 23°C. After excitation of CFP at 433 nm both the CFP fluorescence at 475 nm and the YFP fluorescence at 525 nm were measured in a 96-well fluorescence plate reader (Molecular Devices). Dimerization permits FRET from CFP to YFP and causes the emission of CFP at 475 nm to decrease while enhancing the emission of YFP at 525 nm. Compounds that dissociate MycCFP/MaxYFP dimers increase the emission of CFP and decrease the emission of YFP, resulting in a lower ratio of intensities [525 nm/475 nm]. Candidate inhibitors were retested in single cuvettes (Perkin–Elmer LS 50B) to confirm the fluorescence data.
ELISA.
Max (c = 150 ng/μl, in 1× buffer) was adsorbed onto an ELISA plate (Costar) at 4°C for 15 h. After washing (1× buffer, three times 5 min), the plates were blocked with 3% BSA in 0.5× buffer for 1 h at 37°C and washed with 1× buffer twice for a total of 1 h. Stock solutions of screening compound were added to MycGFP (c = 6.5 ng/μl, 170 nM, in 0.5× buffer), and the mixtures were then added to the plate. The candidate compounds and the negative control were tested at the following concentrations in 25-μM increments: IIA4B20, 0–75 μM; IIA6B17, 0–125 μM; IIA4B11, 0–250 μM; IA4B11, 0–250 μM; and IA4B6, 0–250 μM. The bHLHZip domain of Max was used as a positive control. All test mixtures contained 5% DMSO. The plates were incubated for 40 min at 37°C and 20 min at 23°C, then washed four times with 0.5× buffer and incubated with 10 ng/μl anti-GFP-horseradish peroxidase conjugate (CLONTECH) for 1 h at 37°C. After washing (0.5× buffer, four times 5 min), 2,2′-azino-di-3-ethylbenzthiazoline sulfonate (Roche Molecular Biosciences) was added to measure enzyme activity, and the absorption at 405 nm was read in a microplate reader (BioRad). Concentrations of compounds at which 50% inhibition of Myc/Max dimerization was obtained (IC50) were estimated by plotting percent inhibition against inhibitor concentration.
Electrophoretic Mobility-Shift Assay (EMSA).
MycGFP (c = 6.5 ng/μl, 170 nM) and Max (c = 1.1 ng/μl, 90 nM) were mixed and incubated with stock solutions of the screening compounds for 1 h at 23°C. The candidate compounds and the negative control were tested at the following concentrations in 25-μM increments: IIA6B17, 0–125 μM; IIA4B11, 0–200 μM; IA4B11, 0–200 μM; and IA4B6, 0–200 μM. All test mixtures contained 12% DMSO. A double-stranded DNA oligonucleotide with the consensus binding site of c-Myc/Max dimers (26) was then added (5′AGTTGACCACGTGGTCTGGG3′). The DNA–protein interaction was allowed to proceed for 15 min. Final concentrations were: 100 nM MycGFP, 55 nM Max, 200 mM Hepes (pH 7.0), 500 mM KCl, 30 mM MgCl2, 2 mM DTT, 10 mM EDTA, 5% glycerol, 40 ng/μl salmon testis DNA (Sigma), and 60 pg/μl 32P-labeled oligonucleotide probe. For specificity control, a mutated probe was used (5′AGTTGACTACGTAGTCTGGG3′). For the supershift experiment, 60 ng/μl anti-GFP antibody (CLONTECH) was added for 10 min after the formation of the DNA–protein complex. Protein–DNA complexes were resolved on 4% acrylamide gels (45 mM Tris-borate, 1 mM EDTA), and gels were dried before autoradiography. Concentrations of compounds at which 50% inhibition of Myc/Max dimerization was obtained (IC50) were calculated by extrapolating the inhibition values recorded in the experiments.
Focus Assays.
CEF were seeded at 1.5 × 105 cells per 24-well tissue culture plate in HAM's F10 containing 10% FBS. One day after seeding, the cells were infected with 10-fold serial dilutions of oncogenic retroviruses. These viruses were (i) the RCAS viral vector expressing chicken cellular Myc (27), (ii) the Prague strain of Rous sarcoma virus coding for the Src oncoprotein (28), and (iii) the avian sarcoma viruses 17 and 31 expressing the oncoproteins Jun (29, 30) and Qin (31), respectively. The cultures were then overlaid with nutrient agarose consisting of 57.5% (vol/vol) of media (75% F10 2×, 5% FBS, 2% chicken serum, 15% tryptose-phosphate broth, 1.5% of l-glutamine/penicillin/streptomycin solution, screening compounds in DMSO, final concentration of DMSO:1%) and 42.5% (vol/vol) of 1.5% Sea Plaque Agarose. For focus counts, cultures were stained with 2% crystal violet.
Results
A FRET Assay Identifies Candidate Inhibitors of Myc/Max Dimerization.
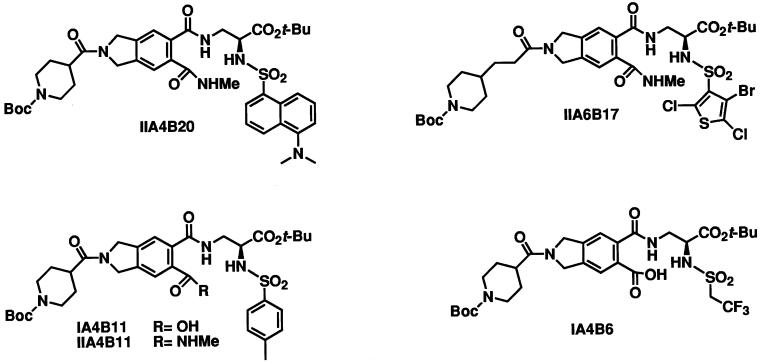
We have screened combinatorial chemical libraries encompassing approximately 7,000 small organic molecules for their ability to interfere with dimerization between Myc and Max. To this end, the bHLHZip domains of Myc and of Max were fused to the N termini of CFP (MycCFP) and YFP (MaxYFP), respectively. The fusions were expressed in E. coli, purified, and allowed to dimerize, followed by excitation of CFP at wave 433 nm. Dimerization generated a FRET spectrum characterized by a strong emission signal of YFP at 525 nm and a weaker emission signal of CFP at 475 nm. In control experiments, the ratios of fluorescence intensities at 525 nm over 475 nm were 1.7 at complete dimerization of MycCFP with MaxYFP and 0.4 for the monomeric state of MycCFP. One hundred percent inhibition of MycCFP/MaxYFP dimerization was achieved by addition of 100× molar excess of the bHLHZip domain of Max, functioning as a competitive inhibitor of Myc-Max dimerization, resulting in a 525 nm/475 nm ratio of slightly more than 0.4. The MycCFP/MaxYFP heterodimers were incubated with individual compounds of the chemical libraries. The ability of the compounds to dissociate the dimer in its monomeric components was followed by monitoring the fluorescence intensities of CFP and YFP upon excitation of CFP. The presence of inhibitors of Myc/Max dimers resulted in a decrease of the ratio of intensities [525 nm/475 nm]. The initial round of screening identified four inhibitory compounds from a peptidomimetic library (IIA4B20, IIA6B17, IIA4B11, and IA4B11) (25). At 25 μM, these compounds caused up to 38% of the dimer to dissociate (Fig. 1, Table 1). An inactive member of this library, IA4B6, was chosen as a negative control compound.
Figure 1.
Small molecule inhibitors of Myc/Max dimers and control compound.
Table 1.
Inhibition of Myc/Max dimerization: Comparison of FRET, ELISA, and EMSA
| Compound | FRET assay inhibition at 25 μM, % | ELISA IC50, μM | EMSA IC50, μM |
|---|---|---|---|
| IIA4B20 | 26 | 75 ± 15 | No effect at 125 μM |
| IIA6B17 | 38 | 125 ± 25 | 50 ± 25 |
| IIA4B11 | 14 | 210 ± 25 | 125 ± 25 |
| IA4B11 | 12 | 40% inhibition at 250 μM | 162 ± 25 |
| IA4B6 | 2 | No effect at 250 μM | No effect at 200 μM |
For ELISA and EMSA, concentrations of compounds at which 50% inhibition of Myc/Max dimerization was obtained (IC50) are given. Data represent typical experiments.
Inhibition of Myc/Max Dimerization Is Confirmed by ELISA.
Candidate inhibitors of Myc/Max dimerization identified by FRET were further investigated by an ELISA assay. The bHLHZip domain of Max was immobilized on an ELISA plate; MycGFP was incubated with serial dilutions of a candidate compound and then added to the plate. Compound IA4B6 served as negative control, and 100× molar excess of the bHLHZip domain of Max was used as a positive control. The inhibition of Myc/Max binding by the screening compounds was determined colorimetrically by using a horseradish peroxidase-conjugated GFP antibody. The four candidate inhibitors interfered with Myc/Max dimerization as measured by ELISA (Table 1). IIA4B20 had the strongest effect with an IC50 value of 75 μM. The negative control compound IA4B6 showed no inhibition even at 250 μM.
Inhibitors Interfere with Myc/Max DNA Binding.
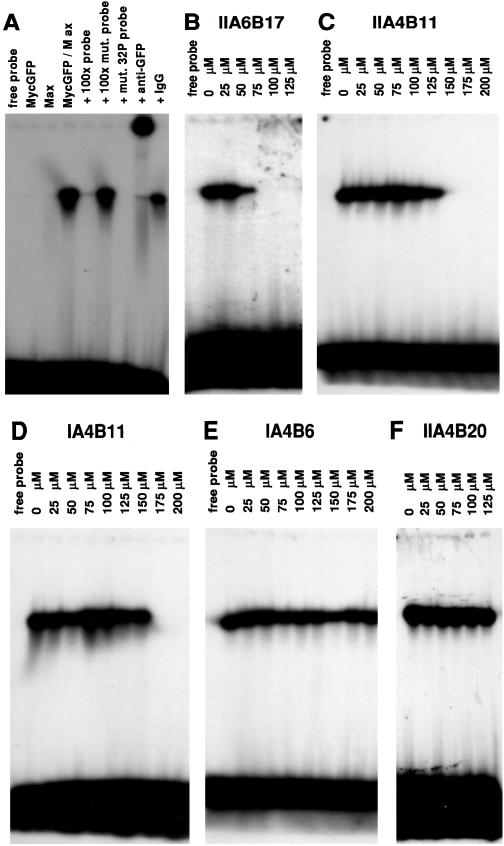
For a test of the inhibitors in EMSA, MycGFP and the bHLHZip domain of Max were mixed and incubated with the compounds, and a 32P-labeled oligonucleotide containing the consensus binding site of Myc/Max heterodimers was added. Under the assay conditions, Myc/Max dimers, but not Max/Max homodimers efficiently bound DNA, resulting in a single retarded band in electrophoresis (Fig. 2A) (26). Three of the candidate compounds were found to abolish DNA binding by MycGFP/Max, and significant inhibition was demonstrated in EMSA at lower compound concentrations than in ELISA (Table 1, Fig. 2 B–D). The negative control compound IA4B6 was ineffective in EMSA (Table 1, Fig. 2E). The relative trends for the test compounds were similar in all three in vitro assays. Compound IIA4B20 that performed well in FRET and ELISA represents an exception as it did not inhibit the formation of the protein dimer in EMSA (Table 1, Fig. 2F). It is possible that the binding site for IIA4B20 is too close to the basic region of the bHLHZip proteins and therefore this binding site may be inaccessible in the presence of DNA. Further studies will be necessary to investigate this point.
Figure 2.
EMSA. (A) Neither MycGFP nor Max alone shift the 32P-labeled probe 5′AGTTGACCACGTGGTCTGGG3′ (lanes 2 and 3). The MycGFP/Max dimer binds to the probe (lane 4). Excess (100-fold) of the correct binding site (lane 5), but not of the mutated binding site 5′AGTTGACTACGTAGTCTGGG3′ (lane 6), inhibits binding of the MycGFP/Max dimer to the radiolabeled probe. MycGFP/Max does not bind to the mutated radiolabeled probe (lane 7). Addition of 60 ng/μl anti-GFP antibody supershifts the dimer/DNA complex (lane 8), but addition of 60 ng/μl IgG does not affect the dimer/DNA complex (lane 9). (B–D) Inhibition of MycGFP/Max-DNA complex by compounds IIA6B17 (B), IIA4B11 (C), and IA4B11 (D). (E and F) Compounds IA4B6 and IIA4B20 do not inhibit formation of the MycGFP/Max-DNA complex.
Biological Effects of the Inhibitors.
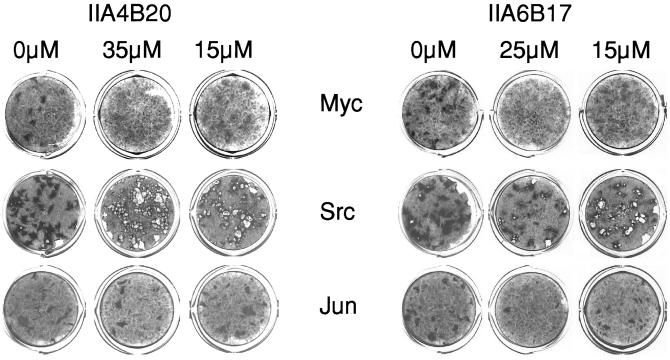
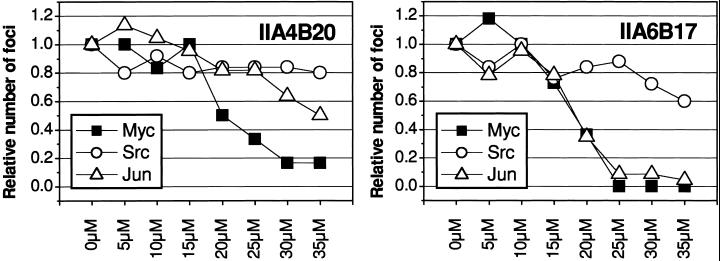
Myc induces the formation of oncogenically transformed cell foci in cultures of CEF. Because this transforming activity of Myc depends on the dimerization with Max, we examined the candidate inhibitors identified in vitro for possible effects on oncogenicity in cell cultures. Two of the candidate compounds, IIA4B20 and IIA6B17, inhibited focus formation by the RCAS-Myc construct. The IC50 for both compounds was 20 μM (Figs. 3 and 4). Compounds IIA4B11 and IA4B11, which were less effective in the in vitro tests, and the negative control compound IA4B6 had no effect on focus formation at 35 μM. At the highest concentrations tested (30 and 35 μM), IIA4B20 and especially IIA6B17 also significantly reduced cell growth. Therefore, the specificity of the in vivo inhibition was examined by testing the compounds in CEF focus assays with two oncoproteins unrelated to Myc, namely Src, for a nonreceptor tyrosine kinase and Jun, a component of the AP-1 transcription factor complex. Src-induced transformation was not significantly affected by IIA4B20, but IIA6B17 reduced focus counts moderately at the higher concentrations tested (Fig. 3). Focus formation by RCAS-Jun was inhibited by IIA6B17 almost to the same extend as that of RCAS-Myc, whereas IIA4B20 reduced the numbers of RCAS-Jun foci only at the highest concentrations. The negative control IA4B6 did not affect RCAS-Src- or RCAS-Jun-induced transformation. A third unrelated oncoprotein, the forkhead/winged helix transcription factor Qin, showed the same response as Src (data not shown).
Figure 3.
The effect of IIA4B20 and IIA6B17 on focus formation by Myc, Scr, or Jun in CEF. Both Myc- and Jun-induced transformation are inhibited by the higher compound concentrations. Src-induced foci consist of poorly adherent cells that are often washed off during the staining process, leaving a hole in the cell sheet.
Figure 4.
Dose–response curves for compounds IIA4B20 and IIA6B17 in CEF assays for transformed focus formation by Myc, Src, or Jun. Shown are the relative numbers of foci from a representative experiment.
Discussion
The structure of the Max/Max homodimer complexed to DNA has been determined (32, 33) and shows the importance of the helix–loop–helix and the leucine zipper domains in dimerization. The bHLHZip domain of each monomer extends for 65 aa and consists of two α-helical segments, one encompassing the basic region and helix 1, the other one composed of helix 2 and the coiled-coil leucine zipper. The two segments are linked by a loop. The basic region contacts the major groove of DNA and introduces a bend of about 25°. The structure of the Myc/Max heterodimer has not yet been determined but probably shares basic properties with the Max/Max homodimer. Heterodimers consisting of the leucine zippers of Myc and Max are less stable than homodimers and heterodimers formed of Jun and Fos or GCN4 (34–36). Although dimerization of Myc and Max is also mediated by the helix–loop–helix domains, it is conceivable that the intrinsic instability of the Myc/Max leucine zipper facilitated the identification of small molecules that interfere with dimerization. The discovery of such inhibitors contradicts the widely shared assumption that large protein interfaces are unlikely to be disturbed by small molecules. However, some recent observations suggest that at least in certain instances, only a minor part of the protein dimer interfaces contributes to the affinity between the proteins (37–40). Targeting these “hot spots” may be sufficient for inhibiting protein–protein interactions. Known inhibitors of protein–protein interactions are either designed peptides derived from the sequence of one of the dimerization partners or small organic molecules found in specific screens (41). For example, HIV protease is catalytically active only as a homodimer and can be inhibited by breaking up the dimeric structure. A tetrapeptide derived from the carboxyl terminus of the protease inhibits the enzyme with a Ki of 45 μM through a dissociative mechanism (42). The natural product Didemnaketal A from Ascidian didemnum sp. and a synthetic analogue inhibit dimerization of the HIV protease with an IC50 of 2.1 μM (43). The heterodimeric herpes virus ribonucleotide reductase is inhibited with an IC50 of 0.3 nM by a peptidomimetic derived from a subunit of the enzyme (44).
The molecular mechanism by which the inhibitors interfere with Myc/Max dimerization is not known. We assume that they bind to a site in either the helix–loop–helix domain or the leucine zipper of either Myc or Max, but the exact docking target remains to be determined. It will be interesting to determine whether the inhibitors also interfere with Max homodimerization. It is reasonable to assume that the inhibition of Myc/Max dimerization results in the observed reduction in oncogenic transformation. However, the available data are insufficient to prove this assumption. Additional work is required to elucidate the mechanism of focus inhibition. The effective concentrations of the candidate inhibitors are still high, but these are first-generation compounds, and libraries of derivatives based on these candidates are being prepared. Complete inhibition of Myc/Max dimerization may be difficult to achieve, and it may not be desirable, considering the normal function of the Myc protein. Partial inhibition may be more easily obtainable, is biologically more desirable, and may be effective in neutralizing Myc gain of function.
The inhibition of oncogenic transformation by the candidate molecules is not entirely specific and also extends to Jun. Absent any knowledge on the mechanisms of the antioncogenic effect for Myc, one can only speculate about this broader range of activity. One possibility is that the small molecule inhibitors target leucine zippers in general that are found in Myc and Jun. Myc is a component of a network that regulates transcription by switching dimerization partners (23, 45). In this network, there are several points at which control of dimerization could have an effect on transcriptional activity. For instance, whereas Myc does not form homodimers, Max readily does so, and stabilizing these homodimers with small molecules could reduce availability of Max for dimerization with Myc. Screens for such stabilizers could be set up in analogous fashion to the FRET assays used in the current experiments.
The ultimate goal of this research is to counteract the oncogenic activity of Myc. Because cancer cells harbor multiple genetic changes, control of a single oncoprotein may not be sufficient to induce reversion of the neoplastic phenotype. However, at least some tumors induced by Myc show long-term dependence on increased Myc function and regress when Myc is deactivated (15, 46). In human cancer cells with several mutations, compensation for one of the oncogenic changes is also sufficient to induce reversion to a nonmalignant phenotype (47).
Our work provides proof of principle for the inhibition of protein–protein interactions between bHLHZip proteins by small molecules. We have demonstrated that a high-throughput screen for inhibitors of Myc/Max interactions in vitro can identify small nonpeptidic molecules that interfere with c-Myc-induced transformation of CEF. The extension of this approach using derivative chemical libraries could result in the identification of more potent inhibitory molecules.
Acknowledgments
We are grateful to Dr. William E. Balch for providing access to laboratory equipment and Dr. Laurent Magnenat for experimental help. Jeff Ludwig provided competent technical assistance. This work was supported by Grant POI CA 78045 from the National Cancer Institute. S.B.C. is the recipient of a fellowship from the National Institutes of Health (Training Grant 5 T32 DK07022). This is manuscript 14235-MEM at The Scripps Research Institute.
Abbreviations
- FRET
fluorescence resonance energy transfer
- EMSA
electrophoretic mobility-shift assay
- CEF
chicken embryo fibroblasts
- bHLHZip
basic helix–loop–helix leucine zipper
- CFP
cyan fluorescent protein
- GFP
green fluorescent protein
- YFP
yellow fluorescent protein
References
- 1.Duesberg P H, Bister K, Vogt P K. Proc Natl Acad Sci USA. 1977;74:4320–4324. doi: 10.1073/pnas.74.10.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duesberg P H, Vogt P K. Proc Natl Acad Sci USA. 1979;76:1633–1637. doi: 10.1073/pnas.76.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kung H J, Boerkoel C, Carter T H. Curr Top Microbiol Immunol. 1991;171:1–25. doi: 10.1007/978-3-642-76524-7_1. [DOI] [PubMed] [Google Scholar]
- 4.Nesbit C E, Tersak J M, Prochownik E V. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 5.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo R C, Croce C M. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bange J, Zwick E, Ullrich A. Nat Med. 2001;7:548–552. doi: 10.1038/87872. [DOI] [PubMed] [Google Scholar]
- 7.Escot C, Theillet C, Lidereau R, Spyratos F, Champeme M H, Gest J, Callahan R. Proc Natl Acad Sci USA. 1986;83:4834–4838. doi: 10.1073/pnas.83.13.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao D J, Dickson R B. Endocr Relat Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 9.Little C D, Nau M M, Carney D N, Gazdar A F, Minna J D. Nature (London) 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 10.Erisman M D, Rothberg P G, Diehl R E, Morse C C, Spandorfer J M, Astrin S M. Mol Cell Biol. 1985;5:1969–1976. doi: 10.1128/mcb.5.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 12.Coller H A, Grandori C, Tamayo P, Colbert T, Lander E S, Eisenman R N, Golub T R. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotsis T, Breit S, Lutz W, Rossler J, Hatzi E, Schwab M, Schweigerer L. Eur J Biochem. 1999;263:757–764. doi: 10.1046/j.1432-1327.1999.00575.x. [DOI] [PubMed] [Google Scholar]
- 14.Meitar D, Crawford S E, Rademaker A W, Cohn S L. J Clin Oncol. 1996;14:405–414. doi: 10.1200/JCO.1996.14.2.405. [DOI] [PubMed] [Google Scholar]
- 15.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 16.Coppola J A, Cole M D. Nature (London) 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 17.Hanson K D, Shichiri M, Follansbee M R, Sedivy J M. Mol Cell Biol. 1994;14:5748–5755. doi: 10.1128/mcb.14.9.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriksson M, Luscher B. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 19.Dani C, Blanchard J M, Piechaczyk M, El Sabouty S, Marty L, Jeanteur P. Proc Natl Acad Sci USA. 1984;81:7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hann S R, Eisenman R N. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory M A, Hann S R. Mol Cell Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackwood E M, Eisenman R N. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 23.Grandori C, Cowley S M, James L P, Eisenman R N. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 24.Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Nature (London) 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 25.Boger D L, Lee J K, Goldberg J, Jin Q. J Org Chem. 2000;65:1467–1474. doi: 10.1021/jo9916481. [DOI] [PubMed] [Google Scholar]
- 26.Solomon D L, Amati B, Land H. Nucleic Acids Res. 1993;21:5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petropoulos C J, Givol I, Hughes S H. Oncogene. 1996;12:2611–2621. [PubMed] [Google Scholar]
- 28.Vogt P K. Virology. 1971;46:939–946. doi: 10.1016/0042-6822(71)90092-4. [DOI] [PubMed] [Google Scholar]
- 29.Cavalieri F, Ruscio T, Tinoco R, Benedict S, Davis C, Vogt P K. Virology. 1985;143:680–683. doi: 10.1016/0042-6822(85)90412-x. [DOI] [PubMed] [Google Scholar]
- 30.Maki Y, Bos T J, Davis C, Starbuck M, Vogt P K. Proc Natl Acad Sci USA. 1987;84:2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Vogt P K. Proc Natl Acad Sci USA. 1993;90:4490–4494. doi: 10.1073/pnas.90.10.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brownlie P, Ceska T, Lamers M, Romier C, Stier G, Teo H, Suck D. Structure (London) 1997;5:509–520. doi: 10.1016/s0969-2126(97)00207-4. [DOI] [PubMed] [Google Scholar]
- 33.Ferre-D'Amare A R, Prendergast G C, Ziff E B, Burley S K. Nature (London) 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 34.Muhle-Goll C, Nilges M, Pastore A. Biochemistry. 1995;34:13554–13564. doi: 10.1021/bi00041a035. [DOI] [PubMed] [Google Scholar]
- 35.O'Shea E K, Rutkowski R, Kim P S. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- 36.Thompson K S, Vinson C R, Freire E. Biochemistry. 1993;32:5491–5496. doi: 10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- 37.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 38.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 39.Wells J A. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells J A, de Vos A M. Annu Rev Biochem. 1996;65:609–634. doi: 10.1146/annurev.bi.65.070196.003141. [DOI] [PubMed] [Google Scholar]
- 41.Cochran A G. Chem Biol. 2000;7:R85–R94. doi: 10.1016/s1074-5521(00)00106-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z Y, Poorman R A, Maggiora L L, Heinrikson R L, Kezdy F J. J Biol Chem. 1991;266:15591–15594. [PubMed] [Google Scholar]
- 43.Fan X, Flentke G R, Rich D H. J Am Chem Soc. 1998;120:8893–8894. [Google Scholar]
- 44.Liuzzi M, Deziel R, Moss N, Beaulieu P, Bonneau A M, Bousquet C, Chafouleas J G, Garneau M, Jaramillo J, Krogsrud R L, et al. Nature (London) 1994;372:695–698. doi: 10.1038/372695a0. [DOI] [PubMed] [Google Scholar]
- 45.Amati B, Land H. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 46.Felsher D W, Bishop J M. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 47.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]