Abstract
Considerable effort has been expended to determine whether the gene for angiotensin I–converting enzyme (ACE) confers susceptibility to cardiovascular disease. In this study, we genotyped 13 polymorphisms in the ACE gene in 1,343 Nigerians from 332 families. To localize the genetic effect, we first performed linkage and association analysis of all the markers with ACE concentration. In multipoint variance-component analysis, this region was strongly linked to ACE concentration (maximum LOD score 7.5). Likewise, most of the polymorphisms in the ACE gene were significantly associated with ACE (P<.0013). The two most highly associated polymorphisms, ACE4 and ACE8, accounted for 6% and 19% of the variance in ACE, respectively. A two-locus additive model with an additive × additive interaction of these polymorphisms explained most of the ACE variation associated with this region. We next analyzed the relationship between these two polymorphisms (ACE4 and ACE8) and blood pressure (BP). Although no evidence of linkage was detected, significant association was found for both systolic and diastolic BP when a two-locus additive model developed for ACE concentration was used. Further analyses demonstrated that an epistasis model provided the best fit to the BP variation. In conclusion, we found that the two polymorphisms explaining the greatest variation in ACE concentration are significantly associated with BP, through interaction, in this African population sample. Our study also demonstrates that greater statistical power can be anticipated with association analysis versus linkage, when markers in strong linkage disequilibrium with a trait locus have been identified. Furthermore, alllelic interaction may play an important role in the dissection of complex traits such as BP.
Introduction
The genes of the renin-angiotensin system (RAS) have been subjected to the most intense molecular scrutiny of any regulatory pathway in cardiovascular disease (CVD). The discovery early on that an Alu element was strongly associated with circulating levels of ACE (MIM 106180) provided a convenient genetic marker (Rigat et al. 1990; Tiret et al. 1992). Several studies of CVD phenotypes have suggested that the D allele from the insertion/deletion (I/D) polymorphism within the ACE gene confers increased risk for CVD (Evans et al. 1994, Soubrier et al. 1994). At the same time, considerable negative evidence exists on this question. An analysis of 11,000 cases and controls showed no relationship between myocardial infarction and the I/D polymorphism (Keavney et al. 2000). The I/D marker was likewise unassociated with myocardial infarction in Italian and French population samples (Arca et al. 1998; Ferrieres et al. 1999). O’Malley et al. (1999) summarized the association between the I/D polymorphism and CVD risk, grouping studies by geographical region and disease prevalence. In this analysis the ACE I/D polymorphism did not appear to be a clinically useful indicator of risk for myocardial infarction.
Hypertension is an important risk factor for CVD. As has been the case for myocardial infarction, conflicting results have been reported regarding the association between the I/D polymorphism and blood pressure (BP) (Schmidt et al. 1993; Morise et al. 1994; Barley et al. 1996; Vassilikioti et al. 1996; Borecki et al. 1997). Two large population-based studies found, for BP and the I/D polymorphism, marginally significant association and linkage evidence, which were restricted to males (Fornage et al. 1998; O’Donnell et al. 1998). Recently, a genome-scan analysis by The Framingham Heart Study found strong evidence for a quantitative-trait locus (QTL) on chromosome 17, which was close to the ACE gene and was linked to BP (Levy et al. 2000). Whether the ACE polymorphisms account for BP variation is, therefore, very much undecided.
Negative findings in prior studies may be due to the inadequate power to detect the modest contribution from an individual gene to a trait such as BP. Furthermore, analyses based on single polymorphisms in candidate genes can lead to false-negative outcomes for complex traits. Therefore, a more thorough understanding of the total genomic variation that exists in a region, as well as of the effect that these variants have on the intermediate pathways, may be necessary. Only after all the genetic and physiological information has been exhausted can definitive conclusions be made regarding a candidate’s role in complex phenotypes such as hypertension and heart disease. To this end, we genotyped an informative set of 13 single-nucleotide polymorphisms (SNPs), across the entire ACE gene, in a sample of Nigerian families. A series of models were fitted, to identify the polymorphisms causing the variation in plasma ACE concentration. These polymorphisms were then used in further analyses, to assess the contribution that the ACE gene makes to BP variation.
Subjects and Methods
Recruitment of Subjects
The sampling frame for this study was provided by the International Collaborative Study on Hypertension in Blacks, as described in detail elsewhere (Cooper et al. 1997; Guo et al. 1999; Rotimi et al. 1999). During the first phase of the study, sample surveys were conducted in residential communities in western Africa (Nigeria and Cameroon), the Caribbean (Barbados, St. Lucia, and Jamaica) and the United States (metropolitan Chicago) (Cooper et al. 1997). In the second phase, nuclear families were identified through middle-aged probands, and all available first-degree relatives were enrolled (Guo et al. 1999; Rotimi et al. 1999). In the present article, we report the findings from Nigeria. The enrolled families included 1,993 persons; analyses were restricted to the set of 1,343 individuals in 332 families for whom both measurements of ACE concentration and genotype data were available. Study protocols were reviewed and approved by the review boards of the participating institutions.
ACE Measurement
Determination of ACE concentration was performed by a previously published “sandwich” enzyme-linked immunosorbent assay (Danilov et al. 1996), with some modifications. In brief, plasma samples diluted 1/10 were incubated in polystyrene microtiter plates coated with monoclonal antobody to ACE 9B9. After unbound human plasma was washed off, the ACE plates were incubated with sheep anti-ACE IgG (kindly provided by Dr. S. Pokrovsky, National Cardiology Research Center, Moscow) conjugated with peroxidase. The plates were washed again, and bound peroxidase was quantified by diaminobenzidine (Sigma) at 492 nm on a microplate reader. The interassay coefficient of variation was 5.6%, and the intraassay coefficient was 6.2%, on the basis of 60 samples. Some concern arose about the possibility that Nigerian samples had defrosted because electrical power there had been intermittent when they were collected; however, freeze-thaw experiments did not suggest that values of ACE would have been falsely elevated under those conditions.
For the majority of samples, DNA was extracted from buffy coats at the University of Utah, by a standard phenol-chloroform procedure, and was brought to a standard concentration of 20 μg/ml. For the remaining samples, DNA was extracted in Oxford, by the Masterpure™ genomic DNA–purification kit, which uses a rapid desalting process to remove contaminating macromolecules.
Genotyping
The 11 diallelic SNPs listed in table 1 were all typed by PCR amplification, followed by restriction-enzyme digestion, with, where necessary, an artificial restriction site introduced by primer mismatch (see table 1). PCR products were digested with the appropriate restriction enzyme (table 1), according to the manufacturer’s protocol. Digested products were run on agarose gels, stained with ethidium bromide, and scored by UV visualization. With the exception of ACE4 and ACE5, all SNPs were amplified by the same procedure. Genomic DNA was amplified in a 15-μl reaction containing 25 ng of DNA, 0.3 mM of each primer, 0.125 mM of each dNTP, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), and 0.25 U of AmpliTaq Gold™ (PE Biosystems), with MgCl2 concentrations as in table 1. Cycling conditions were 95°C for 15 min; followed by 35 cycles of 94°C for 30 s, annealing at the appropriate temperature (TA) (see table 1) for 30 s, and 72°C for 30 s; and, finally, a 10-min extension at 72°C.
Table 1.
Marker Positions and PCR-RFLP Conditions for Detection of 13 SNPs in the ACE Gene
|
Position |
|||||||
| Marker (Location) | Villard et al. (1996) | Present Study | Primer Sequencesa(5′→3′) | TA(°C) | MgCl2(mM) | RFLP Enzyme | T(°C) |
| ACE1 (5′ UTR) | T−5491C | T−5529C | ACE1F:b TACAACCATCACTACTAATGTCACE1R:b TATAATATATGTGACATGGCCTG | 55 | 2 | BstNI | 60 |
| ACE2 (5′ UTR) | A−5466C | A−5499C | ACE2F:b GCCATGTCACATATATTATAGGAACE2R:b CGTCTTTGGAAACTTGTCTGC | 50 | 2.5 | EcoRV | 37 |
| ACE3 (5′ UTR) | T−3892C | T−3925C | ACE3F:b ATAGTGTATATAGGGCTTGGTACACE3R:b AGAAGATATTTGCAAAGTATGTACTG | 55 | 2 | PstI | 37 |
| ACE4 (5′ UTR) | A−240T | A−262T | ACE4F: TGTCACTCCGGAGGCGGGAGGCTACE4R:b GAGAAAGGGCCTCCTCTCTCT | 55 | 1 | XbaI | 37 |
| ACE5 (5′ UTR) | C−93T | C−115T | ACE5F2: ACCATGGCCTGGTGAAGAAGCACE5R:b CGGCTCTGCCCCTTCTCCTGCGC | TDc | 2.5 | HinfI | 37 |
| ACEs2.1 (intron 7) | C5144T | ACEs1F: GGGCGGGAAGTGGTGTGCACEs1AR: CTGAAAGCAAGGAAGGAGGAG | 62 | 2 | PstI | 37 | |
| ACEs1.1 (Intron 7) | G5170A | ACEs1CF: GCAGTGAGCTGAGATTGTGCCACEs1AR: CTGAAAGCAAGGAATGAGGAG | 62 | 2 | PstI | 37 | |
| ACE6 (exon 8) | C1237T | C5467T | ACE6F:b AGTGCACACGGGTCACGATGACE6R-1: CACCAAGTAGCCAAAGGGCAG | 62 | 1.5 | BsmBI | 55 |
| ACENEW 6 (intron 8) | A5967G | ACENEW6F: CAGGGTTCGGGATCCTCCTAGAACE6R-1: CCACCAAGTAGCCAAAGGGCAG | 62 | 1.5 | BsmBI | 55 | |
| ACE7 (exon 15) | A2215G | A9596G | ACE7F:b CACACCCTGAAGTACGGCACACE7R: TCCTCCAGCTCCTGGGCAG | 60 | 1 | HaeII | 37 |
| ACE I/D (intron 16) | ACEIDF: CTGGAGACCACTCCCATCCTTTCTACEIDR: GATGTGGCCATCACATTCGTCAGAT | 62 | 3 | ||||
| ACE8 (exon 17) | A2350G | A11860G | ACE8F:b CTGACGAATGTGATGGCCGCACE8R:b TTGATGAGTTCCACGTATTTCG | 58 | 2 | BstUI | 60 |
| ACE9 (3′ UTR) | 4656 (TC)2/3 | 21288 (CT)2/3 | ACE9FB: GACTCTGGGAAGCAGACATACE9RA: AGCTAATCCCTGTGCAGGT | (See text) | (See text) | (See text) | (See text) |
Underlined nucleotides represent the deliberate primer mismatches, designed to introduce artificial restriction sites.
Described by Keavney et al. (1998).
Touchdown PCR.
The following procedure was used to amplify ACE4 and ACE5: The region spanning both markers was amplified by primers ACE5F2 and ACE5R, in a 50-μl reaction containing 100 ng of genomic DNA, 0.3 mM of each primer, 0.125 mM of each dNTP, 50 mM (NH4)2 SO4, 2.5 mM MgCl2, 10% glycerol, 4 U of AmpliTaq (PE Biosystems). Cycling conditions were 95°C for 1min 30 s; then 10 cycles of 95°C for 1 min, 65°C (−0.5°C/cycle) for 1 min, and 72°C for 3 min; and then 30 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 3 min. A 1:25 dilution of the resulting product was then used as a template to amplify the region containing ACE4, in a 25-μl reaction containing 0.3 mM of each primer, 0.125 mM of each dNTP, 50 mM (NH4)2SO4, 1 mM MgCl2, 1 U of AmpliTaq (PE Biosystems). Cycling conditions were 95°C for 1min 30 s; then 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 3 min; and then 72°C for 10 min (table 1).
For the I/D polymorphism, the primers listed in table 1 were used to amplify the region of Alu insertion, with the Alu element's presence or absence being detected by running the product on an agarose gel. PCR amplification was performed in a 15-μl reaction containing 25 ng of DNA, 0.45 mM of each primer, 0.125 mM of each dNTP, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.25 U of AmpliTaq Gold™ (PE Biosystems), and 3 mM MgCl2. The cycling conditions were 95°C for 15 min; then 35 cycles of 94°C for 1 min, 62°C for 45 s, and 72°C for 1 min; and, finally, 72°C for 10 min.
The dinucleotide polymorphism 21288(TC)2/3, ACE9, was typed by a quasimultiplex strategy. Seven different-sized, fluorescently labeled PCR products were first generated by the following distinct combinations of six primers: (1) 333 bp, ACE9FB and ACE9RA (see table 1); (2) 305 bp, ACE9FB and ACE-VIL-9R (GCTACACTCCAGCGTCTGAGG); (3) 296 bp, ACE9FA (CATTTCCACTGGCAGTGGA) and ACE9RA; (4) 268 bp, ACE9FA and ACE-VIL-9R; (5) 205 bp, ACE-VIL-9F (GAGTACCTTGGAGGGCCTGCT) and ACE9RA; (6) 177 bp, ACE-VIL-9F and ACE-VIL-9R; and (7) 130 bp, ACE-COL-9F (TTGGCTCCTGCTGTACCAG) and ACE9RA. The PCR protocol used was that described above for all the SNPs, except ACE4 and ACE5, and size refers to the larger allele—for example, 21288(TC)3. A mixture of the seven different-sized products, plus a size standard, were then run on an ABI 377 automated sequencer (seven products per lane), with allele sizes scored by GENOTYPER software.
Statistical Analyses
Descriptive statistics were calculated by means of SAS (SAS Institute). The allele frequencies for each SNP were calculated by the allele-counting method. Hardy-Weinberg equilibrium at each locus was assessed by the χ2 test with 1 df (Weir 1996). We used the variance-component approach in GENEHUNTER to test for linkage between a genetic location and ACE concentration, systolic BP (SBP), and diastolic BP (DBP). The variance-component method specifies the expected genetic covariances between relatives, as a function of their identity-by-descent (IBD) relationships at a marker locus (Pratt et al. 2000). The IBD probabilities are derived from available genotyped marker loci. The likelihood-ratio test is applied to test the null hypothesis of no additive genetic variance due to the QTL. Sex, age, and age2 were incorporated as covariates, and their effects were estimated simultaneously by the maximum-likelihood method. The multipoint linkage analysis is dependent on the marker distances and the marker order—which, if there are any genotype errors in such a short genome region, can be inconsistent with the distances transformed on the basis of base pairs and the sequence order. We then conducted single-point linkage analysis using SIBPAL2. In this program, the mean-corrected trait product is linearly regressed on the estimated proportion of alleles that the sib pair shares IBD at each marker locus (Elston et al. 2000).
Measured genotype analysis accounting for familial aggregation and covariate effects was performed by the computer program ASSOC, available in the Statistical Analysis for Genetic Epidemiology package (S.A.G.E. 2000). The ASSOC program is designed to evaluate the association between a continuous trait and a discrete genetic marker, by means of a mixed-effect model proposed by George and Elston (1987). The model treats the genetic marker and covariates as fixed effects and treats the familial effect among relatives as a random effect, together with a random individual environmental effect. The familial effect is modeled on the assumption that the residual correlations between pairs of family members are due entirely to polygenic effects. Heritability is estimated without the use of genetic marker information. We used the delta method to estimate the standard error of the heritability.
To test the association between a genetic marker and a phenotype, ASSOC uses the maximum-likelihood statistic: 2(maximum loge likelihood under the general model − maximum loge likelihood under the null hypothesis). This statistic, under the assumptions of transformability to normality and the null hypothesis, is asymptotically distributed as a χ2 distribution. The appropriate number of degrees of freedom is given by the difference between the numbers of parameters estimated when the likelihoods are maximized under the null and alternative hypotheses.
In this study, ACE was first analyzed under the assumption of an additive, dominant, or recessive mode of inheritance at each marker locus. Since we had 13 markers, adjustment for multiple comparisons was necessary; a value of P<.0013 was conservatively considered to be genetically significant, being equivalent to the nominal value .05 if all 39 tests were independent. The most significant marker associated with ACE was identified, and this marker was then considered as a covariate in the model, to identify the second most significant marker. We then adjusted for these two markers, to test whether the other markers could account for any further variation in ACE. The best two-marker model, including possible interaction between the two markers, was identified, and this model was used to test association with SBP and DBP. Because, to test for association with BP, we decided beforehand to use the best model associated with ACE, a value of P<.05 was considered to be statistically significant. Finally, an epistasis model was defined for BP, to evaluate possible interactions between markers.
Results
The demographic characteristics of the participants are reported in table 2. No individuals were taking medication that might have modified BP levels. The mean ages of men and women were similar, but women had nominally significantly higher levels of BMI, SBP, and DBP, compared with men. ACE concentrations were not significantly different between the two genders. Correlations among the phenotypes (i.e., ACE, BMI, SBP, and DBP) are presented in table 3; no correlation between ACE and SBP was observed, and only a weak correlation between ACE and DBP was observed (|D|<.1).
Table 2.
Demographic Characteristics of Study Participants
| Male | Female | |
| No. (%) | 696 (51.8) | 647 (48.2) |
| Mean age ± SE (years) | 41.6 ± 20 | 40.1 ± 18.5 |
| Mean BMI ± SE (kg/m2) | 20.3 ± 3.6a | 22.1 ± 4.8 |
| Mean ACE ± SE | 612.6 ± 223.4 | 605.3 ± 209.8 |
| Mean SBP ± SE | 126.3 ± 24.7a | 129.4 ± 29.6 |
| Mean DBP ± SE | 75.8 ± 17.0a | 78.4 ± 18.4 |
Significantly different (P<.05) between males and females, when subjects are assumed to be independent.
Table 3.
Pearson Correlation Coefficients among Phenotype Measures
|
Pearson Correlation Coefficient (SE) for |
|||
| Trait | BMI | SBP | DBP |
| ACE | −.103 (.0002) | −.041 (.14) | −.085 (.002) |
| BMI | .265 (.0001) | .349 (.0001) | |
| SBP | .836 (.0001) | ||
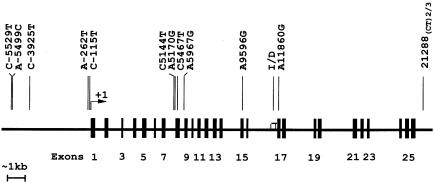
Polymorphisms at 13 sites were genotyped across a 27-kb segment in the ACE gene (fig. 1). No significant deviations from Hardy-Weinberg equilibrium were found among the sites tested, except at sites ACEs2.1 and ACE6 (P=.05 and P=.03, respectively). The genetic analyses were performed with sex, age, and age2 included as covariates. The overall heritability (h2) values for ACE, SBP, and DBP were estimated to be .65±.01, .42±.02, and .46±.05, respectively, after the significant covariates were taken into account. Multipoint linkage analysis was conducted in this region for the three phenotype measures. For ACE, a maximum LOD score of 7.5 was observed close to marker ACE8; a second peak, of 7.2, was seen at the site close to ACE5. The multipoint LOD score for both SBP and DBP was always <0.1. The single-point linkage analysis by SIBPAL2 showed weak evidence of linkage between SBP and ACE2 and between SBP and ACE7 (P=.06 and P=.08, respectively); no significant evidence of linkage between DBP and ACE polymorphisms was found (P=.29). These results thus provide strong evidence that, in this region, there is at least one ACE-related QTL but minimal evidence for linkage between genetic variation and BP.
Figure 1.
Schematic representation of the human ACE (DCP1) gene. Lines in the upper portion of the graph represent the polymorphisms typed in this study, with positions characterized by use of “+1” to represent the first nucleotide of the first codon (AGT) in exon 1.
Association with ACE Concentration
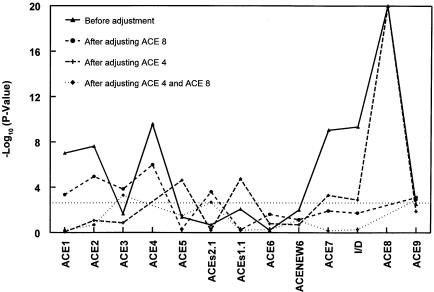
We then performed association analysis at each of the 13 markers, with age and gender as covariates and under the assumption of an additive, recessive, or dominant mode of inheritance, in separate analyses. Figure 2 presents the P values from the likelihood-ratio tests. Only the smallest P values among the three different modes of inheritance are reported. The vertical axis gives the −log10(P) for testing no association between ACE concentration and the marker. The dotted horizontal line represents P=.0013 (−log[.0013] = 2.886 ), the level at which comparisons become significant at the 5% level after adjustment for multiple comparisons. The polymorphisms meeting this criterion included ACE1, ACE2, ACE4, ACE7, I/D, and ACE8 (P<.0013) (fig. 2, line with black triangles). The highest peaks were observed at markers ACE8 and ACE4, explaining 19% and 6% of the total variance, respectively. To determine whether the association with ACE, which was found at the six polymorphisms, was due to linkage disequilibrium between them, we repeated the analysis after adjusting for the effect at ACE8. With this adjustment, the association at markers ACE7 and I/D was no longer significant, but it did remain so at markers ACE1, ACE2, ACE3, and ACEs2.1 (fig. 2, dashed line with black circles). Similar results were found in an analysis stratified by genotype, at ACE8. Similarly, we performed the same analysis, adjusting for ACE4; the association with ACE was then no longer significant for markers ACE1, ACE2, and ACE3 but did remain significant for markers ACE7, I/D, ACE8, and ACE9 (fig. 2, dashed line with crosses). In essence, the association disappears at the markers close to the adjusted polymorphism but remains at the more distant markers. We then adjusted for polymorphisms ACE4 and ACE8 simultaneously. The resulting −log10(P) fell below the level of significance, everywhere except at ACE3, ACEs2.1, and ACE9 (fig. 2, dashed line with black squares). In addition, the −log10(P) increased for ACE3, ACEs2.1, and ACE9, after adjustment for ACE4 and ACE8; this result may have occurred because ACE4 and ACE8 may not be the functional mutations themselves but, instead, are in linkage disequilibrium (LD) with functional variants. In addition, it is possible that variants that are not in strong LD with either ACE4 or ACE8 may have effects on ACE concentrations. Overall, these findings suggest that most of the ACE variation in this region can be explained by variants in the gene, at sites close to the ACE4 and ACE8 polymorphisms.
Figure 2.
Results of association analysis relating ACE concentration to variants in the ACE gene. Age and gender were entered as covariates; additive, recessive, or dominant modes of inheritance were assumed in separate analyses, but only the smallest P value among the three different modes of inheritance is reported. P values are from the likelihood-ratio test with 1 df. The dotted horizontal line represents P=.0013 (-log[.0013]=2.886), the 5%-significance level after adjustment for multiple comparisons.
We then defined the best ACE model as the one that accounted for the greatest proportion of the variance when ACE4, ACE8, and their interaction were considered together. After the testing of all the combinations of additive, dominant, and recessive modes of inheritance, for ACE4 and ACE8 jointly, the model that included ACE4 and ACE8 as additive, together with the “additive × additive” interaction term, was identified as the best ACE model, explaining 22% of the total variance. The best model also agreed with that selected on the basis of Aikake’s information criterion (AIC) (AIC = −2 ln likelihood + 2[no. of independently estimated parameters]). The A allele of the ACE4 polymorphism was associated with increased ACE concentration, as was the G allele of the ACE8 polymorphism.
Association with BP
To test the hypothesis that ACE QTLs also affect SBP and/or DBP, we performed association analyses, using the best ACE model described above. The regression results from the program ASSOC are presented in table 4. (We omitted age2 from the model for SBP because it was not significant.) With gender, age, and BMI as covariates, both ACE4 and ACE8 were significantly associated with SBP (P<.05); The average level of SBP increased 2.3 and 3.2 mmHg with each copy of allele T and G present, respectively. Only the ACE4 polymorphism was significantly associated with DBP (P<.05), yielding a 1.5-mmHg increase for each copy of the allele T. Surprisingly, the alleles at the ACE4 polymorphism had the opposite effect with regard to ACE concentration and BP—the T allele was associated with a decrease in ACE concentration and with an increase in BP.
Table 4.
Regression Coefficients for SBP and DBP, for the Best Genetic Model, on the Basis of ACE Concentration
|
Regression Coefficient (SE) for |
||||||||
| Sex | Age | Age2 | BMI | ACE4 (Ta) | ACE8 (Ga) | ACEA × ACE8 | AIC | |
| SBP (mmHg) | 2.91 (1.41)b | .68 (.04)c | … | .99 (.17)c | 2.34 (1.14)b | 3.2 (1.36)b | 3.4 (2.0) | 10,192.4 |
| DBP (mmHg) | 1.23 (.90) | .41 (.02)c | −.009 (.001)c | .82 (.12)c | 1.50 (.73)b | .99 (.87) | 1.35 (1.29) | 9,192.2 |
Allele increases SBP or DBP.
Statistically significant at P<.05.
Statistically significant at P<.01.
Since the T allele at ACE4 and the G allele at ACE8 were associated with increasing BP, we defined an epistasis model as one in which BP will be increased only when both the T and G alleles are present (table 5). This model was significant for both SBP and DBP (P=.005 and P=.04, respectively; see table 5), predicting ∼5-mmHg-higher and ∼2-mmHg-higher values for SBP and DBP, respectively, in persons with both the T and G alleles, compared with those with neither of these alleles. Because the best ACE model according to ACE concentrations and the epistasis model are not nested within each other, we used the AIC to compare them. From tables 4 and 6, we conclude that the epistasis model is slightly better than the best ACE model derived directly from the analysis of ACE concentrations.
Table 5.
Epistasis Model[Note]
| ACE8 |
|||
| ACE4 | AA | AG | GG |
| AA | 0 | 0 | 0 |
| AT | 0 | 1 | 1 |
| TT | 0 | 1 | 1 |
Note.— The model indicates that BP levels are increased when both the T allele at ACE4 and the G allele at ACE8 are present.
Table 6.
Regression Coefficients and Wald Test P Values for the Epistasis Model, for SBP and DBP
|
Regression Coefficient (Wald Test P Value) for |
||||||
| Sex | Age | Age2 | BMI | βa | AIC | |
| Entire sample: | ||||||
| SBP | 2.88 (1.41)b | .67 (.04)c | … | .99 (.17)c | 5.1 (1.82)c | 10,189.1 |
| DBP | 1.23 (.90) | .41 (.02)c | −.009 (.001)c | .82 (.12)c | 2.4 (1.16)b | 9,188.1 |
| Males: | ||||||
| SBP | … | .49 (.04)c | … | 1.77 (.25)c | 4.4 (2.43) | … |
| DBP | … | .34 (.03)c | −.009 (.002)c | 1.25 (.17)c | 2.7 (1.55) | … |
| Females: | ||||||
| SBP | … | .94 (.06)c | … | .47 (.23)b | 7.1 (2.65)c | … |
| DBP | … | .49 (.04)c | −.008 (.002)c | .57 (.16)b | 2.66 (1.73) | … |
As defined by the epistasis model in table 5. A positive value indicates the increment of BP level when the T allele at ACE4 and the G allele at ACE8 are present.
Statistically significant at P<.05.
Statistically significant at P<.01.
We performed the same analyses separately, by gender, using the epistasis model. A significant association with SBP was found among women (P=.007) but not among men, whereas no association with DBP was found in either gender. Of course, this last null finding could be due to the reduction of the sample size. We also conducted the analysis with only a single SNP in the model and found that ACE8 is associated with SBP (P=.04). Finally, we tested the remaining sites and found no associations with either SBP or DBP (P>.05; data not shown). We repeated all the analyses of SBP and DBP, after including ACE as an additional covariate, and obtained essentially the same results.
Discussion
We have conducted linkage and association analyses based on markers in the ACE candidate region on chromosome 17 in a large population-based sample of families. Unlike previous studies, which, for genetic information, relied solely on the I/D polymorphism, we genotyped 13 polymorphisms in the ACE gene simultaneously. In a sequential analytic strategy, we used ACE concentration as the first phenotype and BP as the second. Two SNPs were identified that accounted for the variation of ACE concentration. The polymorphism in intron 17, ACE8, had the most significant effect, accounting for 19% of the total variance in ACE. The second polymorphism, ACE4, was located in the 5′ section of the gene and accounted for 6% of the variance. The effects of both of these polymorphisms best fitted an additive model. After adjustment for the effect of ACE8, the I/D polymorphism was no longer associated with ACE, indicating that it is in LD with ACE8 and unlikely to be a functional mutation. Our study therefore provides positive evidence that there are two single-nucleotide variants, separated by 12 kb, which define a genomic region influencing ACE concentration. This result is consistent with previous reports in a European population (Villard et al. 1996), in which functional variants at the 3′ and 5′ ends of the ACE gene were reported. Zhu et al. (2000) did not test the 5′ polymorphisms in Europeans and Afro-Caribbeans, but their significant finding in the 3′ region agrees with the results of the present study. We also found that the A allele at ACE4 increased ACE concentration in our sample but has been reported to be associated with decreased ACE concentration in British and French populations (Villard et al. 1996; Keavney et al. 1998). These discrepant findings suggest that ACE4 is not the functional polymorphism but is in strong LD with the relevant ACE variants. In fact, in our sample, with regard to ACE concentration, all the alleles at polymorphisms ACE1–ACE4 have an effect that is opposite to what was seen in the European sample (data not shown), suggesting that ACE1–ACE4 themselves are unlikely to be the functional mutants affecting ACE concentration.
When the best-fitting model for ACE concentration—a model based on the markers designated “ACE4” and “ACE8”—was used, a significant association with BP was also found. An epistasis model—that is, one taking into account the potentially interactive effect of the two genetic loci—best fits the data. When both the G allele at ACE8 and the T allele at ACE4 were present, SBP and DBP increased, on average, by 5 and 2 mmHg, respectively. The size of the allelic effect derived from this model is substantial, relative to most genotype-phenotype relationships thus far described for BP; on the other hand, despite both the large amount of information available from the 13 polymorphisms and the LOD score of >7 for ACE concentration, the evidence of linkage between the ACE locus and BP was hardly significant. In addition, we found more linkage evidence by using single-point linkage analysis than by using multipoint linkage analysis. Because we studied a small chromosomal region, genotyping errors leading to inconsistent physical distances or marker order could potentially reduce the sensitivity of multipoint linkage analysis and dilute the linkage evidence. Furthermore, the polymorphism that we typed explained only 1% of the variance in BP, yet the association result was reasonably powerful. This result is consistent with the greater statistical power that can be anticipated from association analysis compared with linkage analysis, when markers that are in LD with the trait locus have been identified. However, replication of these results is required to confirm that our finding is not due to type I error.
Two recent epidemiological studies have also reported a relationship between the I/D marker and BP, although the findings were marginally significant and restricted to males (Fornage et al. 1998; O’Donnell et al. 1998). Fornage et al. studied a young adult white population sample that was untreated for hypertension (Fornage et al. 1998). The disadvantage of a young sample, of course, is the possible reduction in power to detect hypertension genes when the trait has not been fully expressed. O’Donnell et al. (1998) performed linear regression analyses of SBP and DBP by adjusting for antihypertensive treatment in a large sample from The Framingham Heart Study. Considering antihypertensive treatment as a covariate for adjustment may create potential bias, however, because of the correlation between the treatment and the genes affecting BP variation. We did not replicate these male-specific findings. In our study, a significant association was found between the interaction of A−240T and G2350A and BP, but this result was significant in females only when gender-specific analyses were performed. The null finding in men may be due to the decrease in sample size, since all the P values were close to .05.
The RAS has always been an attractive model system for the study of the genetics of hypertension, because the intermediate phenotypes can be measured directly. On the basis of the hypothesis that, over the course of a lifetime, even small increases in RAS activity elevate risk of hypertension in some individuals, ACE concentration should be a guide to the genetic makeup that conditions that risk. This model system is therefore well suited to a sequential approach that first uses linkage analysis to define the informative variants on the basis of the intermediate phenotype and then uses association analysis with BP as the ultimate phenotype. Although, in the present study, this strategy has appeared to work well for ACE, most candidate genes do not have a readily assayed intermediate phenotype. As a further corollary of this argument, linkage alone may be an ineffective strategy if the impact that other loci have on BP is as small as that which we have observed for ACE.
As noted above, the allelic effect for ACE4 is opposite in direction for ACE concentration and BP. This finding is obviously inconsistent with the simple “overactivity” hypothesis put forward above. We have no immediate explanation for this result. These complex interrelationships could, in part, explain the low correlation between ACE and BP, and could suggest, if confirmed, that there are common—but functionally different—ACE variants affecting both ACE and BP variation. This result may also help explain the conflicting findings in previous studies (Schmidt et al. 1993; Morise et al. 1994; Barley et al. 1996; Vassilikioti et al. 1996), since only the I/D polymorphism was considered in the association with hypertension; for example, we found ACE8 marginally associated with SBP when we tested each marker singly.
In general, the power to detect a gene will increase with information on multiple loci and their interactions. Our study is the first to consider more than one ACE polymorphism in the analysis of BP variation. The result confirms the need to consider multiple loci and allelic interactions—and, possibly, gene × environmental interactions, if they can be characterized—in the study of complex traits such as hypertension.
The families studied here were recruited as part of an international collaborative project examining the genetic and environmental factors that have resulted in the large population shifts in BP that have been seen across the African diaspora (Cooper et al. 1997, 1999). Despite the lower prevalence of hypertension observed in rural Nigeria, several advantages for family studies in that society are present. Nuclear families tend to be larger, and many of them live close together in adulthood. Furthermore, at the first screening encounter, no antihypertensive treatment was observed, given the low level of screening and detection, and BP could be used as a quantitative trait for all individuals. (Within the limitations of a research project, treatment subsequently was provided for newly detected cases.)
Whether these findings will apply to industrialized societies such as the United States remains an open question. In previous analyses, we have shown that the relationships among BP, environmental exposures, and the components of the RAS vary substantially across African-origin populations in the United States, the Caribbean, and West Africa (Cooper et al. 1998, 1999, 2000; Kaufman et al. 1997). In general, the relationship between BP and both the environmental and physiological factors is stronger in the low-risk, Nigerian setting, suggesting that multiple exposures destabilize the regulatory systems. This general pattern could also apply to genetic relationships, throwing them into sharper relief. Whatever the modifying effects, it is still reasonable to assume that the variation in the nature of these relationships, from low- to high-risk environments, is one of degree and not one of kind.
In conclusion, in our large population-based sample, we found strong evidence of two SNPs in the ACE gene that are associated with circulating-ACE concentration. These two polymorphisms also significantly affect BP level through interaction. If confirmed, these results would support the “RAS hypothesis” of hypertension predisposition and would help to reconcile previous, inconsistent findings.
Acknowledgments
We wish to acknowledge the facilities provided by the Wellcome Trust Centre for Human Genetics, under the direction of Dr. G. M. Lathrop, and, specifically, the assistance provided by Roger Cox and his laboratory, especially the work by Alison Hughill. This work was supported by an internal grant from the University of Oxford, and by National Heart, Lung, and Blood Institute grants HL45508 and HL 47910, National Institute of General Medical Sciences grant GM28356, and National Center for Research Resources grant RR03655.
Electronic-Database Information
The accession number and URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for ACE [MIM 106180])
References
- Arca M, Pannitteri G, Campagna F, Candeloro A, Montali A, Cantini R, Seccareccia F, Campa PP, Marino B, Ricci G (1998) Angiotensin-converting enzyme gene polymorphism is associated with coronary atherosclerosis and myocardial infarction in a sample of Italian patients. Eur J Clin Invest 28:485–490 [DOI] [PubMed] [Google Scholar]
- Barley J, Blackwood A, Miller M, Markandu ND, Carter ND, Jeffer S, Cappuccio FP, MacGregor GA, Sagnella GA (1996) Angiotensin converting enzyme gene I/D polymorphism blood pressure and the renin-angiotensin system in Caucasian and Afro-Caribbean peoples. J Hum Hypertens 10:31–35 [PubMed] [Google Scholar]
- Borecki IB, Province MA, Ludwig EH, Ellison RC, Folsom AR, Heiss G, Lalouel JM, Higgins M, Rao DC (1997) Association of candidate loci angiotensinogen and angiotensin-converting enzyme with severe hypertension: the NHLBI Familiy Heart Study. Ann Epidemiol 7:13–21 [DOI] [PubMed] [Google Scholar]
- Cooper R, Forrester T, Ogunbiyi O, Mufunda J (1998) Angiotensinogen levels and obesity in four population samples. J Hypertens 16:571–576 [DOI] [PubMed] [Google Scholar]
- Cooper RS, Guo X, Rotimi CR, Luke A, Ward R, Adeyemo A, Danilov S (2000) Heritability of angiotensinogen and ACE: a comparison of Nigerian and African-American family sets. Hypertension 35:1141–1147 [DOI] [PubMed] [Google Scholar]
- Cooper R, Rotimi C, Ataman S, McGee D, Osotimehin B, Kadiri S, Muna W, Kingue S, Fraser H, Forrester T, Bennett F, Wilks R (1997) Hypertension prevalence in seven populations of African origin. Am J Public Health 87:160–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RS, Rotimi CN, Ward R (1999) The puzzle of hypertension in African Americans. Sci Am 280:56–63 [DOI] [PubMed] [Google Scholar]
- Danilov S, Savoie F, Lenoir B, Jeunemaitre X, Azizi M, Tarnow L, Alhenc-Gelas F (1996) Development of enzyme-linked immunoassays for human angiotensin I converting enzyme suitable for large-scale studies. J Hypertens 14:719–727 [DOI] [PubMed] [Google Scholar]
- Elston RC, Buxbaum S, Jacobs KB, Olson IM (2000) Haseman and Elston revisited. Genet Epidemiol 19:1–17 [DOI] [PubMed] [Google Scholar]
- Evans AE, Poirier O, Kee F, Lecerf L, McCrum E, Falconer T, Crane J, O’Rourke DF, Cambien F (1994) Polymorphisms of the angiotensin-converting-enzyme gene in subjects who die from coronary heart disease. Q J Med 87(4): 211–214 [PubMed] [Google Scholar]
- Ferrieres J, Ruidavets JB, Fauvel J, Perret B, Taraszkiewicz D, Fourcade J, Nieto M, Chap H, Puel J (1999) Angiotensin I-converting enzyme gene polymorphism in a low-risk European population for coronary artery disease. Atherosclerosis 142:211–216 [DOI] [PubMed] [Google Scholar]
- Fornage M, Amos CI, Kardia S, Sing CF, Turner ST, Boerwinkle E (1998) Variation in the region of the angiotensin-converting enzyme gene influences interindividual differences in blood pressure levels in young white males. Circulation 97:1773–1779 [DOI] [PubMed] [Google Scholar]
- George VT, Elston RC (1987) Testing the association between polymorphic markers and quantitative traits in pedigrees. Genet Epidemiol 4:193–201 [DOI] [PubMed] [Google Scholar]
- Guo X, Rotimi C, Cooper R, Luke A, Elston R, Ward R, Ogunbiyi O (1999) Evidence of a major gene effect for angiotensinogen among Nigerians. Ann Hum Genet 63:293–300 [DOI] [PubMed] [Google Scholar]
- Kaufman JS, Asuzu MC, Sparks H, Mufunda J, Luke A, Rotimi CN, Cooper RS (1997) The relationship between blood pressure and body mass index in lean populations. Hypertension 30:1511–1517 [DOI] [PubMed] [Google Scholar]
- Keavney B, McKenzie C, Connell JMC, Julier C, Ratcliffe PJ, Sobel E, Lathrop M, Farrall M (1998) Measured haplotype analysis of the angiotensin-1 converting enzyme (ACE) gene. Hum Mol Genet 7:1745–1751 [DOI] [PubMed] [Google Scholar]
- Keavney B, McKenzie C, Parish S, Palmer A, Clark S, Youngman L, Delepine M, Lathrop M, Peto R, Collins R (2000) Large-scale test of hypothesised associations between the angiotensin-converting-enzyme insertion/deletion polymorphism and myocardial infarction in about 5000 cases and 6000 controls: International Studies of Infarct Survival (ISIS) Collaborators. Lancet 355:434–442 [DOI] [PubMed] [Google Scholar]
- Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples AC, Myers RH (2000) Evidence for a gene influencing blood pressure on chromosome 17: genome scan linkage results for longitudinal blood pressure phenotypes in subjects from The Framingham Heart Study. Hypertension 36:477–483 [DOI] [PubMed] [Google Scholar]
- Morise T, Takeuchi Y, Takeda R (1994) Angiotensin-converting enzyme polymorphism and essential hypertension. Lancet 343:125 [DOI] [PubMed] [Google Scholar]
- O’Donnell CJ, Lindpaintner K, Larson MG, Rao VS, Ordovas JM, Schaefer EJ, Myers RH, Levy D (1998) Evidence for association and genetic linkage of the angiotensin-converting enzyme locus with hypertension and blood pressure in men but not women in The Framingham Heart Study. Circulation 97:1766–1772 [DOI] [PubMed] [Google Scholar]
- O’Malley JP, Maslen CL, Illingworth DR (1999) Angiotensin-converting enzyme and cardiovascular disease risk. Curr Opin Lipidol 10:407–415 [DOI] [PubMed] [Google Scholar]
- Pratt SC, Daly MJ, Kruglyak L (2000) Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am J Hum Genet 66:1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F (1990) An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest 86:1343–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotimi CN, Cooper RS, Cao G, Ogunbiyi O, Ladipo M, Owoaje E, Ward R (1999) Maximum-likelihood generalized heritability estimate for blood pressure in Nigerian families. Hypertension 33:874–878 [DOI] [PubMed] [Google Scholar]
- S.A.G.E. (2000) Statistical analysis for genetic epidemiology, release 4.0. Department of Epidemiology and Biostatistics, Rammelkamp Center for Education and Research, MetroHealth Campus, Case Western Reserve University, Cleveland [Google Scholar]
- Schmidt S, Van Hooft IM, Grobbee DE, Ganten D, Ritz E (1993) Polymorphism of the angiotensin I converting enzyme gene is apparently not related to high blood pressure: Dutch Hypertension and Offspring Study. J Hypertens 11:345–348 [DOI] [PubMed] [Google Scholar]
- Soubrier F, Nadaud S, Williams TA (1994) Angiotensin I converting enzyme gene: regulation, polymorphism and implications in cardiovascular diseases. Eur Heart J 15:24–29 [DOI] [PubMed] [Google Scholar]
- Tiret L, Rigat B, Visvikis S, Breda C, Corvol P, Cambien F, Soubrier F (1992) Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet 51:197–205 [PMC free article] [PubMed] [Google Scholar]
- Vassilikioti S, Doumas M, Douma S, Petidis K, Karagiannis A, Balaska K, Vyzantiadis A, Zamboulis C (1996) Angiotensin converting enzyme gene polymorphism is not related to essential hypertension in a Greek population. Am J Hypertens 9:700–702 [DOI] [PubMed] [Google Scholar]
- Villard E, Tiret L, Visvikis S, Rakotovao R, Cambien F, Soubrier F (1996) Identification of new polymorphisms of the angiotensin I-converting enzyme (ACE) gene, and study of their relationship to plasma ACE levels by two-QTL segregation-linkage analysis. Am J Hum Genet 58:1268–1278 [PMC free article] [PubMed] [Google Scholar]
- Weir BS (1996) Genetic data analysis II. Sinauer Associates, Sunderland, MA [Google Scholar]
- Zhu X, McKenzie CA, Forrester T, Nickerson DA, Broeckel U, Schunkert H, Doering A, Jacob HJ, Cooper RS, Rieder MJ (2000) Localization of a small genomic region associated with elevated ACE. Am J Hum Genet 67:1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]