Abstract
Insulin resistance and hyperinsulinemia are strong correlates of obesity and type 2 diabetes, but little is known about their genetic determinants. Using data on nondiabetics from Mexican American families and a multipoint linkage approach, we scanned the genome and identified a major locus near marker D6S403 for fasting “true” insulin levels (LOD score 4.1, empirical P<.0001), which do not crossreact with insulin precursors. Insulin resistance, as assessed by the homeostasis model using fasting glucose and specific insulin (FSI) values, was also strongly linked (LOD score 3.5, empirical P<.0001) with this region. Two other regions across the genome were found to be suggestively linked to FSI: a location on chromosome 2q, near marker D2S141, and another location on chromosome 6q, near marker D6S264. Since several insulin-resistance syndrome (IRS)–related phenotypes were mapped independently to the regions on chromosome 6q, we conducted bivariate multipoint linkage analyses to map the correlated IRS phenotypes. These analyses implicated the same chromosomal region near marker D6S403 (6q22-q23) as harboring a major gene with strong pleiotropic effects on obesity and on lipid measures, including leptin concentrations (e.g., LODeq for traits-specific insulin and leptin was 4.7). A positional candidate gene for insulin resistance in this chromosomal region is the plasma cell-membrane glycoprotein PC-1 (6q22-q23). The genetic location on chromosome 6q, near marker D6S264 (6q25.2-q26), was also identified by the bivariate analysis as exerting significant pleiotropic influences on IRS-related phenotypes (e.g., LODeq for traits-specific insulin and leptin was 4.1). This chromosomal region harbors positional candidate genes, such as the insulin-like growth factor 2 receptor (IGF2R, 6q26) and acetyl-CoA acetyltransferase 2 (ACAT2, 6q25.3-q26). In sum, we found substantial evidence for susceptibility loci on chromosome 6q that influence insulin concentrations and other IRS-related phenotypes in Mexican Americans.
Introduction
The incidences of obesity and type 2 diabetes have been increasing alarmingly across the globe, especially in the Western world (Burke et al. 1999; Must et al. 1999), for most of the 20th century. Hyperinsulinemia and/or insulin resistance is a strong correlate of obesity and type 2 diabetes in various populations, including Mexican Americans (Haffner et al. 1990; Groop et al. 1991; Ravussin 1995). Insulin resistance and/or hyperinsulinemia often clusters with various metabolic abnormalities—including obesity, dyslipidemia, and hemodynamic traits—and this constellation of metabolic disorders has been referred to as “insulin-resistance syndrome” (IRS) (Reaven 1988; DeFronzo 1995). The IRS has been shown to be a predictor of coronary heart disease (CHD) events as well (Lempiainen et al. 1999). Not only do Mexican Americans have high rates of type 2 diabetes and obesity, compared with non-Hispanic whites (Stern and Haffner 1990), but nondiabetic Mexican Americans have been found to have higher insulin levels and to be more insulin-resistant than nondiabetic non-Hispanic whites (Haffner et al. 1990). Also, among Mexican Americans, individuals who later developed multiple metabolic abnormalities were found to have higher insulin levels at baseline than those who developed only a single abnormality (Haffner 1999).
Although several rare genetic syndromes (e.g., leprechaunism and type A IRS) have been found to have severe insulin resistance (Flier 1992; Kahn et al. 1996), knowledge about genetic regions that influence insulin concentrations and the various constituents of IRS in the general population is limited. In this study, therefore, we conducted a genomewide scan and employed a multipoint variance components approach to identify susceptibility loci influencing fasting specific insulin (FSI) concentrations in nondiabetic Mexican Americans distributed across 27 extended families. In contrast to levels of conventional fasting immunoreactive insulin or nonspecific insulin (FNI), FSI levels do not crossreact with insulin precursors (Hales 1995; Haffner et al. 1997). Because the major chromosomal regions of interest in this study provided strong evidence for linkage not only to measures of insulin concentration and insulin resistance, as calculated by the homeostatic model assessment measure of insulin resistance (i.e., HOMA %S), (Matthews et al. 1985; Levy et al. 1998) but also to other IRS-related phenotypes, including body-mass index (BMI), leptin (LEPT), sum of skin folds (SS), and ln triglycerides (ln TG), a bivariate multipoint linkage technique was used to determine whether mapping of these correlated traits to the same chromosomal regions was due to pleiotropy.
Material and Methods
San Antonio Family Diabetes Study
Phenotypic data
The San Antonio Family Diabetes Study (SAFADS) includes 32 low-income Mexican American extended families, which were ascertained on type 2 diabetic probands, containing 579 (140 diabetics) examined individuals, aged 18–98 years. Metabolic, anthropometric, demographic, and medical-history information was obtained on all the examined individuals. A subset of 440 individuals (116 diabetics) from the 27 largest pedigrees were selected for genotyping. For the present analyses, we used phenotypic information from nondiabetic subjects only from these families (∼310 individuals depending on availability of data). All procedures were approved by the institutional review board of the University of Texas Health Science Center at San Antonio, and all subjects gave informed consent.
Blood samples were obtained after a 12-h fast for assessment of various metabolic traits. Fasting plasma glucose (FG) concentrations were measured using an Abbott Bichromatic Analyzer (Abbott). Serum immunoreactive insulin or FNI levels were measured using a radioimmunoassay (Diagnostic Products). Triglyceride (TG) levels were measured by enzymatic procedures as described elsewhere (Stern et al. 1984). Fasting specific insulin (FSI) concentrations were measured using a monoclonal antibody-based two-site immunoradiometric assay (kindly performed by C. N. Hales; see Sobey et al. [1989]). Unlike conventional insulin immunoassays, insulin measured by this assay does not crossreact with insulin precursors. LEPT levels were measured by a commercial radioimmunoassay (Linco Research; see Ma et al. [1996]).
Using FG and FSI concentrations, estimates of pancreatic β-cell function (HOMA %β) and insulin sensitivity (HOMA %S, an inverse measure of insulin resistance) were derived by means of the homeostasis model assessment (HOMA) method (Matthews et al. 1985; Levy et al. 1998; Hermans et al. 1999a, 1999b). This method is based on a structural computer model of a glucose/insulin feedback system that incorporates mathematical descriptions of the function and interactions of different organs involved in FG control on the basis of empirical data. The model allows the use of either immunoreactive (or nonspecific) insulin or specific (or true) insulin assays since proinsulin is included in the model. The estimates of HOMA %β and HOMA %S are derived from FG and FSI concentrations of an individual and are expressed relative to values in a young, lean reference population. The measures of HOMA %S and HOMA β% have been shown to perform well in comparison with other direct measures of insulin sensitivity and β-cell function, respectively (Levy et al. 1998; Hermans et al. 1999a, 1999b), particularly in nondiabetics. However, we acknowledge that we do not have data on direct measures of either insulin secretion or insulin resistance. For the present study, log-transformed HOMA %β (ln HOMA %β) and HOMA %S (ln HOMA %S) were used.
Blood samples were obtained again 2 h after a standardized oral glucose load (World Health Organization Expert Committee 1985) for plasma glucose. Diabetes was diagnosed according to the World Health Organization (WHO) plasma glucose criteria (World Health Organization Expert Committee 1985). Individuals who did not meet the WHO criteria but reported a history of diabetes and that they were under treatment with insulin or oral antidiabetic agents were also considered to have diabetes. Individuals with diabetes were excluded from the present analyses. The anthropometric data were collected using standardized anthropometric protocols (Haffner et al. 1987). BMI was calculated as weight (in kg) divided by height (in m) squared. The sum of eight skin-fold measures (SS) was used as a measure of overall subcutaneous adiposity (Duggirala et al. 1996). Trait-specific distinct outliers were excluded from the analyses (e.g., TG values >800 mg/dl). For the present study, log-transformed triglyceride (ln TG) and FNI (ln FNI) values were used, since each of their untransformed values exhibited high nonnormality.
Genotypic data
An ∼10–15-cM map, which has been described in detail elsewhere (Duggirala et al. 1999b, 2000), was utilized for the multipoint linkage analysis. In brief, genotypes were collected primarily by polymerase chain reaction (PCR) assays with radiolabeled oligonucleotide primers. However, data for some of the markers were collected by use of fluorescence-labeled primers purchased from Research Genetics. The latter were PCR amplified and were loaded onto an Applied Biosystems Model 373 sequencer, and the data were analyzed with Applied Biosystems Genotyper software. The genotypic data were analyzed for discrepancies (i.e., violations of Mendelian inheritance), by means of the program INFER (Dyke 1996). Such discrepancies were checked in the laboratory for mistyping, and markers for discrepant individuals were either corrected or blanked out prior to analysis. A total of 419 markers have been typed, and all markers were used for two-point linkage analysis. As described elsewhere (Duggirala et al. 2000), since our multipoint linkage approach yields optimum results when similar numbers of individuals are genotyped at all loci, markers with <80% of the sample genotyped were not used for multipoint analysis, unless their absence would result in a map gap of ⩾20 cM. The multipoint analyses were thus conducted using genotypic information from 301 markers. The sex-averaged maps were constructed using the SAFADS marker data and the program CRI-MAP (Green et al. 1990), supplemented by the map information in the Cooperative Human Linkage Center (Murray et al. 1994), the Genome Database (Fasman et al. 1996), the Genetic Location Database (Collins et al. 1996), and the Marshfield Medical Research Foundation (Broman et al. 1998).
Multipoint Linkage Analyses
Univariate multipoint linkage analysis
A variance-components approach was used to test for linkage of a genetic location with a given phenotype (i.e., univariate) using maximum-likelihood methods (Amos 1994; Almasy and Blangero 1998). This method utilizes information from all possible pedigree relationships simultaneously, to examine the genetic architecture of a quantitative trait, and is based on specifying the expected genetic covariances between relatives as a function of their identity by descent (IBD) relationships at a marker locus (which is hypothesized to be linked to a locus influencing the quantitative trait—i.e., a “quantitative-trait locus” [QTL]). It allows for locus-specific effects (h2q = heritability attributed to the QTL), residual additive genetic effects (h2 = heritability attributed to the residual genetic effects), covariate effects (e.g., age and sex), and individual-specific random environmental factors: e2=1-(h2q+h2).
In the present study, in addition to the variance components, mean, and standard deviation of the phenotype and covariate effects (i.e., sex and age terms) were estimated, simultaneously, using maximum-likelihood techniques. Likelihood-ratio tests were used to test various hypotheses. The hypothesis of no linkage (i.e., additive genetic variance caused by the QTL being 0) is tested by comparing the likelihood of this restricted model with that of a model in which the additive genetic variance due to the QTL is estimated. Twice the difference in ln likelihoods of these two models yields a test statistic (Λ) that is asymptotically distributed as a 1/2:1/2 mixture of a χ21 and a point mass at zero (Self and Liang 1987). LOD scores are obtained by converting the ln likelihood values into values of log to the base 10. After obtaining locus-specific IBD information for pairs of relatives using the program SOLAR (Almasy and Blangero 1998), the multipoint mapping technique, as implemented in the program SOLAR, was used to carry out multipoint linkage analyses. To verify the findings from the multipoint linkage analyses, we conducted simulation analyses to determine the trait-specific empirical P values using the following procedure. A fully informative marker was simulated, which was not linked to a given phenotype. For this marker, IBD information was calculated, and then linkage analysis was performed. For a given phenotype, we generated the empirical P values on the basis of LOD-score distribution obtained from the 10,000 replicates.
Bivariate multipoint linkage analysis
In contrast to the above phenotype-specific multipoint linkage analysis, a bivariate (i.e., two-phenotype) multipoint linkage was used to exploit the additional information embedded in the correlation pattern between two quantitative traits. Given the theoretical background for exploitation of the genetic basis of multivariate quantitative phenotypes (Lange and Boehnke 1983; Boehnke et al. 1986), the variance-components approach has been extended to the bivariate situation to test whether covariances between a given set of phenotypes are due to pleiotropic effects of a major gene in the chromosomal region of interest (Almasy et al. 1997; Williams et al. 1999a, 1999b).
After the initial findings of correlated traits that map to the same chromosomal location, the bivariate linkage technique was used to estimate the correlation caused by a major gene (ρq), the correlation caused by residual additive genetic effects (ρg), and the correlation caused by random environmental effects (ρe). In each of the bivariate analyses, in addition to these three correlation estimates, two trait-specific means, variance components relating to major-gene effects, residual additive genetic effects, random environmental effects, and covariate effects (i.e., sex and age terms) were estimated simultaneously using maximum-likelihood techniques. The hypothesis tests were performed by likelihood-ratio tests. The hypothesis of no linkage for either trait (i.e., additive genetic variance caused by the QTL is 0 for both traits) was tested by comparison of the likelihood of this restricted model with that of a model in which the additive genetic variance caused by the QTL for both traits was estimated. The bivariate LOD score obtained with this method involves 2 df. Hence, it is not directly comparable to the LOD score obtained from the univariate linkage analysis. Twice the difference in ln likelihoods of these two models yields a Λ that is asymptotically distributed as a 1/4:1/2:1/4 mixture of χ22, χ21, and χ20 (Self and Liang 1987). The bivariate LOD score with 2 df can be adjusted to 1 df, denoted as LODeq score, by requiring it to provide the same P value as is provided by the true bivariate LOD score (Almasy et al. 1997; Williams et al. 1999a). Thus, the LODeq score can be considered equivalent to the classical univariate LOD score.
Likelihood-ratio tests were used to test the hypotheses of complete pleiotropy (i.e., the same major gene in the chromosomal region of interest affects both traits) and coincident linkage (i.e., no shared major gene effects in the chromosomal region of interest on the two traits). The test of complete pleiotropy is carried out by comparing the likelihood of a restricted model in which ρq is constrained to 1 (or −1) to that of a model in which ρq is estimated. It should be noted that a positive value of 1 for ρq is chosen for trait pairs involving FSI and other phenotypes because all of the overall polygenic correlations between these trait pairs were found to be positive. In the case of trait pairs relating to ln HOMA %S (i.e., an inverse measure of insulin resistance) and other traits, however, a negative value of 1 for ρq is chosen since all of the overall polygenic correlations between the examined trait pairs involving ln HOMA %S were found to be negative. Since the hypothesis of ρq equals 1 (or −1) involves a boundary condition, twice the difference in likelihoods of these two models yields a Λ that is asymptotically distributed as a 1/2:1/2 mixture of a χ21 and a point mass at 0 (Self and Liang 1987). In the case of test of coincident linkage (i.e., ρq=0), twice the difference in likelihoods of the two competing models yields a Λ that is asymptotically distributed as a χ2 with 1 df. The bivariate linkage analyses were carried out using a modified version of the program SOLAR. For a given phenotype pair, we used the simulation strategy already described to generate the empirical P values based on the bivariate LOD score distribution obtained from the 10,000 replicates.
Results
The characteristics of the nondiabetic subjects used for the present study are reported in table 1. All genetic analyses included sex and age terms as covariates. Prior to conducting the linkage analyses, we determined heritabilities for all the phenotypes used in this study. The overall additive genetic heritabilities ranged from 26% (ln TG) to 65% (BMI). Following the findings that all of these phenotypes were significantly heritable, we examined the nature of genetic correlations between the trait pairs analyzed in this study. The additive genetic correlations between FSI and other phenotypes were positive, as expected, and ranged from .47 (FSI–ln TG) to .80 (FSI–SS and FSI–LEPT). Since HOMA %S is an inverse measure of insulin resistance, the additive genetic correlations between ln HOMA %S and others traits were negative, ranging from −.46 (ln HOMA %S–ln TG) to −.81 (ln HOMA %S–LEPT). These correlations suggest strong common genetic influences on the examined trait pairs, especially the ones involving FSI or ln HOMA %S and obesity measures.
Table 1.
Characteristics of Nondiabetic SAFADS Subjects Distributed by Sex across 27 Families
| Variable and Sexa | N | Mean ± SD or % |
| Sex: | ||
| M | 137 | 42% |
| F | 187 | 58% |
| Age (years): | ||
| M | 137 | 39.5 ± 16.9 |
| F | 187 | 37.5 ± 14.2 |
| FG (mg/dl): | ||
| M | 136 | 91.0 ± 11.7 |
| F | 187 | 87.1 ± 10.3 |
| Fasting specific insulin (pmol/L): | ||
| M | 124 | 120.4 ± 85.1 |
| F | 169 | 133.4 ± 87.0 |
| ln HOMA %S: | ||
| M | 121 | 3.8 ± 0.6 |
| F | 169 | 3.8 ± 0.7 |
| ln HOMA %β: | ||
| M | 124 | 5.0 ± 0.5 |
| F | 169 | 5.2 ± 0.4 |
| ln Fasting nonspecific insulin: | ||
| M | 130 | 2.4 ± 0.7 |
| F | 180 | 2.4 ± 0.7 |
| ln Triglycerides: | ||
| M | 132 | 5.1 ± 0.6 |
| F | 179 | 4.8 ± 0.5 |
| Leptin (ng/ml): | ||
| M | 121 | 10.7 ± 7.8 |
| F | 171 | 30.3 ± 18.1 |
| BMI (kg/m2): | ||
| M | 134 | 29.4 ± 5.8 |
| F | 183 | 29.4 ± 7.1 |
| Sum of skin folds (mm): | ||
| M | 135 | 145.3 ± 47.8 |
| F | 184 | 195.1 ± 52.6 |
M = male; F = female.
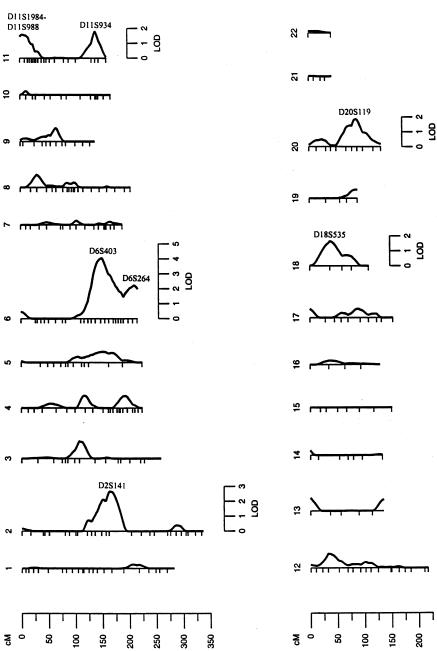
The results of our multipoint genome scan for FSI susceptibility loci are reported in figure 1. The highest LOD score that we observed was 4.1 (empirical P value <.0001), corresponding to a major susceptibility locus for FSI on chromosome 6q at 150 cM from p-ter. This genetic location is 2 cM telomeric to the marker D6S403. The multipoint LOD score profile obtained for FSI is shown in figure 2. As can be seen from this figure, the chromosomal region containing the putative locus for FSI levels is rather broad, and the 1-LOD support interval surrounding the peak extends from 135 to 161 cM from p-ter. After accounting for the covariate effects, this major susceptibility locus (h2q) explains 44%±8% (P=7.5×10-6) of the phenotypic variation in FSI levels. In addition to the major genetic location near marker D6S403, six other regions on five separate chromosomes were found to exhibit some evidence for linkage, with LOD scores >1.5 (fig. 1). Specifically two genetic regions, one on chromosome 2q near marker D2S141 (LOD score 2.7; empirical P value .0006) and another on chromosome 6q near marker D6S264 (LOD score 2.2; empirical P value .0018), are suggestively linked to FSI concentrations. For the remaining chromosomal regions, the LOD scores ranged from 1.6 (empirical P value .0072) to 1.9 (empirical P value .0038) as follows: a genetic location between markers D11S1984-D11S988 (LOD score 1.6) and another near marker D11S934 (LOD score 1.8) on chromosome 11, a genetic location near marker D18S535 (LOD score 1.7) on chromosome 18, and a genetic location near marker D20S119 (LOD score 1.9) on chromosome 20.
Figure 1.
Summary of the multipoint linkage analyses of FSI: peak multipoint LOD score by chromosome.
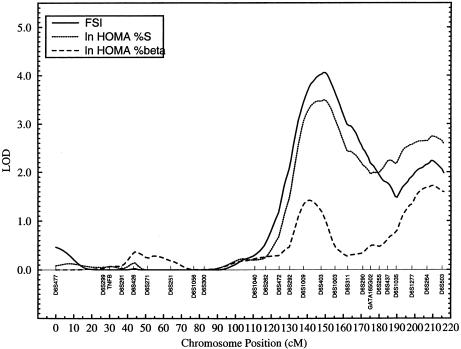
Figure 2.
Plot of LOD scores obtained for measures of insulin (FSI), HOMA %S, and HOMA %β, against map positions on chromosome 6.
The major chromosomal region of interest for FSI, the genetic location near D6S403, showed varying degrees of evidence for linkage to several other IRS-related phenotypes including nonspecific insulin (ln FNI) and other IRS-related traits (BMI, SS, LEPT, and ln TG) (table 2). Similar patterns of linkage profile for these phenotypes could also be seen at the genetic region near marker D6S264. Since hyperinsulinemia could result from abnormalities of either insulin action or secretion, we derived measures of insulin sensitivity (HOMA %S) and β-cell function (HOMA %β) to evaluate the extent of their linkage to the two regions on chromosome 6q. As shown in figure 2, both regions of interest on chromosome 6q exhibit strong evidence for linkage of ln HOMA %S with LOD scores of 3.5 (empirical P value <.0001) and 2.7 (empirical P value .0002), respectively (table 2). By contrast, ln HOMA %β, reflecting insulin secretion, appears to be only weakly linked to these regions (table 2 and fig. 2). On the basis of the trait-specific linkage profiles, the two peaks on chromosome 6q, which are >50 cM apart, appear to represent two distinct genetic locations influencing FSI and other IRS-related phenotypes (fig. 2 and table 2).
Table 2.
Linkage of Various IRS-Related Phenotypes to the Regions of Interest on Chromosome 6q with Variable Degrees of Evidence
|
Region Covered by Markers D6S1009, D6S403, and D6S1003 |
RegionContainingMarkerD6S264 |
|||
| Phenotype | cMa | LOD | cMa | LOD |
| FSI | 150 | 4.1 | 209 | 2.2 |
| LEPT | 139 | 2.2 | 210 | 2.8 |
| BMI | 138 | 1.5 | 207 | .7 |
| SS | 144 | 1.6 | 210 | 1.6 |
| ln TG | 156 | .8 | 207 | 1.3 |
| ln FNI | 142 | 1.9 | 206 | 1.1 |
| ln HOMA %S | 149 | 3.5 | 210 | 2.7 |
| ln HOMA %β | 141 | 1.4 | 210 | 1.7 |
Distance from the p-ter.
Following the above findings of linkage of multiple correlated phenotypes to the same chromosomal region involving the ordered markers D6S1009-D6S403-D6S1003, we extended our analytical approach to the bivariate situation to exploit the additional information underlying the patterns of covariation between pairs of quantitative phenotypes. Table 3 presents the results from multipoint bivariate linkage analyses of various pairs of IRS-related traits. As shown in this table for the region near marker D6S403, for the trait pairs involving FSI and other traits, the bivariate LOD scores with 2 df range from 3.9 (FSI-BMI) to 5.4 (FSI-LEPT). The Λ obtained for each of these bivariate linkage tests is distributed as a mixture of χ2 distributions, as shown in table 3. Thus, the P values range from 4.8×10-5 (FSI-BMI) to 1.5×10-6 (FSI-LEPT). Likewise, the LODeq score, which is the equivalent 1-df LOD score corresponding to the stated bivariate LOD score with 2 df, ranges from 3.3 (FSI-BMI) to 4.7 (FSI-LEPT). All of the LODeq scores for the marker D6S403 region are >3.0, adding further significance to this putative locus in affecting IRS-related phenotypes. The bivariate linkage profiles of the measure of insulin resistance (ln HOMA %S) and other IRS-related phenotypes are similar to the findings from the bivariate analyses of FSI and other IRS traits (figs. 3 and 4). Evidence for bivariate linkage is stronger for FSI and other traits, compared with the ln HOMA %S and other traits (table 3). For example, the LODeq scores ranged from 2.8 (ln HOMA %S-BMI) to 4.1 (ln HOMA %S-LEPT). For the trait pairs relating to FSI and other traits, the empirical P values range from .0006 (FSI-BMI) to <.0001 (FSI-ln TG). The empirical P values range from .0004 (ln HOMA %S-ln TG) to .0002 (ln HOMA %S-BMI) for trait pairs involving ln HOMA %S and other phenotypes.
Table 3.
Multipoint Bivariate Linkage Analyses of Selected Pairs of IRS-Related Phenotypes versus the Marker Regions of Interest on Chromosome 6q
| Region and Phenotype Pair | cMa | BivariateLOD | Λb | Pc | LOD eqd |
| Marker D6S403 region: | |||||
| FSI-BMI | 150 | 3.9 | 17.74 | 4.8 × 10−5 | 3.3 |
| FSI-SS | 150 | 4.7 | 21.55 | 6.8 × 10−6 | 4.1 |
| FSI-LEPT | 149 | 5.4 | 24.74 | 1.5 × 10−6 | 4.7 |
| FSI-ln TG | 150 | 4.3 | 19.64 | 1.8 × 10−5 | 3.7 |
| ln HOMA %S-BMI | 150 | 3.3 | 15.17 | 1.8 × 10−4 | 2.8 |
| ln HOMA %S-SS | 150 | 4.2 | 19.13 | 2.4 × 10−5 | 3.6 |
| ln HOMA %S-LEPT | 149 | 4.7 | 21.74 | 6.3 × 10−6 | 4.1 |
| ln HOMA %S-ln TG | 151 | 3.5 | 16.00 | 1.2 × 10−4 | 2.9 |
| Marker D6S264 region: | |||||
| FSI-BMI | 209 | 2.4 | 11.12 | 1.4 × 10−3 | 1.9 |
| FSI-SS | 209 | 3.4 | 15.77 | 1.3 × 10−4 | 2.9 |
| FSI-LEPT | 208 | 4.6 | 21.33 | 7.8 × 10−6 | 4.1 |
| FSI-ln TG | 207 | 3.3 | 15.11 | 1.8 × 10−4 | 2.8 |
| ln HOMA %S-BMI | 209 | 2.6 | 12.08 | 8.5 × 10−4 | 2.1 |
| ln HOMA %S-SS | 209 | 3.7 | 16.89 | 7.4 × 10−5 | 3.1 |
| ln HOMA %S-LEPT | 208 | 4.8 | 22.02 | 5.8 × 10−6 | 4.2 |
| ln HOMA %S-ln TG | 209 | 3.4 | 15.61 | 1.4 × 10−4 | 2.9 |
Distance from p-ter
Likelihood-ratio statistic Λ.
Asymptotic P value, under the assumption that Λ is distributed as a ¼χ22:½χ21:¼χ20 mixture.
LODeq is the equivalent 1-df LOD score corresponding to the reported bivariate LOD score with 2 df.
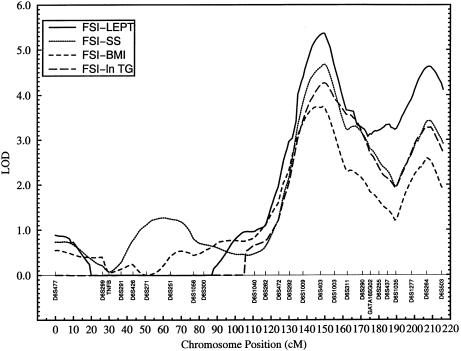
Figure 3.
Bivariate linkage profiles of various trait pairs, including FSI and other IRS-related phenotypes on chromosome 6
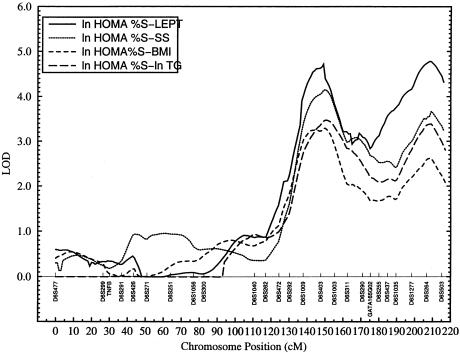
Figure 4.
Bivariate linkage profiles of various trait pairs, including HOMA %S and other IRS-related phenotypes on chromosome 6
Similar patterns of bivariate linkage involving the same trait pairs could also be seen at the D6S264 marker region. For this genetic region, the bivariate LOD scores range from 2.4 (FSI-BMI) to 4.6 (FSI-LEPT), and the corresponding P values range from 1.4×10-3 (FSI-BMI) to 7.8×10-6 (FSI-LEPT) (table 3). One of the four corresponding LODeq scores, involving the trait pair FSI-LEPT, is well above 3.0. Again, the bivariate linkage profiles of ln HOMA %S versus other IRS-related phenotypes are similar to those obtained from the trait-pair analyses involving FSI and other traits (figs. 3 and 4). However, at the D6S264 marker region, evidence for bivariate linkage is slightly stronger for ln HOMA %S and other traits compared to the FSI and other traits (table 3). The empirical P values range from .0039 (FSI-BMI) to .0002 (FSI-ln TG) for trait pairs involving FSI and other phenotypes. For the trait pairs relating to ln HOMA %S and other traits, the empirical P values range from .0005 (ln HOMA %S-BMI) to .0003 (ln HOMA %S-LEPT). Thus, both univariate and bivariate linkage analyses implicate the genetic locations near markers D6S403 and D6S264 as significant regions of colocalization of the correlated IRS phenotypes. As can be seen from figures 3 and 4, the bivariate analyses yielded peaks much sharper than those obtained from the univariate analyses (fig. 2).
Our bivariate linkage technique quantifies the extent to which the chromosomal region containing the major gene affects the correlation between a given pair of traits. As shown in table 4, the correlations between trait pairs FSI and other traits caused by the major-gene effects at the D6S403 region range from .8, for trait pair FSI-ln TG, to 1, for other trait pairs. In the case of ln HOMA %S and other trait pairs, the correlations range from −.6, for trait pair FSI-ln TG, to −1, for other trait pairs (table 4). None of the trait-pair models of complete pleiotropy (i.e., locus-specific correlation between a given pair of traits not significantly different from 1 or −1) could be rejected (table 4), suggesting significant evidence for pleiotropic effects of the genetic location near marker D6S403 on all of the IRS-related trait pairs examined in this study. By contrast, all of the tests of coincident linkage were strongly rejected. For the region near marker D6S264, complete pleiotropy was rejected only for the trait pairs involving ln TG. For the other trait pairs, coincident linkage was strongly rejected in favor of complete pleiotropy (table 4).
Table 4.
Evidence for Pleiotropic Influences of Specific Genetic Regions of Chromosome 6q on IRS-Related Phenotypes
|
Complete Pleiotropy |
Coincident Linkage |
||||
| Phenotype Pair | ρqa | Λb | pc | Λb | pd |
| Marker D6S403 region: | |||||
| FSI-BMI | 1 | .00 | .5000 | 13.74 | .0002 |
| FSI-SS | 1 | .00 | .4955 | 19.65 | <.0001 |
| FSI- LEPT | 1 | .00 | .4990 | 17.22 | <.0001 |
| FSI-ln TG | .8 | .23 | .3151 | 6.39 | .0115 |
| ln HOMA %S-BMI | −1 | .00 | .4877 | 13.25 | .0003 |
| ln HOMA %S-SS | −1 | .00 | .4972 | 18.94 | <.0001 |
| ln HOMA %S-LEPT | −1 | .00 | .4971 | 16.47 | <.0001 |
| ln HOMA %S-ln TG | −.6 | .84 | .1790 | 4.15 | .0416 |
| Marker D6S264 region: | |||||
| FSI-BMI | .9 | .93 | .1680 | 7.60 | .0059 |
| FSI-SS | 1 | .00 | .4901 | 15.25 | <.0001 |
| FSI-LEPT | .9 | .09 | .3844 | 16.77 | <.0001 |
| FSI-ln TG | .5 | 2.77 | .0479 | 3.44 | .0637 |
| ln HOMA %S-BMI | −.9 | .76 | .1917 | 7.91 | .0049 |
| ln HOMA %S-SS | −1 | .00 | .4942 | 15.26 | <.0001 |
| ln HOMA %S-LEPT | −.9 | .61 | .2172 | 16.73 | <.0001 |
| ln HOMA %S-ln TG | −.4 | 2.88 | .0450 | 3.44 | .0636 |
ρq is correlation due to QTL effects.
Likelihood-ratio statistic Λ.
Likelihood for a model in which ρq is estimated, compared with that of another model in which ρq is constrained to 1 (or −1) (i.e., complete pleiotropy). Λ is distributed as a ½χ21:½χ20 mixture.
Likelihood of a model in which ρq is estimated, compared with another model in which ρq is constrained to 0 (i.e., coincident linkage). Λ is distributed as a χ2 with df 1.
Discussion
Hyperinsulinemia and/or insulin resistance is a strong correlate of complex disease conditions, such as obesity and type 2 diabetes, and may play a role in the pathogenesis of CHD as well (DeFronzo 1995; Haffner 1999; Lempiainen et al. 1999). In relation to direct insulin-resistance measures, such as the euglycemic clamp and the intravenous glucose-tolerance test, fasting insulin concentration, a simple noninvasive measure, has been considered a surrogate measure of insulin resistance in nondiabetic subjects (Haffner et al. 1997). It is well established that insulin levels are under substantial genetic influences (Stern and Mitchell 1999) and that insulin levels and other IRS-related phenotypes (e.g., obesity) are influenced by a common set of genes (i.e., pleiotropy) (Mitchell et al. 1996). However, knowledge about specific common genetic determinants of insulin levels or insulin resistance and their correlates, such as obesity and dyslipidemia, is extremely limited.
In this study, using a genomewide scan, we found significant evidence (LOD score 4.1) for a major locus influencing FSI concentrations on chromosome 6q near marker D6S403. The evidence for linkage of FSI to this region is significant at the level of a genomewide scan according to the criteria proposed by Lander and Kruglyak (1995). Furthermore, this genetic region is strongly linked with HOMA %S, suggesting that the genetic location near marker D6S403 is a major determinant of variation in insulin resistance. Our simulation analyses add further strength to these findings. By contrast, this region appears to be weakly linked to HOMA %β, suggesting that the underlying susceptibility gene accounting for the linkage primarily influences insulin resistance, rather than insulin secretion. We acknowledge, however, that since we lack direct measures of insulin resistance and insulin secretion, these interpretations should be considered tentative. The failure of FNI levels to provide evidence for linkage as strong as the FSI concentrations in our study could be due to the fact that, unlike FNI, FSI denotes “true” insulin concentration by not crossreacting with insulin precursors. Given that a high proinsulin:insulin ratio can be considered indicative of a defect in proinsulin processing or insulin secretion, our failure to find evidence for linkage of this ratio to the region near marker D6S403 further strengthens the argument that the linkage near this marker is primarily related to insulin resistance.
We subsequently showed, using a bivariate linkage technique, that the same major gene in the chromosomal region of interest also had significant shared effects on other IRS-related phenotypes (e.g., obesity and triglycerides). The bivariate linkage approach has been shown to substantially improve power to detect susceptibility loci that have pleiotropic effects on correlated traits and can help to localize such genes more precisely (Amos and Liang 1993; Almasy et al. 1997; Allison et al. 1998; Williams et al. 1999a). All of the bivariate analyses implicate the region near marker D6S403 (6q22-q23) as harboring the putative susceptibility locus, which appears to simultaneously affect measures of insulin concentration and insulin resistance, as well as other IRS-related traits, including various measures of obesity/adiposity and triglyceride levels. Since the traits examined in this study are genetically correlated, no attempt has been made to correct for multiple testing.
To date, a number of studies have conducted linkage analyses to find chromosomal regions harboring type 2 diabetes and metabolic syndrome related susceptibility loci across the genome. Elsewhere, we reported, on the basis of sibship analyses, some evidence for linkage of glucose concentrations to different locations on chromosome 6, including the regions near markers D6S292 and D6S290 (Stern et al. 1996). Given that the marker D6S292 is on the same cytogenetic band (i.e., 6q22-q23) as marker D6S403 (fig. 2), Ghosh et al. (2000) found strong evidence for a locus near marker D6S292 influencing the lowest FG concentrations in a subset of Finnish families. In another analysis, involving nondiabetic individuals in the Finnish families, Watanabe et al. (2000) reported several different locations near the D6S292 marker region that were linked to phenotypes such as fasting insulin, 2-h insulin, and measures of both insulin resistance and insulin secretion.
In a genomewide search for genes influencing systolic blood pressure, strong evidence for linkage was found between a location near markers D6S1009 and D6S1003 and systolic blood pressure (Krushkal et al. 1999). As can be seen from figure 2, these markers flank D6S403. In Mexican Americans, some evidence for linkage of cholesterol concentrations in the LDL-3 size fraction to a location near marker D6S1003 has been reported (Rainwater et al. 1999). As part of their genomewide scan for loci linked to obesity measures in a collection of French families, Hager et al. (1998) reported some evidence for linkage of leptin concentrations to a location, telomeric to marker D6S292, that corresponds well to our region of interest. This region, however, in another study involving the Mexican population (Comuzzie et al. 1997), was not implicated as influencing leptin concentrations. Although we failed to find evidence for linkage of diabetes to the region near marker D6S403 in our previous study (Duggirala et al. 1999b), it is worth noting that this region (i.e., markers D6S1009 and D6S1003) has been reported to be linked to age-adjusted diabetes in Pima Indians (Hanson et al. 1998a). Also, the marker region D6S1009 has been reported to be linked to diabetes in Japanese subjects (Iwasaki et al. 1999). Ghosh et al. (2000) reported evidence for a diabetes-susceptibility gene, influencing age at diagnosis and located on chromosome 6q, that is centromeric to the region of interest in this study.
Since a chromosomal region implicated by a linkage analysis may contain a large number of known or unknown genes, an initial step for genetic epidemiological investigations would be to identify and examine potential positional candidate genes in the chromosomal regions of interest. Such a positional candidate gene for insulin resistance in our chromosomal region of interest (i.e., D6S403 region) is the plasma cell membrane glycoprotein PC-1 gene (Maddux et al. 1995), which has been localized to chromosome 6q22-q23 (Buckley et al. 1990). The PC-1 gene, when overexpressed, has been shown to be an inhibitor of insulin receptor tyrosine kinase activity, and its levels are elevated in muscle, fat, and fibroblasts of subjects with insulin resistance (Goldfine et al. 1998; Pizzuti et al. 1999). There is evidence that the PC-1 could inhibit insulin receptor signaling by interacting with the insulin receptor α-subunit (Maddux and Goldfine 2000). Recently, a polymorphism (K121Q) in exon 4 of the PC-1 gene has been shown to be strongly associated with insulin resistance, using data from a sample of white subjects from Sicily (Pizzuti et al. 1999). We acknowledge, however, that as yet we have no direct evidence that functional variants in the PC-1 gene account for our linkage signal. Another potential candidate gene in our chromosomal region of interest relates to transient neonatal diabetes mellitus (TNDM), which is a rare, developmental disorder of insulin production that regresses in postnatal life (Cavé et al. 2000). Although TNDM patients generally recover before 1 year of age, some patients may tend to develop type 2 diabetes later in life (Gardner et al. 1999; Temple et al. 2000). The gene for TNDM has been originally localized to the chromosomal band 6q22-q23 (Temple et al. 1996). Recently, however, it has been proposed that the chromosomal bands 6q24.1-q24.3 contain an imprinted gene for TNDM (Gardner et al. 1999; Cavé et al. 2000).
One of the chromosomal regions that appears to be suggestively linked to FSI (LOD score 2.2) is also on chromosome 6q, but near marker D6S264 (6q25.2-q26). However, our bivariate linkage analyses identified the same region as having significant common effects on various IRS-related phenotypes. On the basis of the map used for this study, this genetic region is ∼58 cM telomeric to D6S403. It is possible that the observed two peaks on chromosome 6q may be related to the same chromosomal region. However, the LOD profile patterns revealed by both univariate and bivariate linkage analyses are more suggestive of two distinct loci on chromosome 6q. In Mexican Americans, evidence was found for linkage of cholesterol concentrations in LDL-3 size fraction to a genetic location very close to the marker D6S1277 (Rainwater et al. 1999), which is ∼8 cM centromeric to marker D6S264 on our map. Using the same Mexican American family data, Mitchell et al. (1999) reported some evidence for linkage of BMI to a location near marker D6S1008, which is ∼2 cM centromeric to marker D6S1277. Recently, syndromal obesity has been shown to be due to paternal duplication of chromosome 6q involving the chromosomal bands 6q24-q27 (Smith et al. 1999). It is worthwhile to note that the human homolog of Obq4, a mouse obesity QTL, could map to the chromosome 6q25-q27 region (Taylor and Philips 1997). A region near marker D6S1035, which is not far from D6S264, has been shown to provide some evidence for linkage with diabetes in Pima Indians (Hanson et al. 1998b) and with the FG concentrations in Mexican Americans (Stern et al. 1996). The marker D6S264 region has also been shown to contain a susceptibility locus for IDDM8 (Luo et al. 1995). This chromosomal region also harbors several positional candidate genes for IRS or obesity, such as the insulin-like growth factor 2 receptor (IGF2R) in the cytogenetic location 6q26 (MIM 147280), and the acetyl-CoA acetyltransferase 2 (ACAT2) in the cytogenetic location 6q25.3-q26 (MIM 100678). It is also worth mentioning that the mu 1 opioid receptor gene (OPRM1; MIM 600018), a candidate gene for obesity, lies in the cytogenetic position 6q24-q25, between the peaks of interest in this study.
The second genetic location exhibiting suggestive linkage to FSI (LOD score 2.7) is on chromosome 2 near marker D2S141 (2q23-24). It has been shown that the genetic marker D2S141 is physically linked to the pancreatic β-cell mitochondrial glycerol-3-phosphate dehydrogenase (GPD2; Ferrer et al. 1996), which is located in the chromosomal band 2q24.1. Mitochondrial glycerol-3-phosphate dehydrogenase plays a major role in glucose-stimulated insulin secretion (Ferrer et al. 1996). Another gene closely linked to D2S141 marker is the inwardly rectifying potassium channel GIRK1 (Kir3.1, KCNJ3) [2q24.1], which is implicated in the regulatory pathways of insulin secretion (Vionnet et al. 1997). These genes have been considered not to be common causes of type 2 diabetes in some populations (MacDonald et al. 1997; Vionnet et al. 1997). However, recently, some evidence has been reported in a French population (Vionnet et al. 2000) for linkage of a diabetes-related phenotype to the D2S2330 marker region, which is close and telomeric to the marker D2S141 that showed suggestive linkage to FSI in our study.
As noted earlier, there are several other chromosomal regions across the genome that exhibited some evidence for linkage (i.e., LOD scores >1.5) to FSI (fig. 1). The location (between markers D11S1984 and D11S988) linked to FSI is ∼10 cM telomeric to the β-hemoglobin (HBB) locus on our map, which we previously reported to be linked with both fasting and 2-h glucose concentrations (Stern et al. 1996). Vionnet et al. (1997) reported suggestive evidence for linkage of a severe form of diabetes to a genetic location which overlaps with this region on chromosome 11p (see also Vionnet et al. 2000). In fact, as we reported elsewhere (Duggirala et al. 1999b), some evidence for linkage of both diabetes and age of diabetes onset was found at a location between markers D11S988 and HBB. Our second region of interest on chromosome 11 is near the marker D11S934, which is located between the markers D11S4464 and D11S912. This region strongly overlaps the genetic location (between markers D11S4464 and D11S912) reported to be significantly linked with BMI in Pima Indians (Hanson et al. 1998a). In fact, this genetic location showed some evidence for linkage to diabetes, but it provided strong evidence for bivariate linkage of BMI and diabetes in the Pima population. Also, this region was reported to be linked to diabetes in a white population (Elbein et al. 1999).
Recently, the region near marker D18S535, where we found some evidence for linkage of FSI, has been shown to be significantly linked to fat-free body mass in the Quebec Family Study (Chagnon et al. 2000). Also, this region has been reported to provide some evidence for linkage to diabetes or its related phenotypes (Ehm et al. 2000; Vionnet et al. 2000). The region linked to FSI on chromosome 20q is near the marker D20S119. A number of linkage studies have implicated this region as influencing type 2 diabetes (Zouali et al. 1997; Ghosh et al. 1999, 2000; Klupa et al. 2000; Vionnet et al. 2000) or its related traits, such as insulin secretion (Ghosh et al. 1999), obesity (Borecki et al. 1994; Lee et al. 1999), and energy metabolism (Norman et al. 1998). Aside from the regions discussed thus far, Pratley et al. (1998) found modest evidence for linkage of fasting plasma insulin concentration and insulin action at physiologic insulin concentrations to the D3S1764-marker region on chromosome 3 in the Pima population. We failed to find evidence for linkage of FSI to this region but found that FSI was linked (LOD 1.2) to the marker D3S2460 region, which is ∼25 cM centromeric to D3S1764. Recently, in another study of Mexican American families, Mitchell et al. (2000) reported a region on chromosome 3 near markers D3S1600-D3S1285 to be strongly linked to fasting insulin concentration. We failed to find any evidence for linkage of FSI to this region. However, the observed LOD curve in the study by Mitchell et al. was broad, and it covered the region around marker D3S2460 (LOD >1.0), where we found some evidence for linkage of FSI.
Our study, in addition to the identification of a major locus near marker D6S403 with significant influence on measures of insulin and HOMA %S, provides strong evidence for pleiotropic effects of the same location on several IRS-related phenotypes. Also, there appears to be another susceptibility locus near marker D6S264, which exerts appreciable influences on various trait pairs related to the IRS. Of the phenotypes examined in this study, the trait pairs leptin-specific insulin and leptin-HOMA %S appear to portray the shared major-gene influences most effectively. Although the relationship between leptin levels and diabetes is uncertain (Haffner et al. 1996; Van Gaal et al. 1999), several studies have reported associations between fasting insulin and leptin levels (e.g., Zimmet et al. 1996; Ruige et al. 1999). In fact, a strong phenotypic correlation between FSI and fasting leptin concentrations (r=0.55, P<.001) has been observed in a population-based study involving nondiabetic Mexican Americans (Haffner et al. 1998). There has been considerable recent interest in the role of insulin in leptin regulation and in the influence of leptin on both insulin resistance and insulin secretion (Kolaczynski et al. 1996; Lönnqvist et al. 1999; Ruige et al. 1999; Keiffer and Habener 2000). It has been shown that insulin resistance, as measured directly by euglycemic-hyperinsulinemic clamp, is significantly correlated to leptin in a sample of nondiabetic men and women (Donahue et al. 1999). According to these authors, the rate of insulin-mediated glucose disposal is significantly inversely associated with leptin concentrations in both men (r=-.83; P<.001) and women (r=-0.59; P<.001).
Although some genes across the genome may be identified as having common influences on correlated phenotypes, such traits can also be influenced by trait-specific loci (Mahaney et al. 1995; Duggirala et al. 2000). Aside from the findings of loci affecting specific traits, several studies have identified genetic regions that influence correlated phenotypes (e.g., Duggirala et al. 1996; Lembertas et al. 1997; Hanson et al. 1998a; Ghosh et al. 1999; Kissebah et al. 2000). In both theory and practice, the genetic causes of correlation between phenotypes through the pleiotropic effects of genes are of particular interest (Falconer 1989). As stated by Falconer (1989, p. 313), “the genetic cause of correlation is chiefly pleiotropy,” which is “simply the property of a gene whereby it affects two or more characters, so that if the gene is segregating it causes simultaneous variation in the characters it affects.” In this study, we first found significant evidence for linkage of both insulin and an indirect measure of insulin resistance to a location near marker D6S403 on chromosome 6q that was subsequently identified to have strong pleiotropic influences on various IRS-related phenotypes. As part of our ongoing efforts to identify other genetic regions that have common influences on IRS-related traits, for example, we have mapped several IRS-related phenotypes (e.g., lipids and obesity measures) to a genetic region on chromosome 7q (Duggirala et al. 1999a).
The pedigree-based variance-component approach (the approach used in the present study) has been shown to be a powerful technique for linkage analysis (Almasy and Blangero 1998; Pratt et al. 2000). However, since this technique is based on the assumption of multivariate normal distribution, there has been continued interest in its sensitivity to violations of this assumption (Allison et al. 1999; Blangero et al. 2000). In general, this approach has been shown to be robust to such violations (Beaty et al. 1985; Amos 1994), although some types of nonnormality (e.g., leptokurtosis) could result in inflated type I–error rates (Allison et al. 1999; Blangero et al. 2000, 2001). Data simulations have shown that trait distributions with coefficient of kurtosis <2 appear not to yield grossly inflated type I error (Blangero et al. 2001). For the phenotypes used in this study, the coefficient of kurtosis ranged from −0.9 (ln HOMA %S) to 1.6 (LEPT). Nonetheless, as an added precaution, in addition to the reported empirical P values to support our findings, we used a robust statistical approach to verify our original univariate linkage findings (Blangero et al. 2000, 2001). For a given phenotype, we obtained the empirical distribution for the LOD scores by simulation as described earlier. A correction constant was obtained by regression of the expected LOD scores on the observed simulated LOD scores to adjust the trait-specific observed LOD scores. All of the phenotypes examined in this study appeared not to require an adjustment to the observed LOD score, with the exception of FSI, which needed a slight adjustment (yielding a robust corrected LOD of 3.6 versus the observed LOD of 4.1), thus indicating that the findings relating to FSI may have been slightly inflated. Given that deviations from multivariate normality are expected for phenotypes that are under the effects of major genes or oligogenes, it is reassuring to note that the findings from our analyses are consistent, despite subtle differences in the distributional properties of the various phenotypes used.
In conclusion, we found strong evidence for a major locus influencing insulin concentrations and insulin resistance in nondiabetic Mexican Americans, which has significant shared effects (i.e., pleiotropy) on various obesity-/IRS-related phenotypes as well. A positional candidate gene for insulin resistance in this region on chromosome 6q is the plasma cell membrane glycoprotein PC-1 gene. There appears to be a second susceptibility locus for insulin on chromosome 6q that also had significant pleiotropic effects on various obesity-/IRS-related phenotypes. Given the focus on the insulin-leptin relationship, our findings suggest that the loci on chromosome 6q have strong shared major-gene effects on variation in both insulin and obesity measures, especially leptin concentrations. Presently, we are in the process of characterizing the plasma cell membrane glycoprotein PC-1 gene in an effort to identify a functional variant.
Acknowledgments
This research was supported by grants from the National Institutes of Health (R01 DK42273, R01 DK47482, R01 DK53889, and MH59490). We are grateful to Professor C. N. Hales of the University of Cambridge, who kindly measured the specific insulin levels used for this study. R.A. was supported by the ADA Mentor-Based Postdoctoral Fellowship Program. We thank Drs. Anthony G. Comuzzie and Jeff T. Williams for helpful suggestions. We also thank Dr. Mary Pat Moyer and Ms. Florence Wall for establishing some of the lymphoblastoid cell lines. We wish to acknowledge Edgardo Benavides, Stefenie Fleming, Rajeswari Cheruvu, Michelle Zavala, and Bonnie Reus for excellent technical support. We warmly thank the families from SAFADS for their support and participation.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for IGF2R [MIM 147280], ACAT2 [MIM 100678], and OPRM1 [MIM 600018])
References
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999) Testing robustness of the likelihood-ratio test in a variance-component-trait loci-mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Thiel B, St. Jean P, Elston RC, Infante MC, Schork NJ (1998) Multiple phenotype modeling in gene-mapping studies of quantitative traits: power advantages. Am J Hum Genet 63:1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Dyer TD, Blangero J (1997) Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol 14:953–958 [DOI] [PubMed] [Google Scholar]
- Amos CI (1994) Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Liang AE (1993) A comparision of univariate and multivariate tests for genetic linkage. Genet Epidemiol 10:671–676 [DOI] [PubMed] [Google Scholar]
- Beaty TH, Self SG, Liang KY, Connolly MA, Chase GA, Kwiterovich PO (1985) Use of robust variance components models to analyze triglyceride data in families. Ann Hum Genet 49:315–328 [DOI] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L (2000) Robust LOD scores for variance component-based linkage analysis. Genet Epidemiol 19:S8–S14 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Variance component methods for detecting complex trait loci. Adv Genet 42:151–181 [DOI] [PubMed] [Google Scholar]
- Boehnke M, Moll PP, Lange K, Weidman WH, Kottke BA (1986) Univariate and bivariate analyses of cholesterol and triglyceride levels in pedigrees. Am J Med Genet 23:775–792 [DOI] [PubMed] [Google Scholar]
- Borecki IB, Rice T, Pérusse L, Bouchard C, Rao DC (1994) An exploratory investigation of genetic linkage with body composition and fatness phenotypes: The Québec Family Study. Obes Res 2:213–219 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley MF, Loveland KA, McKinstry WJ, Garson OM, Goding JW (1990) Plasma cell membrane glycoprotein PC-1: cDNA cloning of the human molecule, amino acid sequence, and chromosomal location. J Biol Chem 265:17506–17511 [PubMed] [Google Scholar]
- Burke JP, Williams K, Gaskill SP, Hazuda HP, Haffner SM, Stern MP (1999) Rapid rise in the incidence of type 2 diabetes from 1987 to 1996-results from the San Antonio Heart Study. Arch Intern Med 159:1450–1456 [DOI] [PubMed] [Google Scholar]
- Cavé H, Polak M, Drunat S, Denamur E, Czernichow P (2000) Refinement of the 6q chromosomal region implicated in transient neonatal diabetes. Diabetes 49:108–113 [DOI] [PubMed] [Google Scholar]
- Chagnon YC, Borecki IB, Pérusse L, Roy S, Lacaille M, Chagnon M, Ho-Kim MA, Rice T, Province MA, Rao DC, Bouchard C (2000) Genome-wide search for genes related to the fat-free body mass in the Qubéc family study. Metabolism 49:203–207 [DOI] [PubMed] [Google Scholar]
- Collins A, Frezal J, Teague J, Morton NE (1996) A metric map of humans: 23,500 loci in 850 bands. Proc Natl Acad Sci 93:14771–14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comuzzie AG, Hixson JE, Almasy L, Mitcell BD, Mahaney MC, Dyer TD, Stern MP, MacCluer JW, Blangero J (1997) A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nat Genet 15:273–276 [DOI] [PubMed] [Google Scholar]
- DeFronzo RA (1995) Insulin resistance and hyperinsulinemia: the link between NIDDM, CAD, hypertension and dyslipidemia. In: Schwartz CL, Born GVR (eds) New horizons in diabetes mellitus and cardiovascular disease. Current Science, London, pp 11–27 [Google Scholar]
- Donahue RP, Prineas RJ, DeCarlo Donahue R, Zimmet P, Bean J, De Courten M, Collier G, Goldberg RB, Skyler JS, Schneiderman N (1999) Is fasting leptin associated with insulin resistance syndrome among nondiabetic individuals? The Miami Community Health Study. Diabetes Care 22:1092–1096 [DOI] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Arya R, O’Connell P, Stern MP (1999a) Bivariate quantitative trait linkage of phenotypes related to the insulin resistance syndrome to a genetic region on chromosome 7 in Mexican Americans. Paper presented at the 2d Research Symposium on the Genetics of Diabetes, San Jose, October 17–19 [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O’Connell P, Stern MP (1999b) Linkage of type 2 diabetes mellitus and age of diabetes onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2000) A major susceptibility locus influencing plasma triglyceride concentrations is located on chromosome 15q in Mexican Americans. Am J Hum Genet 66:1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Stern MP, Mitchell BD, Reinhart LJ, Shipman PA, Uresandi OC, Chung WK, Leibel RL, Hales CN, O’Connell P, Blangero J (1996) Quantitative variation in obesity-related traits and insulin precursors linked to the OB gene region on human chromosome 7. Am J Hum Genet 59:694–703 [PMC free article] [PubMed] [Google Scholar]
- Dyke B (1996) PEDSYS: a pedigree data management system. PGL tech rep no. 2, Population Genetics Laboratory, Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio [Google Scholar]
- Ehm MG, Karnoub MC, Sakul H, Gottschalk K, Holt DC, Weber JL, Vaske D, Briley D, Briley L, Kopf J, McMillen P, Nguyen Q, Reisman M, Lai EH, Joslyn G, Shepherd NS, Bell C, Wagner MJ, Burns DK, American Diabetes Association GENNID Study Group (2000) Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet 66:1871–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC, Hoffman MD, Kui T, Leppert MF, Hasstedt SJ (1999) A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes 48:1175–1182 [DOI] [PubMed] [Google Scholar]
- Falconer DS (1989) Introduction to quantitative genetics. Longman Scientific and Technical, New York [Google Scholar]
- Fasman KH, Letovsky SI, Cottingham RW, Kingsbury DT (1996) Improvements to the GBDTM Human Genome Data Base. Nucleic Acids Res 24:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer J, Aoki M, Behn P, Nestorowicz A, Riggs A, Permutt MA (1996) Mitochondrial glycerol-3-phosphate dehydrogenase: cloning of an alternatively spliced human islet-cell cDNA, tissue distribution, physical mapping, and identification of a polymorphic genetic marker. Diabetes 45:262–266 [DOI] [PubMed] [Google Scholar]
- Flier JS (1992) Syndromes of insulin resistance: from patient to gene and back again. Diabetes 41:1207–1219 [DOI] [PubMed] [Google Scholar]
- Gardner RJ, Mungall AJ, Dunham I, Barber JCK, Shield JPH, Temple IK, Robinson DO (1999) Localization of a gene for transient neonatal diabetes mellitus to an 18.72 cR3000 (∼5.4 Mb) interval on chromosome 6q. J Med Genet 36:192–196 [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Hauser ER, Valle T, Magnuson VL, Erdos MR, Langefeld CD, et al (1999) Type 2 diabetes: Evidence for linkage on chromosome 20 in 716 Finnish affected sib pairs. Proc Natl Acad Sci USA 96:2198–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, et al (2000) The Finland-United States Investigation of Non-Insulin-Dependent Diabetes Mellitus Genetics (FUSION) Study: I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67:1174–1185 [PMC free article] [PubMed] [Google Scholar]
- Goldfine ID, Maddux BA, Youngren JF, Frittitta L, Trischitta V, Dohm GL (1998) Membrane glycoprotein PC-1 and insulin resistance. Mol Cell Biochem 182:177–184 [PubMed] [Google Scholar]
- Green P, Falls K, Crooks S (1990) Documentation for CRI-MAP, version 2.4. Department of Genetics, School of Medicine, Washington University, St. Louis [Google Scholar]
- Groop LC, Saloranta C, Shank M, Bonadonna RC, Ferrannini E, DeFronzo RA (1991) The role of free fatty acid metabolism in the pathogenesis of insulin resistance in obesity and non-insulin-dependent diabetes mellitus. J Clin Endocrinol Metab 72:96–107 [DOI] [PubMed] [Google Scholar]
- Haffner SM (1999) Epidemiology of insulin resistance and its relation to coronary artery disease. Am J Cardiol 84:11J–14J [DOI] [PubMed] [Google Scholar]
- Haffner SM, Gonzalez C, Mykkänen L, Stern M (1997) Total immunoreactive proinsulin, immunoreactive insulin and specific insulin in relation to conversion to NIDDM: the Mexico City Diabetes Study. Diabetologia 40:830–837 [DOI] [PubMed] [Google Scholar]
- Haffner SM, Miettinen H, Mykkänen L, Stern MP (1998) Leptin concentrations are associated with higher proinsulin and insulin concentrations but a lower proinsulin/insulin ratio in non-diabetic subjects. Int J Obes Relat Metab Disord 22:899–905 [DOI] [PubMed] [Google Scholar]
- Haffner SM, Stern MP, Hazuda HP, Mitchell BD, Patterson JK (1990) Cardiovascular risk factors in confirmed prediabetic individuals: does the clock for coronary heart disease start ticking before the onset of clinical diabetes? JAMA 263:2893–2898 [DOI] [PubMed] [Google Scholar]
- Haffner SM, Stern MP, Hazuda HP, Pugh JA, Patterson JK (1987) Do upper-body and centralized adiposity measure different aspects of regional body-fat distribution? Diabetes 36:43–50 [DOI] [PubMed] [Google Scholar]
- Haffner SM, Stern MP, Miettinen H, Wei M, Gingerich RL (1996) Leptin concentrations in diabetic and nondiabetic Mexican-Americans. Diabetes 45:822–824 [DOI] [PubMed] [Google Scholar]
- Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P (1998) A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet 20:304–308 [DOI] [PubMed] [Google Scholar]
- Hales CN (1995) Proinsulin and cardiovascular risk. In: Schwartz CL, Born GVR (eds) New horizons in diabetes mellitus and cardiovascular disease, Current Science, London, pp 146–151 [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998a) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RL, Knowler WC, the Pima diabetes genes group (1998b) Linkage analyses of insulin levels in a genome-wide scan in nondiabetic Pima indians. Diabetes Suppl 47:A170 [Google Scholar]
- Hermans MP, Levy JC, Morris RJ, Turner RC (1999a) Comparison of insulin sensitivity tests across a range of glucose tolerance from normal to diabetes. Diabetoligia 42:678–687 [DOI] [PubMed] [Google Scholar]
- ——— (1999b) Comparison of tests of β-cell function across a range of glucose tolerance from normal to diabetes. Diabetes 48:1779–1786 [DOI] [PubMed] [Google Scholar]
- Iwasaki N, Wang Y-Q, Cox NJ, Ogata M, Iwamoto Y (1999) A genome-wide screen for type 2 diabetes susceptibility genes in Japanese. Paper presented at the 2d Research Symposium on the Genetics of Diabetes, San Jose, October 17–19 [Google Scholar]
- Kahn CR, Vicent D, Doria A (1996) Genetics of non-insulin-dependent (type-II) diabetes mellitus. Annu Rev Med 47:509–531 [DOI] [PubMed] [Google Scholar]
- Keiffer TJ, Habener JF (2000) The adipoinsular axis: effects of leptin on pancreatic β-cells. Am J Physiol Endocrinol Metab 278:E1–E14 [DOI] [PubMed] [Google Scholar]
- Kissebah AH, Sonnenberg GE, Myklebust J, Goldstein M, Broman K, James RG, Marks JA, Krakower GR, Jacob HJ, Weber J, Martin L, Blangero J, Comuzzie AG (2000) Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA 97:14478–14483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupa T, Malecki MT, Pezzolesi M, Linong J, Simon C, Langefeld CD, Rich SS, Warram JH, Krolewski AS (2000) Further evidence for a susceptibility locus for type 2 diabetes on chromosome 20q13.1-q13.2. Diabetes 49:2212–2216 [DOI] [PubMed] [Google Scholar]
- Kolaczynski JW, Nyce MR, Considine RV, Boden G, Nolan JJ, Henry R, Mudaliar SR, Olefsky J, Caro JF (1996) Acute and chronic effect of insulin on leptin production in humans: Studies in vivo and in vitro. Diabetes 45:699–701 [DOI] [PubMed] [Google Scholar]
- Krushkal J, Ferrell R, Mockrin SC, Turner ST, Sing CF, Boerwinkle E (1999) Genome-wide linkage analyses of systolic blood pressure using highly discordant siblings. Circulation 99:1407–1410 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lange K, Boehnke M (1983) Extensions to pedigree analysis. IV. Covariance components models for multivariate traits. Am J Med Genet 14:513–524 [DOI] [PubMed] [Google Scholar]
- Lee HL, Reed DR, Li W-D, Xu W, Joo E-J, Kilker RL, Nanthakumar E, North M, Sakul H, Bell C, Price RA (1999) Genome scan for human obesity and linkage to markers in 20q13. Am J Hum Genet 64:196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembertas AV, Pérusse L, Chagnon YC, Fisler JS, Warden CH, Purcell-Huynh DA, Dionne FT, Gagnon J, Nadeau A, Lusis AJ, Bouchard C (1997) Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage to body fat and insulin on the human homologous region 20q. J Clin Invest 100:1240–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen P, Mykkänen L, Pyorala K, Laakso M, Kuusisto J (1999) Insulin resistance syndrome predicts coronary heart disease events in elderly nondiabetic men. Circulation 100:123–128 [DOI] [PubMed] [Google Scholar]
- Levy J, Matthews DR, Hermans MP (1998) Correct Homeostasis Model Assessment (HOMA) evaluation uses the computer program. Diabetes Care 21:2191–2192 [DOI] [PubMed] [Google Scholar]
- Lönnqvist F, Nordfors L, Schalling M (1999) Leptin and its potential role in human obesity. J Intern Med 245:643–652 [DOI] [PubMed] [Google Scholar]
- Luo DF, Bui MM, Muir A, Maclaren NK, Thomson G, She JX (1995) Affected-sib-pair mapping of a novel susceptibility gene to insulin-dependent diabetes mellitus (IDDM8) on chromosome 6q 25-q27. Am J Hum Genet 57:911–919 [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Gingerich RL, Santiago JV, Klein S, Smith HC, Landt M (1996) Radioimmunoassay of leptin in human plasma. Clin Chem 42:942–946 [PubMed] [Google Scholar]
- MacDonald MJ, Brown LJ, Hasan NM, Stoffel M, Dills DG (1997) Single-stranded conformational polymorphism analysis of the mitochondrial glycerol phosphate dehydrogenase gene in NIDDM. Diabetes 46:1660–1661 [DOI] [PubMed] [Google Scholar]
- Maddux BA, Goldfine ID (2000) Membrane glycoprotein PC-1 inhibitor of insulin receptor function occurs via direct interaction with the receptor α-subunit. Diabetes 49:13–19 [DOI] [PubMed] [Google Scholar]
- Maddux BA, Sbraccia P, Kumakura S, Sasson S, Youngren J, Fisher A, Spencer S, Grupe A, Henzel W, Stewart TA, Reaven GM, Goldfine ID (1995) Membrane glycoprotein PC-1 and insulin resistance in non-insulin-dependent diabetes mellitus. Nature 373:448–451 [DOI] [PubMed] [Google Scholar]
- Mahaney MC, Blangero J, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW (1995) Plasma HDL cholesterol, triglycerides, and adiposity: a quantitative genetic test of the conjoint trait hypothesis in the San Antonio Family Heart Study. Circulation 92:3240–3248 [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RL (1985) Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28:412–419 [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Cole SA, Comuzzie AG, Almasy L, Blangero J, MacCluer JW, Hixson JE (1999) A quantitative trait locus influencing BMI maps to the region of the β-3 adrenergic receptor. Diabetes 48:1863–1867 [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Cole SA, Hsueh W-C, Comuzzie AG, Blangero J, MacCluer JW, Hixson JE (2000) Linkage of serum insulin concentrations to chromosome 3p in Mexican Americans. Diabetes 49:513–516 [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Mahaney MC, Blangero J, Comuzzie AG, Atwood LD, Haffner SM, Stern MP, MacCluer JW (1996) Genetic analysis of the IRS. Pleiotropic effects of genes influencing insulin levels on lipoprotein and obesity measures. Arterioscler Thromb Vasc Biol 16:281–288 [DOI] [PubMed] [Google Scholar]
- Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, Manion F, Quillen J, et al (1994) A comprehensive human linkage map with centimorgan density. Science 265:2049–2054 [DOI] [PubMed] [Google Scholar]
- Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH (1999) The disease burden associated with overweight and obesity. JAMA 282:1523–1529 [DOI] [PubMed] [Google Scholar]
- Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, Garvey WT, Burns D, Knowler WC, Bennett PH, Bogardus C, Ravussin E (1998) Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet 62:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzuti A, Frittita L, Argiolas A, Baratta R, Goldfine ID, Bozzali M, Ercolino T, Scarlato G, Iacoviello L, Vigneri R, Tassi V, Trischitta V (1999) A polymorphism (K121Q) of the human glycoprotein PC-1 gene coding region is strongly associated with insulin resistance. Diabetes 48:1881–1884 [DOI] [PubMed] [Google Scholar]
- Pratley RE, Thompson DB, Prochazka M, Baier L, Mott D, Ravussin E, Sakul H, Ehm MG, Burns DK, Foroud T, Garvey WT, Hanson RL, Knowler WC, Bennett PH, Bogardus C (1998) An autosomal genomic scan for loci linked to prediabetic phenotypes in Pima Indians. J Clin Invest 101:1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt SC, Daly MJ, Kruglyak L (2000) Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am J Hum Genet 66:1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainwater DL, Almasy L, Blangero J, Cole SA, VandeBerg JL, MacCluer JW, Hixson JE (1999) A genome search identifies major quantitative trait loci on human chromosomes 3 and 4 that influence cholesterol concentrations in small LDL particles. Arterioscler Thromb Vasc Biol 19:777–783 [DOI] [PubMed] [Google Scholar]
- Ravussin E (1995) Metabolic differences and the development of obesity. Metabolism Suppl 44:12–14 [DOI] [PubMed] [Google Scholar]
- Reaven GM (1988) Role of insulin resistance in human disease. Diabetes 37:1595–1607 [DOI] [PubMed] [Google Scholar]
- Ruige JB, Dekker JM, Blum WF, Stehouwer CDA, Nijpels G, Mooy J, Kostense PJ, Bouter LM, Heine RJ (1999) Leptin and variables of body adiposity, energy balance, and insulin resistance in a population-based study: The Hoorn Study. Diabetes Care 22:1097–1104 [DOI] [PubMed] [Google Scholar]
- Self SG, Liang K-Y (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Smith A, Jauch A, Slater H, Robson L, Sandanam T (1999) Syndromal obesity due to paternal duplication. Am J Med Genet 84:125–131 [DOI] [PubMed] [Google Scholar]
- Sobey WJ, Beer SF, Carrington CA, Clark PMS, Frank BH, Gary IP, Luzio SD, Owens DR, Schneider AE, Siddle K, Temple RC, Hales CN (1989) Sensitive and specific two-site immunoradiometric assays for human insulin, proinsulin, 65-66 split and 32-33 split proinsulins. Biochem J 260:535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MP, Duggirala R, Mitchell BD, Reinhart LJ, Shivakumar S, Shipman PA, Uresandi OC, Benavides E, Blangero J, O’Connell P (1996) Evidence for linkage of regions on chromosomes 6 and 11 to plasma glucose concentrations in Mexican Americans. Genome Res 6:724–734 [DOI] [PubMed] [Google Scholar]
- Stern MP, Haffner SM (1990) Type II diabetes and its complications in Mexican Americans. Diabetes Metab Rev 6:29–45 [DOI] [PubMed] [Google Scholar]
- Stern MP, Mitchell BD (1999) Genetics of insulin resistance. In: Reaven G, Laws A (eds) Contemporary endocrinology: insulin resistance: the metabolic syndrome X. Humana, New Jersey, pp 3–18 [Google Scholar]
- Stern MP, Rosenthal M, Haffner SM, Hazuda HP, Franco LP (1984) Sex difference in the effects of sociocultural status on diabetes and cardiovascular risk factors in Mexican Americans: the San Antonio Heart Study. Am J Epidemiol 120:834–851 [DOI] [PubMed] [Google Scholar]
- Taylor BA, Phillips SJ (1997) Obesity QTLs on mouse chromosomes 2 and 17. Genomics 43:249–257 [DOI] [PubMed] [Google Scholar]
- Temple IK, Gardner RJ, Mackay DJG, Barber JCK, Robinson DO, Shield JPH (2000) Transient neonatal diabetes: widening the understanding of the etiopathogenesis of diabetes. Diabetes 49:1359–1366 [DOI] [PubMed] [Google Scholar]
- Temple IK, Gardner RJ, Robinson DO, Kibirige MS, Ferguson AW, Baum JD, Barber JCK, James RS, Shield JPE (1996) Further evidence for an imprinted gene for neonatal diabetes localized to chromosome 6q22-q23. Hum Mol Genet 5:1117–1121 [DOI] [PubMed] [Google Scholar]
- Van Gaal LF, Wauters MA, Mertens IL, Considine RV, De Leeuw IH (1999) Clinical endocrinology of human leptin. Int J Obes Relat Metab Disord Suppl 23:29–36 [DOI] [PubMed] [Google Scholar]
- Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, Matos FD, Durand E, Leprêtre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P (2000) Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2 diabetes locus on chromosome 1q21-q24. Am J Hum Genet 67:1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vionnet N, Hani EH, Lesage S, Philippi A, Hager J, Varret M, Stoffel M, Tanizawa Y, Chiu KC, Glaser B, Permutt AM, Passa P, Demenais F, Froguel P (1997) Genetics of NIDDM in France: studies with 19 candidate genes in affected sib pairs. Diabetes 46:1062–1068 [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Ghosh S, Langefeld CD, Valle TT, Hauser ER, Magnuson VL, Mohlke KL, et al (2000) The Finland-United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study: II. an autosomal genome scan for diabetes-related quantitative-trait loci. Am J Hum Genet 67:1186–1200 [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L, Blangero J (1999b) Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet 65:1148–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Van Eerdewegh P, Almasy L, Blangero J (1999a) Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet 65:1134–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Expert Committee (1985) Diabetes mellitus: report of a WHO study group. World Health Organ Tech Rep Ser 727:1–113 [PubMed] [Google Scholar]
- Zimmet P, Hodge A, Nicolson M, Staten M, de Courten M, Moore J, Morawiecki A, Lubina J, Collier G, Alberti G, Dowse G (1996) Serum leptin concentration, obesity, and insulin resistance in western Samoans: cross sectional study. BMJ 313:965–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouali H, Hani EH, Philippi A, Vionnet N, Beckman JS, Demenais F, Froguel P (1997) A susceptibility locus for early-onset non-insulin dependent (type 2) diabetes mellitus maps to chromosome 20q, proximal to the phosphoenolpyruvate carboxykinase gene. Hum Mol Genet 6:1401–1408 [DOI] [PubMed] [Google Scholar]