Abstract
Distal hereditary motor neuronopathy type VII (dHMN-VII) is an autosomal dominant disorder characterized by distal muscular atrophy and vocal cord paralysis. We performed a genomewide linkage search in a large Welsh pedigree with dHMN-VII and established linkage to chromosome 2q14. Analyses of a second family with dHMN-VII confirmed the location of the gene and provided evidence for a founder mutation segregating in both pedigrees. The maximum three-point LOD score in the combined pedigree was 7.49 at D2S274. Expansion of a polyalanine tract in Engrailed-1, a transcription factor strongly expressed in the spinal cord, was excluded as the cause of dHMN-VII.
The peroneal muscular atrophy syndrome, also known as Charcot-Marie-Tooth disease (CMT [MIM 118200]) is the most common inherited disorder of the peripheral nervous system, affecting 1 in 2,500 people (Keller and Chance 1999). The cardinal clinical features are weakness and wasting of the distal limb muscles, hypo- or areflexia, and pes cavus deformity of the foot. Distal sensory loss is present in some cases. Three distinct groups of peroneal muscular atrophy conditions are recognized: (1) a demyelinating form, hereditary motor and sensory neuropathy type I (HMSN I or CMT1); (2) an axonal form, hereditary motor and sensory neuropathy type II (HMSN II or CMT2); and (3) distal hereditary motor neuronopathy (dHMN, also known as distal spinal muscular atrophy, or spinal CMT) (De Jonghe et al. 1998).
dHMN accounts for ∼10% of peroneal muscular atrophy syndrome (Harding and Thomas 1980). Motor and sensory nerve conduction velocities are normal, and electromyography typically shows features of neurogenic atrophy, consistent with degeneration of the anterior horn cell of the spinal cord. On the basis of clinical and genetic criteria, dHMN is divided into seven subtypes (Harding 1993). The genes for several dHMN subtypes and related conditions have been localized (table 1). However, no causative dHMN genes have thus far been isolated.
Table 1.
Distal Hereditary Motor Neuronopathy Conditions for Which Predisposition Genes Have Been Localized
| Disorder | Features | Inheritancea | Locus | Reference |
| dHMN-II | Adult-onset distal wasting and weakness | AD | 12q24.3 | Timmerman et al. (1996) |
| dHMN-V | Upper limb predominance; occasionally pyramidal features | AD | 7p15 | Christodoulou et al. (1995) |
| dHMN-VI | Severe infantile form with respiratory distress | AR | 11q13-21 | Grohmann et al. (1999) |
| dHMN-VII | Adult onset with vocal cord paralysis | AD | 2q14 | Present report |
| dHMN–Jerash typeb | Juvenile onset with pyramidal features | AR | 9p21.1-p12 | Middleton et al. (1999); Christodoulou et al. (2000) |
| Congenital dSMAb,c | Congenital nonprogressive dHMN with contractures | AD | 12q23-q24 | van der Vleuten et al. (1998) |
AD = autosomal dominant; AR = autosomal recessive.
dHMN–Jerash type and congenital dSMA were not included in the original classification of these disorders by Harding (1993).
dSMA = distal spinal muscular atrophy, interchangeable with dHMN.
dHMN-VII (MIM 158580) is an autosomal dominant condition characterized by distal muscular atrophy associated with unilateral or bilateral vocal cord paralysis. Young and Harper (1980) first described the condition in a large Welsh kindred. Other families with similar phenotypes were subsequently reported (Serratrice et al. 1984; Boltshauser et al. 1989; Pridmore et al. 1992). We report the localization of the dHMN-VII gene to chromosome 2q14 by linkage and allelic association analyses in two families with dHMN-VII, including the original family, YH1, described by Young and Harper.
Detailed clinical and pathological features of members of family YH1 have been published elsewhere (Young and Harper 1980). Permission for the current study was obtained from the local research-ethics committee, and all individuals analyzed were reevaluated clinically by one of the authors (M.M.). dHMN-VII typically presented in the second decade, with weakness and wasting of the small muscles of the hand and thenar eminence. Subsequently, weakness and wasting of the distal muscles of the lower limbs occurred. All but one affected individual developed a hoarse voice, which resulted from unilateral or bilateral vocal cord paralysis. The age of subjects at vocal cord involvement varied and preceded the hand changes in some individuals. As muscle wasting progressed, deep-tendon reflexes were lost, but sensation remained intact in all cases. Disease progression was slow and compatible with a normal life span. Motor and sensory nerve conduction velocities were normal, and electromyography was suggestive of chronic partial denervation.
A genomewide linkage search was performed on 10 affected individuals and 3 spouses in family YH1. A total of 230 microsatellite markers, spaced at 20-cM intervals, were selected from the ABI PRISM linkage-mapping set version 2 (Applied Biosystems) and were PCR amplified using standard protocols. Amplified markers were electrophoresed through 4.5% denaturing polyacrylamide gels on an ABI 377 DNA sequencer and were analyzed with GENESCAN and GENOTYPER software (Applied Biosystems).
dHMN-VII was modeled as an autosomal dominant trait with a penetrance of 95% by age 20 years. Unaffected individuals aged <20 years were coded as “unknown.” A disease allele frequency of .0001 and equal recombination fractions in males and females were assumed. Two-point LOD scores were calculated using the MLINK program of LINKAGE, and multipoint (three-point) LOD scores were generated using the VITESSE program. Marker allele frequencies were estimated from unrelated individuals from the pedigrees.
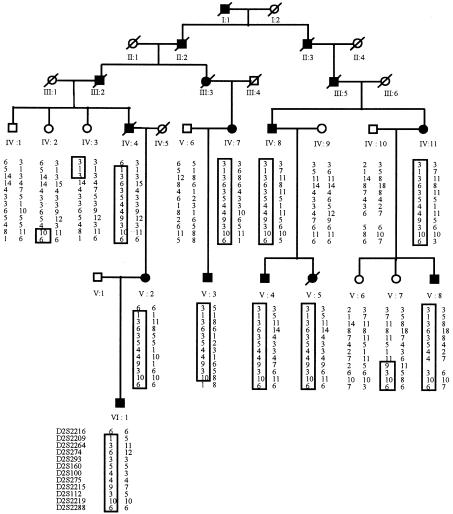
In the linkage search, marker D2S112 generated the highest two-point LOD score, and positive LOD scores at consecutive markers occurred only at D2S160 and D2S112 on chromosome 2q14 (table 2). To confirm and refine the location of the dHMN-VII predisposition gene, 10 additional markers, spanning a 40-cM region on chromosome 2q14, were analyzed. DNA from an additional six unaffected relatives were included in these analyses. The order and distance from the p-telomere of the chromosome 2q14 markers are shown in table 3. Examination of the marker-allele haplotypes, segregating in affected individuals only, placed the dHMN-VII gene in a 36-cM region flanked by D2S2216 and D2S2288 (fig. 1). Meiotic recombinants in unaffected individuals IV:3 and V:7 refined the dHMN-VII region to a 20-cM interval flanked by D2S2264 and D2S2215 (fig. 1). The maximum two-point LOD score was 4.21 at D2S275, and the maximum multipoint LOD score was 5.01 at D2S100.
Table 2.
Two-Point LOD Scores at Chromosome 2q14 Markers in dHMN-VII Pedigrees
|
Two-Point LOD Score at θ= |
||||||
| Marker and Family | 0 | .01 | .05 | .10 | .20 | .30 |
| D2S2216: | ||||||
| YH1 | −3.85 | −1.23 | −.56 | −.30 | −.11 | −.04 |
| P2 | .95 | .93 | .83 | .70 | .47 | .26 |
| D2S2209: | ||||||
| YH1 | 2.02 | 2.01 | 1.92 | 1.75 | 1.30 | .79 |
| P2 | .25 | .24 | .20 | .15 | .08 | .04 |
| D2S2264: | ||||||
| YH1 | 1.83 | 1.80 | 1.67 | 1.48 | 1.08 | .66 |
| P2 | 2.12 | 2.08 | 1.92 | 1.71 | 1.27 | .79 |
| D2S274: | ||||||
| YH1 | 3.66 | 3.59 | 3.29 | 2.91 | 2.11 | 1.29 |
| P2 | 1.79 | 1.76 | 1.61 | 1.42 | 1.02 | .62 |
| D2S293: | ||||||
| YH1 | 4.06 | 3.97 | 3.64 | 3.22 | 2.33 | 1.42 |
| P2 | .38 | .37 | .33 | .28 | .20 | .13 |
| D2S160: | ||||||
| YH1 | .95 | .91 | .75 | .56 | .25 | .06 |
| P2 | 2.09 | 2.05 | 1.89 | 1.67 | 1.23 | .75 |
| D2S100: | ||||||
| YH1 | 3.44 | 3.38 | 3.11 | 2.75 | 2.00 | 1.21 |
| P2 | 2.18 | 2.14 | 1.98 | 1.76 | 1.31 | .82 |
| D2S275: | ||||||
| YH1 | 4.21 | 4.12 | 3.79 | 3.37 | 2.47 | 1.53 |
| P2 | 1.85 | 1.82 | 1.67 | 1.49 | 1.09 | .67 |
| D2S2215: | ||||||
| YH1 | 2.19 | 2.20 | 2.14 | 1.96 | 1.46 | .88 |
| P2 | .21 | .20 | .17 | .13 | .07 | .03 |
| D2S112: | ||||||
| YH1 | 2.36 | 2.37 | 2.29 | 2.10 | 1.56 | .94 |
| P2 | 2.10 | 2.06 | 1.90 | 1.69 | 1.26 | .79 |
| D2S2219: | ||||||
| YH1 | 1.10 | 1.13 | 1.16 | 1.09 | .82 | .49 |
| P2 | 1.68 | 1.64 | 1.50 | 1.32 | .94 | .56 |
| D2S2288: | ||||||
| YH1 | −3.85 | −1.45 | −.55 | −.16 | .10 | .12 |
| P2 | .51 | .50 | .47 | .43 | .35 | .25 |
Table 3.
Microsatellite Marker Alleles Segregating with dHMN-VII in Families YH1 and P2[Note]
|
Disease-Linked Allele |
||||
| Distance from p-tel(cM) | Marker | YH1 | P2 | Two-Point LOD Score in Combined Pedigree atθ=0 |
| 104 | D2S2333 | 10 | 6 | −3.01 |
| 111 | D2S2216 | 6 |
3 |
−2.52 |
| 112 | D2S2209 | 1 | 1 | 2.60 |
| 114 | D2S2264a | 3 | 3 | 4.51 |
| 115 | D2S274 | 6 | 6 | 6.04 |
| 116 | D2S2229 | 6 | 6 | 4.27 |
| 117 | D2S1897 | 3 | 3 | 2.67 |
| 118 | D2S293 | 3 | 3 | 4.96 |
| 120 | D2S1890 | 4 |
4 |
3.56 |
| 122 | D2S160 | 5 | 3 | .19 |
| 124 | D2S308 | 3 | 6 | 1.41 |
| 125 | D2S437 | 4 | 5 | .03 |
| 127 | D2S2970 | 3 |
5 |
2.24 |
| 128 | D2S100 | 4 | 4 | 6.33 |
| 129 | D2S2265 | 4 | 4 | 1.12 |
| 130 | D2S2224 | 1 | 1 | 1.92 |
| 131 | D2S347 | 12 | 12 | 4.05 |
| 132 | D2S2339 | 4 | 4 | 5.18 |
| 132 | D2S275 | 4 | 4 | 6.63 |
| 132 | D2S1273 | 8 | 8 | 2.03 |
| 132 | D2S1272 | 11 | 11 | 3.19 |
| 133 | D2S2271 | 1 | 1 | 3.97 |
| 133 | D2S95 | 6 | 6 | 6.05 |
| 134 | D2S2215a | 9 | 9 | 2.84 |
| 137 | D2S1260 | 9 | 9 | 5.36 |
| 141 | D2S112 | 3 | 3 | 4.95 |
| 142 | D2S2219 | 10 | 10 | 3.18 |
| 145 | D2S1334 | 11 |
11 |
4.59 |
| 147 | D2S2288 | 1 | 6 | −3.06 |
| 152 | D2S151 | 15 | 11 | −6.34 |
Note.— The two intervals in which the disease-linked alleles in families YH1 and P2 are identical are shown in boxes. Genealogy investigations confirmed that the two families are related, and the most likely explanation for the interval between D2S1890 and D2S100, in which the disease-linked alleles differ, is a historic double recombination between the two families.
Recombinants at this marker in family YH1 (individuals IV:3 and V:7) define the minimal dHMN-VII interval.
Figure 1.
Haplotypes for 12 markers from chromosome 2q14 from members of family YH1. Blackened symbols, individuals affected with dHMN-VII. Unblackened symbols, unaffected individuals. Boxes around marker alleles indicate disease haplotypes present in the affected individuals. Boxed alleles in unaffected individuals IV:3 and V:7 define the minimum interval encompassing the dHMN-VII gene to the region between D2S2264 and D2S2215.
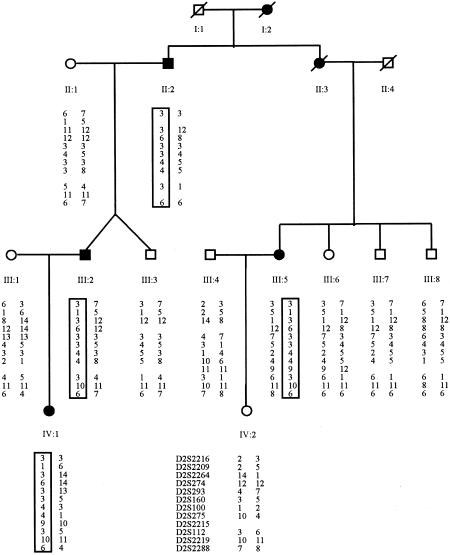
A second pedigree with dHMN-VII (family P2) was examined for linkage to chromosome 2q14. The clinical, pathological, and electrophysiological features in members of family P2 were similar to those in members of family YH1 and have been published elsewhere (Pridmore et al. 1992). All individuals analyzed in this study were reevaluated by one of the authors (H.H.). Twelve microsatellite markers, spanning the 20-cM dHMN-VII interval defined in family YH1, were analyzed. The marker haplotypes demonstrated that affected individuals shared a haplotype of marker alleles that was not present in their unaffected relatives (fig. 2). Positive two-point LOD scores were observed at all markers with a maximum two-point LOD score of 2.18 at D2S100 (table 2). The maximum multipoint LOD score was 2.25 at D2S2265. At a candidate locus, these values provide strong evidence in favor of linkage. The disorder in family P2 is thus highly likely to be due to mutation of the dHMN-VII gene on chromosome 2q14.
Figure 2.
Haplotypes for 12 markers from chromosome 2q14 from members of family P2. Data from at-risk individuals aged <20 years have been omitted. Boxes around marker alleles indicate disease haplotypes present in the affected individuals.
Scrutiny of the marker haplotypes revealed the disease-associated alleles in family P2 to be identical to those segregating in family YH1 for 11 of the 12 markers initially analyzed. This was suggestive of a founder mutation predisposing to the disease in both families. To investigate this further, 30 markers from the dHMN-VII interval were analyzed in both families (table 3). Identical disease-segregating alleles were seen in both families at 22 markers, which supports our conjecture of a common ancestral origin for the two families. Genealogy investigations subsequently confirmed that the families are indeed related, with individual I:1 in family YH1 being the uncle of individual I:2 in family P2. LOD scores calculated with the combined pedigree generated a maximum two-point LOD score of 6.63 at D2S275 and a maximum three-point LOD score of 7.49 at D2S274.
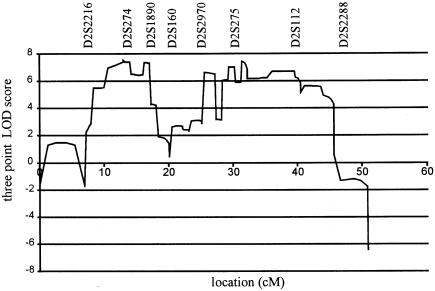
There were two separate regions in the disease-associated haplotypes in families YH1 and P2, at which the segregating alleles were identical in both families (table 3). The centromeric interval, demonstrating allelic association in the two families, was between D2S2216 and D2S160. The telomeric interval, showing allelic association, was between D2S2970 and D2S2288. The multipoint LOD scores were strongly positive within both of these intervals (fig. 3). Between the two regions there was a 7-cM interval, flanked by D2S1890 and D2S100, in which the disease-segregating alleles differed in the two families (table 3) and the multipoint LOD scores were much lower (fig. 3). The location of the markers analyzed within the D2S1890–D2S100 interval could be confidently assigned by reference to the genetic radiation hybrid and physical maps of chromosome 2q14, and they clearly bisect the disease-associated haplotype that segregates in the two families. It is therefore most likely that a double recombination event has occurred between the two families, with one recombination event occurring between markers D2S1890 and D2S160 and the other between D2S2970 and D2S100. The data strongly suggest that the dHMN-VII gene does not lie between markers D2S160 and D2S2970.
Figure 3.
Multipoint LOD score for combined dHMN-VII pedigree.
The combined linkage, allelic association, and recombinant data from families YH1 and P2 localize the dHMN-VII predisposition gene either to the 8-cM region flanked by D2S2264 and D2S160 or the 7-cM region flanked by D2S2970 and D2S2215. The gene has been designated DHMNVP (distal hereditary motor neuronopathy with vocal cord paralysis).
No genes for distal hereditary motor neuronopathy have been identified, and it is therefore difficult to predict what the function of DHMNVP might be. However, it is highly likely that the gene is expressed in the spinal cord. Two possible candidates for the dHMN-VII gene are Engrailed-1 (EN1) and the neuronal PAS domain protein-2 gene (NPAS-2), both of which encode transcription factors that are strongly expressed in the spinal cord of mice (Davis et al. 1988; Zhou et al. 1997). EN1 is a homeobox-containing gene that has been implicated in the regulation of axon pathfinding by association interneurons that project to motor neurons in the spinal cord (Saueressig et al. 1999). To confirm that EN1 is within the dHMN-VII interval(s), a dinucleotide repeat located 3 kb 5′ of EN1 was identified, and amplifying primers were designed and optimized (table 3). The repeat was amplified in families YH1 and P2, and a single allele segregated with the disease in both families, generating a two-point LOD score of 3.58 (data not shown).
The first exon of EN1 contains an imperfect polyalanine repeat of the form alanine(10)-valine-alanine(9). Pathogenic expansions of short polyalanine tracts have been reported in a number of disorders, including oculopharyngeal muscular dystrophy, in which expansions of alanine(6) to alanine(8-13) in the PABP2 gene have been identified (Brais et al. 1998). Primers amplifying the polyalanine tract in EN1 were designed (table 4) and analyzed in one affected and one unaffected individual from families YH1 and P2. No alteration in the size of the amplified fragment was identified in the two individuals affected by dHMN-VII compared with the controls (data not shown). Thus, expansion of the EN1 polyalanine tract is not responsible for dHMN-VII. However, intragenic EN1 mutations were not investigated and may be responsible for dHMN-VII.
Table 4.
Primer Pairs for PCR Amplification of EN1 Repeat and Polyalanine Tract
|
Primer (5′→3′) |
||
| Forward | Reverse | |
| Dinucleotide repeat | GACTAGTGGGGCTGACTTGG | GCTCAGCGGTCTTGAAGAGT |
| Polyalanine tract | GCAGGTAGAGACCCTGTCCA | AAGTAGGATAGCCGGGTTGC |
Hereditary motor and sensory neuropathy type IIC (HMSN-IIC [MIM 158580]) shows considerable overlap with dHMN-VII. HMSN-IIC is an autosomal dominant disorder characterized by motor and sensory involvement of the limbs, with progressive weakness of the vocal cords, diaphragm, and intercostal muscles (Dyck et al. 1994; Yoshioka et al. 1996; Donaghy et al. 1999). The distinguishing feature of HMSN-IIC is sensory involvement, which is not seen in dHMN-VII. The gene for HMSN-IIC has not been localized, but it has been suggested that HMSN-IIC and dHMN-VII may be allelic disorders that are caused by mutations of the same gene (Dyck et al. 1994). There is a precedent for such allelism, as dHMN-V and HMSN-IID have both been linked to chromosome 7p15 (Christodoulou et al. 1995; Ionasescu et al. 1996). Moreover, a kindred in which both phenotypes segregate in association with the same chromosome 7p15 haplotype has been reported (Sambuughin et al. 1998). Linkage analyses at chromosome 2q14 in HMSN-IIC pedigrees should determine whether or not the condition is likely to be due to the dHMN-VII gene on chromosome 2q14.
Other conditions with phenotypic overlap with dHMN-VII would also be worth examining for linkage to chromosome 2q14. A family with dHMN, vocal cord paralysis, and sensorineural deafness has been reported; these features may be the result of a DHMNVP mutation (Bolthauser et al. 1989). However, a family with some individuals having CMT, sensorineural deafness, and vocal cord paralysis has been described, in which a pathogenic PMP22 point mutation was identified (Kovach et al. 1999). Thus, genetic heterogeneity in conditions with peroneal muscular atrophy and vocal cord paralysis is likely to exist.
The identification of a further disease locus for dHMN adds to our growing knowledge of the disease loci involved in this subset of peroneal muscular atrophy. Isolation of genes altered in dHMN will not only improve our understanding of these disorders but will also help to clarify the relationship between dHMN and HMSN.
Acknowledgments
We are grateful to all the family members for their kind cooperation in this research. The authors also wish to thank Dr. Meena Upadhyaya, for providing samples, and Juliet Gayton and Gerry Toop, for carrying out the genealogy searches. This research was supported by the Muscular Dystrophy Campaign.
Electronic-Database Information
Accession numbers and URL for data in this article are as follows:
References
- Boltshauser E, Lang W, Spillman T, Hof E (1989) Hereditary distal muscular atrophy with vocal cord paralysis and sensorineural hearing loss: a dominant form of spinal muscular atrophy? J Med Genet 26:105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brais B, Bouchard JP, Xie YG, Rochefort DL, Chretien N, Tome FM, Lafreniere RG, Rommens JM, Uyama E, Nohira O, Blumen S, Korcyn AD, Heutink P, Mathieu J, Duranceau A, Codere F, Fardeau M, Rouleau GA, Korcyn AD (1998) Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet 18:164–167 [DOI] [PubMed] [Google Scholar]
- Christodoulou K, Kyriakides T, Hristova AH, Georgiou D, Kalaydjieva L, Yshpekova B, Ivanova T, Weber JL, Middleton LT (1995) Mapping of a distal form of spinal muscular atrophy with upper limb predominance to chromosome 7p. Hum Mol Genet 4:1629–1632 [DOI] [PubMed] [Google Scholar]
- Christodoulou K, Zamba E, Tsingis M, Mubaidin A, Horani K, Abu-Sheik S, El-Khateeb M, Kyriacou K, Kyriakides T, Al-Qudah AK, Middleton L (2000) A novel form of distal hereditary motor neuronopathy maps to chromosome 9p21.1-p12. Ann Neurol 48:877–884 [PubMed] [Google Scholar]
- Davis CA, Joyner AL (1988) Expression patterns of the homeo box-containing genes En-1 and En-2 and the proto-oncogene int-1 diverge during mouse development. Genes Dev 2:1736–1744 [DOI] [PubMed] [Google Scholar]
- De Jonghe P, Timmerman V, Van Broeckhoven C, workshop participants (1998) 2nd workshop of the European CMT Consortium: 53rd ENMC international workshop on classification and diagnostic guidelines for Charcot-Marie-Tooth type 2 (CMT2-HMSN-II) and distal hereditary motor neuropathy (distal HMN-spinal CMT), September 26–28, 1997, Naarden, The Netherlands. Neuromuscul Disord 8:426–431 [PubMed] [Google Scholar]
- Donaghy M, Kennett R (1999) Varying occurrence of vocal cord paralysis in a family with autosomal dominant hereditary motor and sensory neuropathy. J Neurol 246:552–555 [DOI] [PubMed] [Google Scholar]
- Dyck JP, Litchy WJ, Minnerath S, Bird TD, Chance PF, Schaid DJ, Arronson AE (1994) Hereditary motor and sensory neuropathy with diaphragm and vocal cord paresis. Ann Neurol 35:608–615 [DOI] [PubMed] [Google Scholar]
- Grohmann K, Wienker TF, Saar K, Rudnik-Schoneborn S, Stoltenburg-Didinger G, Rossi R, Novelli G, Nurnberg G, Pfeufer A, Wirth B, Reis A, Zerres K, Hubner C (1999) Diaphragmatic spinal muscular atrophy with respiratory distress is heterogeneous, and one form is linked to chromosome 11q13-q21. Am J Hum Genet 65:1459–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AE (1993) Inherited neuronal atrophy and degeneration predominantly of lower motor neurons. In: Dyck PJ, Thomas PK (eds) Peripheral neuropathy. Vol. 2. W. B. Saunders, Philadelphia, pp. 1051–1064 [Google Scholar]
- Harding AE, Thomas PK (1980) Hereditary distal spinal muscular atrophy: a report on 34 cases and a review of the literature. J Neurol Sci 45:337–348 [DOI] [PubMed] [Google Scholar]
- Ionasescu V, Searby C, Sheffield VC, Roklina T, Nishimura D, Ionasescu R (1996) Autosomal dominant Charcot-Marie-Tooth axonal neuropathy mapped on chromosome 7p (CMT2D). Hum Mol Genet 5:1373–1375 [DOI] [PubMed] [Google Scholar]
- Keller MP, Chance PF (1999) Inherited peripheral neuropathy. Semin Neurol 19:353–362 [DOI] [PubMed] [Google Scholar]
- Kovach MJ, Lin J, Boyadjiev S, Campbell K, Mazzeo L, Herman K, Rimer LA, Frank W, Llewellyn B, Wang Jabs E, Gelber D, Kimonis VE (1999) A unique point mutation in the PMP22 gene is associated with Charcot-Marie-Tooth and deafness. Am J Hum Genet 64:1580–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton LT, Christodoulou K, Mubaidin A, Zamba E, Tsingis M, Kyriacou K, Abu-Sheik S, Kyriakides T, Neocleous V, Georgiou DM, El-Khateeb M, Al-Quadah A, Horany K (1999) Distal hereditary motor neuronopathy of the Jerash type. Ann NY Acad Sci 883:65–68 [PubMed] [Google Scholar]
- Pridmore C, Baraitser M, Brett EM, Harding AE (1992) Distal spinal muscular atrophy with vocal cord paralysis. J Med Genet 29:197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambuughin N, Sivakumar K, Selenge B, Lee HS, Friedlich D, Baasanjav D, Dalakas MC, Goldfarb LG (1998) Autosomal dominant distal spinal muscular atrophy type V (dSMA-V) and Charcot-Marie-Tooth disease type 2D (CMT2D) segregate within a single large kindred and map to a refined region on chromosome 7p15. J Neurol Sci 161:23–28 [DOI] [PubMed] [Google Scholar]
- Saueressig H, Burrill J, Goulding M (1999) Engrailed-1 and Nectrin-1 regulate axon pathfinding by association interneurons that project to motor neurons. Development 126:4201–4212 [DOI] [PubMed] [Google Scholar]
- Serratrice G, Pellissier JF, Gastaut JL, Desnuelle C (1984) Amyotrophie spinale chronique avec paralysie des cordes vocales: syndrome de Young et Harper [Chronic spinal amyotrophy with paralysis of the vocal cords: Young-Harper syndrome]. Rev Neurol (Paris) 140:657–658 [PubMed] [Google Scholar]
- Timmerman V, De Jonghe P, Simokovic S, Lofgren A, Beuten J, Nelis E, Ceuterick C, Martin JJ, Van Broeckhoven C (1996) Distal hereditary motor neuropathy type II (distal HMN II): mapping of a locus to chromosome 12q24. Hum Mol Genet 5:1065–1069 [DOI] [PubMed] [Google Scholar]
- van der Vleuten AJ, van Ravenswaaij-Arts CMA, Frijns CJ, Smits AP, Hageman G, Padberg GW, Kremer H (1998) Localisation of the gene for a dominant congenital spinal muscular atrophy predominantly affecting the lower limbs to chromosome 12q23-q24. Eur J Hum Genet 6:376–82 [DOI] [PubMed] [Google Scholar]
- Yoshioka R, Dyck PJ, Chance PF (1996) Genetic heterogeneity in Charcot-Marie-Tooth neuropathy type 2. Neurology 46:569–571 [DOI] [PubMed] [Google Scholar]
- Young ID, Harper PS (1980) Hereditary distal spinal muscular atrophy with vocal cord paralysis. J Neurol Neurosurg Psychiatry 43:413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YD, Barnard M, Tian H, Li X, Ring HZ, Francke U, Shelton J, Richardson J, Russell DW, McKnight SL (1997) Molecular characterisation of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc Natl Acad Sci USA 94:713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]