Abstract
Reduced nicotinamide adenine dinucleotide (NADH):ubiquinone oxidoreductase (complex I) is the largest complex of the mitochondrial respiratory chain and complex I deficiency accounts for ∼30% cases of respiratory-chain deficiency in humans. Only seven mitochondrial DNA genes, but >35 nuclear genes encode complex I subunits. In an attempt to elucidate the molecular bases of complex I deficiency, we studied the six most-conserved complex I nuclear genes (NDUFV1, NDUFS8, NDUFS7, NDUFS1, NDUFA8, and NDUFB6) in a series of 36 patients with isolated complex I deficiency by denaturing high-performance liquid chromatography and by direct sequencing of the corresponding cDNA from cultured skin fibroblasts. In 3/36 patients, we identified, for the first time, five point mutations (del222, D252G, M707V, R241W, and R557X) and one large-scale deletion in the NDUFS1 gene. In addition, we found six novel NDUFV1 mutations (Y204C, C206G, E214K, IVS 8+41, A432P, and del nt 989–990) in three other patients. The six unrelated patients presented with hypotonia, ataxia, psychomotor retardation, or Leigh syndrome. These results suggest that screening for complex I nuclear gene mutations is of particular interest in patients with complex I deficiency, even when normal respiratory-chain–enzyme activities in cultured fibroblasts are observed.
Introduction
Reduced nicotinamide adenine dinucleotide (NADH):ubiquinone oxidoreductase (complex I) catalyzes electron transfer from NADH to ubiquinone. This enzyme, the largest complex of the mitochondrial respiratory chain, contains ⩾40 subunits (Fearnley and Walker 1992). It is embedded in the inner mitochondrial membrane and is partly protruding in the matrix. Complex I can be divided into three parts—namely, (1) the flavoprotein fraction, containing the binding sites for NADH, flavin mononucleotide (FMN), and iron-sulfur (Fe-S) clusters, (2) the iron protein fraction with several Fe-S clusters, and (3) the hydrophobic fraction, which binds quinone, in the inner membrane. Complex I deficiency is the most common cause of mitochondrial disorders. It represents largely one-third of all cases of respiratory chain deficiency (von Kleist-Retzow et al. 1998; Kirby et al. 1999) and is responsible for a variety of clinical symptoms, ranging from neurological disorders to cardiomyopathy, liver failure, and myopathy (von Kleist-Retzow et al. 1998; Loeffen et al. 2000).
Most of the 43 complex I subunits are encoded by nuclear genes, and only seven of them are mitochondrially encoded. Recently, mutations in nuclear complex I genes have been identified in patients with Leigh syndrome. The disease-causing genes (NDUFS8 [MIM 602141], NDUFS4 [MIM 602694], NDUFS7 [MIM 601825], and NDUFV1 [MIM 161015]) (Loeffen et al. 2000) are highly conserved across species and play a major role in catalytic activity (Fearnley and Walker 1992). In an attempt to identify the molecular bases of the disease in our patients with complex I deficiency and to contribute to genetic counseling, we developed a systematic search for mutations in the six most-conserved complex I nuclear genes by denaturating high-performance liquid chromatography (D-HPLC) and direct sequencing of their respective cDNA from cultured skin fibroblasts of the probands. In this article, we report on novel NDUFV1 and NDUFS1 mutations in 6/36 patients, thus supporting the high prevalence of catalytic subunit gene mutations in complex I deficiency.
Patients
Family 1
A boy, whose parents were healthy, unrelated, and of French origin, was born after a normal pregnancy and delivery (birth weight 3,560 g). He was first hospitalized at age 1 year for seizures and moderately elevated levels of plasma lactate (2.6 mM). He then presented with cerebellar ataxia with persistent seizures and was given valproate. At age 28 mo, psychomotor regression, strabismus, and ptosis were noted. Magnetic resonance imaging (MRI) showed brain atrophy and multiple symmetric areas of hyperintensity in the brain stem. He died at age 3 years, of an acute episode of metabolic acidosis. Postmortem analysis of the brain confirmed the gray matter degeneration with foci of necrosis in the brain stem. A complex I deficiency was identified in muscle (table 1) and liver. Oxidation of NADH-generating substrates was normal in cultured skin fibroblasts.
Table 1.
Respiratory-Chain Analysis in Skeletal Muscle Mitochondria and Homogenates in Patients and Control[Note]
|
Patient from |
||||
| Source, Activity, and Activity Ratio | Family 3 | Family 5 | Family 6 | 80 Controls |
| Activity(nmol/min/mg protein) |
||||
| Muscle mitochondria: | ||||
| NADH quinone reductase | 15 | 106 | 29 | 47–182 |
| Succinate quinone dichlorophenol indophenol reductase | 116 | 164 | 77 | 69–268 |
| Decylubiquinone cytochrome c reductase | 900 | … | 510 | 421–1,654 |
| Cytochrome c oxidase | 833 | 1376 | 578 | 575–2,419 |
| Pyruvate oxidase | … | 10 | 11 | 32–56 |
| Malate oxidase | 9 | 11 | … | 20–44 |
| Succinate oxidase | 35 | 40 | 46 | 34–92 |
| Activity Ratio |
||||
| Cytochrome c oxidase/NADH quinone reductase | 55.5 | 13 | 19.9 | 9.9±1.6 |
| Cytochrome c oxidase/succinate quinone dichlorophenol indophenol reductase | 7.2 | 8.4 | 7.5 | 8.8±1.5 |
| Cytochrome c oxidase/decylubiquinone cytochrome c reductase | .9 | … | 1.1 | 1.4±.2 |
| Succinate oxidase/pyruvate oxidase |
4.2 |
4.0 |
4.2 |
1.9±.2 |
| Family 1 | Family 2 | Family 4 | 51 Controls | |
| Activity(nmol/min/mg protein) |
||||
| Muscle homogenate: | ||||
| NADH quinone reductase | 14 | 13 | 11 | 8–32 |
| Succinate quinone dichlorophenol indophenol reductase | 55 | 48 | 34 | 18–70 |
| Decylubiquinone cytochrome c reductase | 259 | 357 | 203 | 67–268 |
| Cytochrome c oxidase | 274 | 227 | 167 | 86–342 |
| Activity Ratio |
||||
| Cytochrome c oxidase/NADH quinone reductase | 19.5 | 17.5 | 15.2 | 9.9±.9 |
| Cytochrome c oxidase/succinate quinone dichlorophenol indophenol reductase | 5.0 | 4.7 | 5.0 | 5.4±.4 |
| Cytochrome c oxidase/decylubiquinone cytochrome c reductase | 1.1 | .6 | .8 | 1.2±.2 |
Note.— Substrate oxidation was polarographically measured (Rustin et al. 1994). Abnormal values are in bold italic.
Family 2
A girl, the second child of healthy nonconsanguineous parents of French origin, was born after a normal pregnancy and delivery (birth weight 3,600 g, length 48.5 cm, and head circumference 33 cm). She was hospitalized at age 6 mo for vomiting and floppiness. Recurrent episodes of vomiting, hypotonia, lethargy, and apnea at age 18 mo led to an intubation. Metabolic acidosis (pH 7.24, plasma bicarbonate 16 mM) with high levels of lactate were noted (4 mM) and MRI (T2 sequence) revealed areas of hyperintensity in the basal ganglia. She died at age 18 mo, of an acute episode of metabolic acidosis (bicarbonate 14 mM). A complex I deficiency was identified in muscle (table 1) and liver. Oxidation of NADH-generating substrates was normal in cultured skin fibroblasts.
Family 3
A boy, whose parents were healthy, unrelated, and of French origin, was born after a term pregnancy (weight 2,600 g, length 45 cm, and head circumference 33 cm). At age 5 mo he had ptosis and strabismus. At age 9 mo he was hypotonic and unable to sit. Ataxia, bilateral ptosis, and ophthalmoplegia were noted, and he rapidly developed a metabolic acidosis with high levels of lactate (4.7, N<2.3 mM). MRI showed areas of hyperintensity of the locus niger. A complex I deficiency was identified in muscle (table 1). Oxidation of NADH-generating substrates was normal in both cultured skin fibroblasts and circulating lymphocytes.
Family 4
The fourth-born child of unrelated healthy parents was normal until age 4 mo, when he developed psychomotor retardation with hypotonia. At age 7 mo, he presented nystagmus and bilateral optic atrophy. Leukodystrophy, lactic acidosis (4.4 mM), and hyperlactatorachia were also noted (3.2 mM). He died at age 10 mo, from an acute episode of bradycardia. His older sister also presented with hyperlactatemia, hypotonia, pyramidal syndrome, and leukodystrophy; she died at age 7 mo. His older brother developed two episodes of ataxia and mild psychomotor retardation at age 2 years. A complex I deficiency was identified in muscle (table 1) and liver. Decreased NADH-generating substrates oxidation in fibroblasts suggested a complex I deficiency. Lymphocytes were normal.
Family 5
The fourth child (weight 3,100 g, height 49 cm, and head circumference 34 cm) of unrelated healthy parents was normal until age 2 mo, when he presented with growth retardation, axial hypotonia, hepatomegaly, and persistent hyperlactatemia (5 mM). An MRI showed hyperintensity of basal ganglia. The child then developed macrocytic anemia and dystonia. He died suddenly, at age 5 mo. His older sister presented with growth retardation, macrocytic anemia, and metabolic acidosis, at age 3 mo. Shortly thereafter she died, after an acute episode of hyperlactatemia. Oxidation of NADH-generating substrates was low in muscle mitochondria (table 1), in fibroblasts, and in circulating lymphocytes.
Family 6
The third boy (weight 2,900 g, height 48.5 cm, and head circumference 37 cm) of unrelated healthy Portuguese parents presented, shortly after birth, with failure to thrive with hypotonia, microcephalia, and pyramidal syndrome. At age 5 mo, he developed hyperlactatemia (5.3 mM) and hyperlactatorachia (4 mM). An MRI showed structural abnormalities of the corona radiata, suggestive of Leigh syndrome, and permanent anemia necessitated blood transfusions. A complex I deficiency was identified in muscle (table 1). Cultured skin fibroblasts normally oxidized NADH-generating substrates.
Methods
Polarographic tests and/or spectrophotometric assays of respiratory-chain enzymes were performed on skeletal muscle mitochondria, muscle homogenate, liver homogenate, cultured skin fibroblasts, or circulating lymphocytes, as described elsewhere (Rustin et al. 1994). Total RNAs were extracted from cultured skin fibroblasts, by means of the Rnasin kit (Qiagen), and were reverse transcribed with random hexamer primers (GenAmp RNA PCR core kit, Applied Biosystems). The reverse transcriptase (RT)–PCR amplification of specific RNA was performed in overlapping fragments spanning the entire coding region of the NDUFV1 (MIM 161015), NDUFS8 (MIM 602141), NDUFS7 (MIM 601825), NDUFS1 (MIM 157655), NDUFA8 (MIM 603359), and NDUFB6 (MIM 603322) genes. Table 2 shows the sequence of primers as well as PCR and D-HPLC conditions. Amplification products were mixed with control PCR products. This mixture was denatured for 10 min at 96°C and then gradually reannealed by decreasing sample temperature from 96°C to 30°C. The annealed specimens (4–7 μl) were loaded onto a DNA sep column (Transgenomic) by means of a Dynamax automatic sample injector model AI-1A. PCR products were then separated over a 4.5-min period through a linear acetonitrile gradient (flow rate: 0.9 ml/min), the values of which were based on the size and G/C content of the PCR products (table 2). The column mobile phase consisted of a mixture of 0.1 M triethylamine acetate (TEEA), pH 7.0 with (buffer A) or without 25% acetonitrile (buffer B). The mobile-phase temperatures required for optimal resolution of homoduplexes were empirically determined by injecting the product of each amplified fragment at increasing temperatures until a significant decrease in sample retention time was observed. The temperatures resulting in 75% of double-strand DNA in the melting profile curve were considered as the optimal ones. Specific values for the gradient ranges and mobile-phase temperatures used are reported in table 2. PCR fragments displaying abnormal profiles were further characterized by direct sequencing using the BigDye terminator cycle sequencing kit (ABI Prism).
Table 2.
Oligonucleotides Used for D-HPLC and Sequence Analysis of Complex I Subunits
| PrimerDesignationa | Primer Sequences5′→ 3′ | Annealing Temperature(°C) | AcetonitrileGradient(% Buffer B) | Mobile-Phase Temperature(°C) |
| NDUFV1 (AF053070):b | ||||
| F1 Ex 1–4 | ATCGCGCCAGTTCCTCAGCCCTGCAGATTGGAGGCCTCATT | 60 | 59–68 | 64 |
| F2 Ex 4–7 | CGCGCTGCCTATATCTACATCCATGACGATCACCGCAGCTGT | 60 | 52–72 | 63 |
| F3 Ex 7–10 | CTGGTGCAGGCACAGACAGGGTGGGCAGCACTCGCTTTATT | 60 | 58–67 | 65 |
| NDUFV1 (AF053069):c | ||||
| Int 4-F | AGTTATAGGCTGACTCCTGGG | 60 | … | … |
| Int 5-R | TAGCCAGATCCCGGGTGTCA | 60 | … | … |
| Ex 6-F | GTGGCCAACGTGGAGACAGT | 60 | … | … |
| Ex 6-R | TACCTCCACGGCGGCAGATT | 60 | … | … |
| Ex 7-F | CTGGTGCAGGCACAGACAGG | 60 | … | … |
| Ex 7-R | CATGACGATCACCGCAGCTGT | 60 | … | … |
| Int 7-F | GCTGAGGCCCAGGCTTCTGT | 60 | … | … |
| Int 8-R | GGGCTGCAGGCCACGTGG | 60 | … | … |
| Ex 9-F | GACTGGATGAACAAGGTGAT | 60 | … | … |
| Ex 10-R | CCGAAAGTGGCGGATCAGAC | 60 | … | … |
| Ex 10-R.2 | GTGGGCAGCACTCGCTTTATTG | 60 | … | … |
| NDUFS1 (NM005006):b | ||||
| F1 Ex 1–7 | TAGCACAACACCCTCCGCGGAATCGGCTCCTATGATTTCCAAA | 58 | 55–64 | 60 |
| F2 Ex 6–10 | AATCACCCATTGGACTGTCCTGGCAAATCTGGTTTTATCAGA | 62 | 58–67 | 59 |
| F3 Ex 9–13 | AGAACTGGAGAAGTGATGAGGATAAGTCATTATGCAGGGAGCTC | 61 [5 mM MgCl2] | 56–65 | 59 |
| F4 Ex 12–15 | GGCAGATGTTGTTCTTCTGGTTTCCATCTGCTCCCAGGAGAAA | 60 [2.5% DMSO] | 59–68 | 59 |
| F5 Ex 15–19 | CTTGGCTATAAGCCTGGGGTGAACTGGGATCCTAGTAGAAGCT | 60 | 59–68 | 59 |
| NDUFS1 (AC007383):d | ||||
| Ex 2 | CCTACAATAATATAATTCTTTGTAAGGTCTAATATCCACGAATGC | 56 | … | … |
| Ex 3 | ATAATAAACTGTTAATCTGTTTTTCGATTTACTAATAATCAGTATAAAGGAAA | 56 | … | … |
| Ex 4 | ATAACAGCAGTGAAGTCAGGTTATTAGTTTGCTGCAAGATTTAAAGTTTT | 60 | … | … |
| Ex 5 | TTGTAATCACAAATAAATACAAATACATAAATTCTTAATCTATGGGAAGGTC | 56 | … | … |
| Ex 6–7 | GTCAGTGTGTGTGTATTTGAGAGATTGTGACTATTAAGTATACAATTATA | 60 | … | … |
| Ex 8 | GGACTTGACCATTGATGACACTTGAATAGTAATGAATTAAGGACAA | 60 | … | … |
| Ex 9 | AGCAACCATAGTAAACACCGTATACTGTAAAAGTTAAGGAAAACAATA | 60 | … | … |
| Ex 10 | TATAGTTACTTCTTTAGCAAGATTCCTTGATAATCACAAACACTATTTAAC | 60 | … | … |
| Ex 11 | TGCTGCGTGTAGGTTTCTTGGACATTTTATACATAACTTGTAACTAA | 60 | … | … |
| Ex 12 | ATGGATTCTCTGTATGTCTTAATTCACTACCACTAACACTATTAGGA | 60 | … | … |
| Ex 13 | GTAAATAGATGTATCACTTAGGATTCAACAAGAGTAGATACACATAAG | 62 | … | … |
| Ex 14 | ATATGGTTTGGTGTTCAGCTTACACATATACACAACATTACTTGAATT | 62 | … | … |
| Ex 15 | ACATTGAAAATACTGCAGTTATGGCTAACATACATAATGTACTACTCTC | 60 | … | … |
| Ex 16 | GATATATACTATCTTCATGTACAGTAGAGGCTAAAATATCAAATATGCCTTTA | 60 | … | … |
| Ex 17–18 | TGTTGTTTTTGGGGACCTGACACTTACCTTTCGTATTTGGCAGAG | 60 | … | … |
| Ex 19 | AGTTGTATTATCAAAATGACATTTTCAACTGGGATCCTAGTAGAAGCT | 60 | … | … |
| NDUFS8 (U65579):c | ||||
| TGGCCGAATGGCAGCGTCCTCGATGTCATAGCGGGTGGTCC | 64 | 56–65 | 67 | |
| CATTGCCTGCAAGCTCTGCGTTTTATTGGGCAGCAGGGGCT | 66 | 59–68 | 65 | |
| NDUFS7 (AC005329):c | ||||
| CTGAAGGCCGAGGCCAAGGTTGACGAGGTCATCCAG | 60 | 52–61 | 68 | |
| AGAGCCGTGGCTCCCAAACCATGTCCACGGGCACGATGCG | 60 | 54–63 | 67 | |
| CTATTCCTACTCGGTGGTCCTCACGGGACACAAGCA | 60 | 51–60 | 68 | |
| NDUFA8 (NM014222):c | ||||
| GGGAGTTCAAGGAGACGGGGGCAATGCAAGTCCAATATTCTG | 60 | 54–63 | 62 | |
| TTAGGCAGATAAAACGTCACTCTGAGCACATGACCGAGTGTGG | 60 | 55–64 | 62 | |
| NDUFB6 (NM002493):c | ||||
| GGTGCGCCGTGTCCTTTTTGGTAATATAGGAACAAACTT | 60 | 57–66 | 57 |
GenBank accession numbers are indicated.
cDNA sequence analysis.
Genomic sequence analysis, performed by means of nested primers.
Genomic sequence analysis.
For genotyping, microsatellite markers of the Généthon database were used (Dib et al. 1996). PCR products were loaded on a polyacrylamide gel and transferred onto a positive nylon membrane (Nytran plus, Schleicher and Schuell). Membranes were hybridized with a (CA)12 probe. The labeling of the probe and the revelation of the blots were performed by means of the ECLTM direct nucleic acid labeling and detection system (Amersham Pharmacia Biotech). The blots were autoradiographed with Kodak-X-OMAT films for 2–30 min.
Results
NDUFV1 Gene Mutations
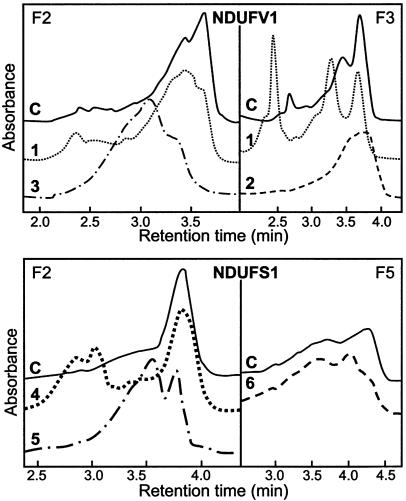
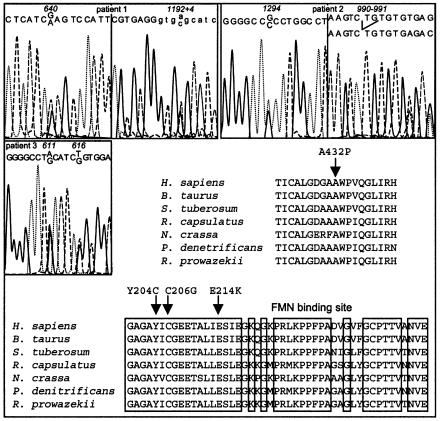
The entire coding sequence of the NDUFV1, NDUFS8, NDUFS7, NDUFS1, NDUFA8, and NDUFB6 genes was submitted to D-HPLC analysis in 36 unrelated complex I–deficient patients. Abnormal D-HPLC patterns in the NDUFV1 gene (i.e., additional peaks caused by the reduced retention time of heteroduplex DNAs) were found in 3/36 patients (fig. 1). In patient 1, a seemingly homozygous G→A transition at nucleotide (nt) 640 in exon 5 changed a highly conserved glutamic acid into a lysine (E214K inherited from the father, fig. 2, nt 1 is the “A” of the ATG translation initiation codon). Since the parents were unrelated, we sequenced the NDUFV1 genomic DNA of the patient and found that he was in fact compound heterozygous for the E214K mutation and had an A→C transversion in the donor splice-site of intron 8, inherited from the mother (fig. 2), resulting in the skipping of exon 8 and in an unstable RNA (not shown). In patient 2, a seemingly homozygous G→C substitution in exon 9 at nt 1294 of the cDNA changed a highly conserved alanine into a proline (A432P, fig. 2). Again, the patient was in fact compound heterozygous for the A432P mutation inherited from the father and a 2-bp deletion in exon 7 inherited from the mother, resulting in a frameshift and a premature stop codon (fig. 2). This mRNA was unstable, as it was almost undetectable at the cDNA level (not shown). In patient 3, sequencing the abnormal PCR fragment detected compound heterozygosity for two exon 5 mutations. An A→G transition at nt 611 inherited from the father changed a tyrosine into a cysteine in the NDUFV1 protein (Y204C; fig. 2). The second mutation, a T→G transversion at nt 616, inherited from the mother, changed a cysteine into a glycine (C206G; fig. 2). None of the missense mutations identified in the patients were found in 100 controls.
Figure 1.
Abnormal D-HPLC patterns of NDUFV1 and NDUFS1 RT-PCR products. “F2,” “F3,” and “F5” refer to the different PCR fragments (table 2). C = control; 1–5 = patients 1–5.
Figure 2.
Molecular analysis of the NDUFV1 gene. Sequence analysis was performed on genomic DNA for patients 1 and 2, and on cDNA derived from skin fibroblast for patient 3. Sequence alignment of the NDUFV1 proteins from various species is presented. Boxes indicate the FMN binding site. Arrows show the mutated amino acids (patient 1: E214K; patient 2: A432P; patient 3: Y204C and C206G).
Interestingly, all three NDUFV1 mutations (Y204C, C206G, and E214K) altered the FMN binding site of the NDUFV1 protein (Fearnley and Walker 1992) (fig. 2). Yet, the patients failed to express complex I deficiency in their cultured skin fibroblasts, since pyruvate-malate and glutamate-malate oxidations were in the normal range (not shown). We hypothesized that the high concentration of riboflavin (the FMN precursor) in the culture medium might have restored normal complex I activity, and we grew the patients' skin fibroblasts in Rosswell Park Memorial Institute riboflavin-free medium supplemented with 10% fetal-calf serum. However, we failed to reveal an enzyme deficiency in cultured cells in these conditions (not shown), presumably caused by the presence of traces of riboflavin in the serum, but we still decided to put patient 3, the only patient alive, on a riboflavin treatment (60 mg/day). At age 10 mo, his severe ptosis had improved on a low-carbohydrate diet. At age 2 years, he received oral idebenone for 7 mo, followed by a 3-mo riboflavin treatment, which resulted in the disappearance of his ptosis and ophthalmoplegia and the recovery of a normal growth. However, ataxia and pyramidal syndrome persisted. The boy is now 3 years of age; he is unable to walk unaided and is still mentally retarded.
NDUFS1 Gene Mutations
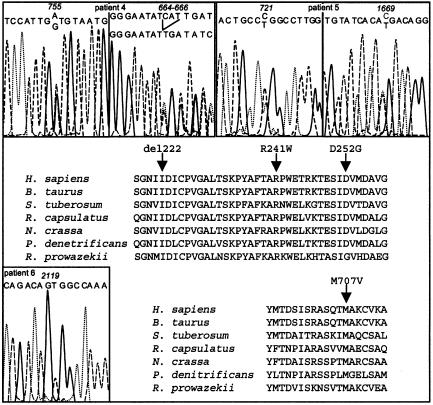
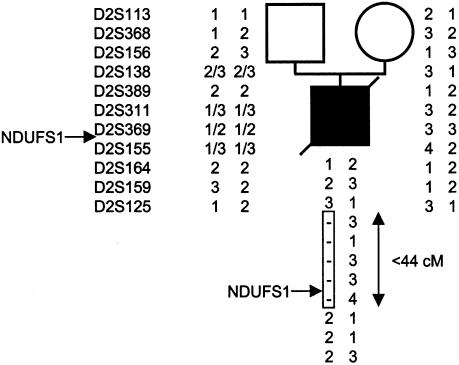
Abnormal D-HPLC patterns in the NDUFS1 gene were found in 3/36 patients (fig. 1). Sequencing the abnormal PCR fragments revealed that patient 4 was compound heterozygous for a 3-bp deletion in exon 8 at nt 664–666, resulting in an in-frame codon 222 deletion (fig. 3) and an A→G substitution at nt 755 in exon 9, changing a highly conserved asparagine into a glycine in the protein (D252G; fig. 2). In patient 5, a seemingly homozygous C→T transition at nt 721 of the cDNA changed a highly conserved arginine into a tryptophan (R241W; fig. 3). We determined the exon-intron boundaries of the gene by combining BLAST searching with the cDNA sequence and PCR-sequence analysis, which allowedus to identify a genomic clone containing the entire NDUFS1 coding sequence. Primers used for exon amplification on genomic DNA are presented in table 2. The patient was, in fact, compound heterozygous for the R241W and a R557X mutation (inherited, respectively, from the father and the mother) detected on genomic DNA (fig. 3). In patient 6, an apparently homozygote A→G transition at nt 2119 in exon 19 changed a highly conserved methionine into a valine (M707V; fig. 3). His mother was heterozygous for the M707V mutation. Surprisingly however, his father was wild-type homozygous. Segregation analysis of microsatellite markers flanking the NDUFS1 gene on chromosome 2q revealed that patient 6 failed to inherit polymorphic markers from his father at loci D2S138–D2S155 (44 cM), providing evidence of a de novo deletion of the paternal NDUFS1 allele (fig. 4).
Figure 3.
Molecular analysis of the NDUFS1 gene. Sequence analysis was performed on cDNA for patient 4 and on genomic DNA for patients 5 and 6. Sequence alignment of the NDUFS1 proteins from various species is presented. Arrows show the mutated amino acids (patient 4: D252G and del 222; patient 5: R241W; patient 6: M707V).
Figure 4.
Haplotype analysis of family 6. Haplotypes are given (top to bottom) for loci D2S113, D2S368, D2S156, D2S138, D2S389, D2S311, D2S369, D2S155, D3S164, D2S159, and D2S125. The box indicates the deleted region between 2q32.1 and q34.
No mutations could be identified in the NDUFS8, NDUFS7, NDUFA8, and NDUFB6 genes. In several patients, neutral polymorphisms were detected either in the coding or in the noncoding regions of the genes (not shown).
Discussion
In this article, we report on a large-scale NDUFS1 deletion and 11 novel mutations in the NDUFV1 and NDUFS1 genes of complex I in 6/36 unrelated complex I–deficient children. All the mutations reported in this study involved highly conserved amino acids, and three of them altered the FMN binding site of NDUFV1, possibly hampering FMN binding to complex I and reducing electron transfer through the respiratory chain. Nonsense mutations have been reported elsewhere in the NDUFV1, NDUFS8, NDUFS7, and NDUFS4 genes (Schuelke et al. 1999; Smeitink et al. 1999; Loeffen et al. 2000). To our knowledge, however, neither NDUFS1 mutations nor nuclear DNA deletion have been reported in respiratory-chain deficiency. This study not only demonstrates the autosomal recessive mode of inheritance of the disease in most patients but also suggests that de novo deletions can be occasionally involved. The direct measurement of complex I in cultured skin fibroblasts is hampered by a contaminating microsomal NADH cytochrome c reductase activity. This feature prevents the reliable quantitation of complex I deficiency in cultured fibroblasts. For this reason, only polarographic assays of pyruvate or malate oxidation were performed to estimate the activity of NADH oxidation by complex I, and only fibroblasts showing severely decreased pyruvate or malate oxidation can be reliably considered as complex I deficient. As enzymological diagnosis of complex I deficiency in chorionic villi or amniotic fluid is hazardous, the identification of the mutant genotypes allowed us to offer prenatal diagnosis for the next pregnancies (Amiel et al., in press). In mutations altering the FMN binding site, the presence of the FMN precursor (riboflavin) in the culture medium might have restored a normal complex I activity.
Several of the complex I 43 subunits are highly conserved across species, from mammals to bacteria (Fearnley and Walker 1992). This high degree of conservation suggests that these subunits play a key role in catalytic activity, especially as they encompass the NADH, FMN, or Fe-S binding sites. The most conserved subunits and those of functional importance for electron transfer and substrate oxidation represent, therefore, obvious candidates for other cases of complex I deficiency, but the large number of genes possibly underlying complex I deficiency requires rapid mutation-screening techniques such as D-HPLC or microarrays. Until now, complex I deficiency has only been ascribed to nuclear genes encoding structural complex I subunits. However, it is conceivable that complex I deficiency might result from an abnormal complex I subunit assembly, as observed in complex IV deficiency (Tiranti et al. 1998; Zhu et al. 1998; Valnot et al. 2000a, 2000b). Until now, only two Neurospora crassa genes encoding chaperones have been shown to govern assembly of complex I (Kuffner et al. 1998). The human homologues of these genes will represent obvious candidate genes for complex I deficiency.
Although complex I subunit genes are ubiquitously expressed, the patients reported in this study presented mostly neurological disorders—namely, hypotonia, ataxia, psychomotor retardation, or Leigh syndrome. It has been claimed that the variable clinical expression of mitochondrial disorders could be ascribed to mitochondrial DNA mutations, the expression of the deficiency paralleling the level of heteroplasmy in the corresponding tissue (Munnich et al. 2001). Yet, an increasing number of ubiquitously expressed nuclear genes have been shown to account for mitochondrial disorders affecting specific organs. Indeed, thymidine phosphorylase, SURF1, SCO1, SCO2, COX10, frataxin, DDP, OPA1, and ABC7, which are all housekeeping genes, are involved in MNGIE (Nishino et al. 1999), Leigh syndrome (Tiranti et al. 1998; Zhu et al. 1998), hepatic failure (Valnot et al. 2000a, 2000b), cardioencephalomyopathy (Papadopoulou et al. 1999), tubulopathy (Valnot et al. 2000a, 2000b), Friedreich ataxia (Campuzano et al. 1996), deafness-dystonia syndrome (Koehler et al. 1999), optic atrophy (Delettre et al. 2000), and sideroblastic anemia-ataxia (Allikmets et al. 1999). This suggests that other as yet unknown mechanisms play an important role in the variable clinical expression of the disease. Noticeably, neither our patients nor those reported elsewhere presented Leber hereditary optic atrophy (LHON), whereas mitochondrial complex I gene mutations most often result in LHON (MITOMAP). The identification of other genes responsible for complex I deficiency will, it is hoped, help in the elucidation of the variable clinical expression of complex I deficiency. Still, the increasing number of disease genes will require fast and cost-effective mutation screening techniques for rapid identification of disease-causing mutations, genetic counseling, and prenatal diagnosis.
Acknowledgments
This research was supported, in part, by the Association Française contre les Myopathies (7145-AO99).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www2.ncbi.nlm.nih.gov/genbank/query_form.html (for NDUFV1 [AF053070, AF053069], NDUFS1 [NM005006, AC007383], NDUFS8 [U65579], NDUFS7 [AC005329], NDUFA8 [NM014222], and NDUFB6 [NM002493])
- Généthon, http://www.genethon.fr
- MITOMAP Human mitochondrial genome database, http://www.gen.emory.edu/mitomap.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NDUFV1 [MIM 161015], NDUFS1 [MIM 157655], NDUFS8 [MIM 602141], NDUFS7 [MIM 601825], NDUFA8 [MIM 603359], and NDUFB6 [MIM 603322])
References
- Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM (1999) Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum Mol Genet 8:743–749 [DOI] [PubMed] [Google Scholar]
- Amiel J, Gigarel N, Benacki A, Bénit P, Valnot I, Parfait B, von Kleist-Retzow JC, Raclin V, Hadj-Rabia S, Dumez Y, Rustin P, Bonnefont JP, Munnich A, Rötig A. Prenatal diagnosis of respiratory chain deficiency by direct mutation screening. Prenatal Diagnosis (in press) [DOI] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423–1427 [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet 26:207–210 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Fearnley IM, Walker JE (1992) Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim Biophys Acta 1140:105–134 [DOI] [PubMed] [Google Scholar]
- Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR (1999) Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology 52:1255–1264 [DOI] [PubMed] [Google Scholar]
- Koehler CM, Leuenberger D, Merchant S, Renold A, Junne T, Schatz G (1999) Human deafness dystonia syndrome is a mitochondrial disease. Proc Natl Acad Sci USA 96:2141–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffner R, Rohr A, Schmiede A, Krull C, Schulte U (1998) Involvement of two novel chaperones in the assembly of mitochondrial NADH:Ubiquinone oxidoreductase (complex I). J Mol Biol 283:409–417 [DOI] [PubMed] [Google Scholar]
- Loeffen JL, Smeitink JA, Trijbels JM, Janssen AJ, Triepels RH, Sengers RC, van den Heuvel LP (2000) Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum Mutat 15:123–134 [DOI] [PubMed] [Google Scholar]
- Munnich A, Rötig A, Cormier V, Rustin P (2001) Clinical presentation of respiratory chain deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 2261–2274 [Google Scholar]
- Nishino I, Spinazzola A, Hirano M (1999) Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science 283:689–692 [DOI] [PubMed] [Google Scholar]
- Papadopoulou LC, Sue CM, Davidson MM, Tanji K, Nishino I, Sadlock JE, Krishna S, Walker W, Selby J, Glerum DM, Coster RV, Lyon G, Scalais E, Lebel R, Kaplan P, Shanske S, De Vivo DC, Bonilla E, Hirano M, DiMauro S, Schon EA (1999) Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet 23:333–337 [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, Gérard B, Rötig A, Saudubray JM, Munnich A (1994) Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta 228:35–51 [DOI] [PubMed] [Google Scholar]
- Schuelke M, Smeitink J, Mariman E, Loeffen J, Plecko B, Trijbels F, Stockler-Ipsiroglu S, van den Heuvel L (1999) Mutant NDUFV1 subunit of mitochondrial complex I causes leukodystrophy and myoclonic epilepsy. Nat Genet 21:260–261 [DOI] [PubMed] [Google Scholar]
- Smeitink J, van den Heuvel L (1999) Human mitochondrial complex I in health and disease. Am J Hum Genet 64:1505–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez JA, Uziel G, Bertini E, Dionisi-Vici C, Franco B, Meitinger T, Zeviani M (1998) Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am J Hum Genet 63:1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnot I, Osmond S, Gigarel N, Mehaye B, Amiel J, Cormier-Daire V, Munnich A, Bonnefont JP, Rustin P, Rötig A (2000a) Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase (COX) deficiency with neonatal-onset hepatic failure and encephalopathy. Am J Hum Genet 67:1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rötig A (2000b) A mutation in the human heme A:farnesyl transferase gene (COX10) causes cytochrome c oxidase deficiency. Hum Mol Genet 9:1245–1249 [DOI] [PubMed] [Google Scholar]
- von Kleist-Retzow JC, Cormier-Daire V, de Lonlay P, Parfait B, Chretien D, Rustin P, Feingold J, Rötig A, Munnich A (1998) A high rate (20%–30%) of parental consanguinity in cytochrome oxidase deficiency. Am J Hum Genet 63:428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 20:337–343 [DOI] [PubMed] [Google Scholar]