Abstract
To obtain more information of the functional domains of the NPC1 protein, the mutational spectrum and the level of immunoreactive protein were investigated in skin fibroblasts from 30 unrelated patients with Niemann-Pick C1 disease. Nine of them were characterized by mild alterations of cellular cholesterol transport (the “variant” biochemical phenotype). The mutations showed a wide distribution to nearly all NPC1 domains, with a cluster (11/32) in a conserved NPC1 cysteine-rich luminal loop. Homozygous mutations in 14 patients and a phenotypically defined allele, combined with a new mutation, in a further 10 patients allowed genotype/phenotype correlations. Premature-termination–codon mutations, the three missense mutations in the sterol-sensing domain (SSD), and A1054T in the cysteine-rich luminal loop all occurred in patients with infantile neurological onset and “classic” (severe) cholesterol-trafficking alterations. By western blot, NPC1 protein was undetectable in the SSD missense mutations studied (L724P and Q775P) and essentially was absent in the A1054T missense allele. Our results thus enhance the functional significance of the SSD and demonstrate a correlation between the absence of NPC1 protein and the most severe neurological form. In the remaining missense mutations studied, corresponding to other disease presentations (including two adults with nonneurological disease), NPC1 protein was present in significant amounts of normal size, without clear-cut correlation with either the clinical phenotype or the “classic”/“variant” biochemical phenotype. Missense mutations in the cysteine-rich luminal loop resulted in a wide array of clinical and biochemical phenotypes. Remarkably, all five mutant alleles (I943M, V950M, G986S, G992R, and the recurrent P1007A) definitively correlated with the “variant” phenotype clustered within this loop, providing new insight on the functional complexity of the latter domain.
Introduction
Niemann-Pick type C (NPC [MIM 257220]) disease is a neurodegenerative lysosomal lipid storage disease, with autosomal recessive transmission, characterized by lysosomal/late-endosomal accumulation of endocytosed unesterified cholesterol (Pentchev et al. 1995; Patterson et al. 2001). The clinical manifestations of NPC are heterogeneous. Most patients have a progressive neurologic disease, but both age at onset, which ranges from early infancy to adulthood, and subsequent course vary (Vanier and Suzuki 1996; Patterson et al. 2001). We have also documented a wide variation in severity of the cellular cholesterol lesion, with typical severe alterations described as the “classic” biochemical phenotype and mild alterations as the “variant” phenotype (Vanier et al. 1991b).
Although the exact location and nature of the trafficking defect(s) are still under investigation (Liscum and Munn 1999; Blanchette-Mackie 2000; Cruz and Chang 2000; Davies et al. 2000; Ory 2000), substantial advances have occurred in our knowledge of NPC. Cell-hybridization studies and linkage analysis (Steinberg et al. 1994; Vanier et al. 1996) demonstrated the existence of two complementation groups. NPC1, the disease-causing gene in >95% of NPC patients, has been located to 18q11-q12 and is now fully characterized (Carstea et al. 1997; Morris et al. 1999). Very recently, a previously identified gene, HE1, mapped to chromosome 14q24.3, was recognized as the gene mutated in the minor NPC2 complementation group (Naureckiene et al. 2000). The finding of identical clinical, cellular, and biochemical phenotypes in patients belonging to the two groups (Vanier et al. 1996; Christomanou et al. 2000) led to the conclusion that both gene products may function in tandem or sequentially (Vanier et al. 1996; Carstea et al. 1997).
The NPC1 cDNA sequence (see Genbank) predicts a protein of 1,278 amino acids. Topological analysis of NPC1 has revealed that the protein contains 13 transmembrane domains, three large and four small luminal loops, six small cytoplasmic loops, and a cytoplasmic tail (Davies and Ioannou 2000). The region located between amino acid residues 615 and 797 shows strong homology to the sterol-sensing domain (SSD) identified in several other integral membrane proteins that respond to endoplasmic reticulum cholesterol (Carstea et al. 1997). This putative SSD has the same orientation as those in HMG-CoA reductase and SCAP (sterol regulatory element binding protein [SREBP] cleavage activation protein) (Davies and Ioannou 2000) and appears to have important functional significance (Watari et al. 1999). The mature protein appears glycosylated, with a size of 170–190 kilodaltons (kD) (Higgins et al. 1999; Watari et al. 1999), and there is convincing evidence that NPC1 resides in late endosomes and interacts transiently with lysosomes and the trans-Golgi network (Higgins et al. 1999; Neufeld et al. 1999). A number of studies point toward a key role for the NPC1 protein in modulating vesicular trafficking of cholesterol and of glycolipids (Neufeld et al. 1999; Blanchette-Mackie 2000; Ory 2000; Zhang et al. 2001), but recent data suggest that NPC1 is a permease acting as a transmembrane efflux pump (Davies et al. 2000).
There are still only limited published studies on NPC1 mutations (Carstea et al. 1997; Greer et al. 1998, 1999; Millat et al. 1999; Yamamoto et al. 1999, 2000). The mutation prevalent in patients with Acadian Nova Scotian origins (Niemann-Pick type D) was identified as G992W (Greer et al. 1998), and that prevalent in Hispanic patients from the Upper Rio Grande area of the United States was identified as I1061T (Millat et al. 1999). The latter allele was further shown to be the most common NPC1 mutation among individuals of western European descent (Millat et al. 1999). Greer et al. (1999) pointed out that a majority of mutations were clustered in a luminal cysteine-rich loop, highlighting the potential functional importance of this domain. On the other hand, the study by Yamamoto et al. (2000), including mutational and immunoblotting studies, as well as information on the clinical presentation, disclosed a much wider distribution of the mutations. Preliminary results from the present study were presented in abstract form (Vanier et al. 1999; Vanier and Millat 2000).
Identification of mutations and studies of the mutated protein in cultured skin fibroblasts of selected patients with mutations in NPC1 was initiated primarily to obtain more information on functional domains of the NPC1 protein. Although we attempted to cover the entire spectrum of clinical and biochemical phenotypes, we were particularly interested in studying two subsets of patients. Children with infantile neurological onset invariably showed pronounced cellular cholesterol abnormalities (Vanier et al. 1988, 1991a); they clearly corresponded to the most severe form of the disease and therefore were best suited to pinpoint the most critical regions of the protein. Among patients with other clinical phenotypes, a majority share the same “classic” biochemical pattern, but about one-fifth show moderate alterations of cellular cholesterol trafficking, described as the “variant phenotype” (Vanier et al. 1991b). The biochemical subtype has been shown to be constant within a sibship (Vanier et al. 1991b) and thus is most likely defined by the genotype. Whether the variant phenotype is correlated with alterations of a specific domain of the protein is not yet known. To address this question, we have studied patients with late onset showing either severe or mild cholesterol-trafficking abnormalities. Since no biochemical marker has been found, so far, to correlate with a given clinical phenotype, the second aim of this study has been to evaluate correlations between genotype and clinical phenotype. Overall, our findings allow assignment of existing or new allelic combinations to specific phenotypes and provide new information on the functional significance of two particular domains of the NPC1 protein.
Subjects and Methods
Subjects
Cultured skin fibroblasts were obtained from 30 unrelated NPC patients from various ethnic backgrounds, including French (13), German (4), British (4), Dutch (1), Gypsy (1), Tunisian (3), Turkish (2), Pakistani (1), and African American (1). Close consanguinity was certain or likely in 10 families. The methods used for diagnosis by evaluation of cellular cholesterol by filipin staining and of LDL-induced cholesteryl ester formation were those described by Vanier et al. (1991b). Genetic-complementation analysis was performed prior to molecular studies to ensure that all patients belonged to the main (NPC1) complementation group (Vanier et al. 1996). Classification of patients with respect to their clinical and biochemical characteristics (table 1) was as proposed by us elsewhere (Vanier et al. 1988, 1991a, 1991b; Vanier and Suzuki 1996; Millat et al. 1999). In summary, patients with neurological symptoms were categorized, by type and age at onset of first neurological symptoms, as having either a severe infantile form (onset at age <2 years), a late infantile form (onset at age 3–5 years), a juvenile form (onset at age >5–16 years), or an adult form (onset at age >16 years). The denomination “rapidly fatal cholestatic form” was applied to the disease of patients who died from liver failure in the first months of life. On the basis of the severity of impairment in intracellular cholesterol processing in cultured fibroblasts (Vanier et al. 1991b), patients were also classified into either a classic or a variant biochemical phenotype. The classic phenotype refers to patients with a striking accumulation of free cholesterol in lysosomes assessed by filipin staining, together with a severe block in LDL-induced cholesteryl ester formation. In cells from biochemically variant patients, cholesterol storage may be evidenced only after challenge with pure LDL, and the rate of cholesteryl ester formation may not be affected. Cultured skin fibroblasts or peripheral blood samples were also available from the parents of 11 patients. Genomic DNA was obtained from a control population of 95 unrelated unaffected subjects and from 30 additional patients with mutations in NPC1, of whom 24 showed a variant biochemical phenotype.
Table 1.
Clinical Summary of the 30 Families Included in the Study[Note]
| Family | CellLineCode | BiochemicalPhenotypea | ClinicalPhenotypeb | Age atDeath(years) | Age atLastFollow-up(years) | Ethnicity | Consanguinity | ParentsAvailable | Reference |
| 1 | 98119 | Classic | Infantile | 2.83 | African American | No | No | Dawson et al. 1971 | |
| 2 | 80001 | Classic | A: Infantile | 3.83 | Gypsy | Yes | No | Vanier et al. 1988, case 2 | |
| B: Infantile | 3.33 | ||||||||
| C: Cholestatic | .30 | ||||||||
| 3 | 87024 | Classic | A: Infantile | 2.58 | Tunisian | Yes | Yes | Kanoun et al. 1989 | |
| B: Cholestatic | .25 | ||||||||
| 4 | 91029 | Classic | A: Infantile | 5.16 | French | Yes | Yes | ||
| B: Infantile | 4.92 | ||||||||
| 5 | 81057 | Classic | A: Cholestatic | .04 | French | Yes | No | Vanier et al. 1988, case 10 | |
| B: Infantile | 3.16 | ||||||||
| 6 | 83024 | Classic | Infantile | 5.08 | Turkish | Likely | No | ||
| 7 | 83049 | Classic | Infantile | 5.33 | French/West Indies | No | No | Vanier et al. 1988, case 13 | |
| 8 | 82052 | Classic | Infantile | 3.5 | French | No | No | Vanier et al. 1988, case 8 Vanier et al. 1999 | |
| 9 | 79011 | Classic | Infantile | 5.0 | Tunisian | No | No | Vanier et al. 1988, case 14 | |
| 10 | 96115 | Classic | Severe late infantile | 4.50 | French/West Indies | No | Yes | ||
| 11 | 82042 | Classic | Late infantile | 5.75 | French | No | No | Vanier et al. 1988, case 16 | |
| 12 | 95085 | Classic | A: No neurological signs (cardiac problem) | 1.00 | Pakistani | Yes | No | ||
| B: Late infantile | 6.00 | ||||||||
| C: Late infantile | 5.75 | ||||||||
| 13 | 90104 | Variant | Late Infantile | 10 | German | No | Yes | ||
| 14 | 93094 | Classic | A: Late infantile | 11.66 | French | No | Yes | Millat et al. 1999, family 8 | |
| B: Late infantile | 8.00 | ||||||||
| 15 | 91048 | Classic | A: Juvenile | 24 | French | No | Yes | Millat et al. 1999, family 1 | |
| B: Juvenile | 20 | ||||||||
| C: Juvenile | 20 | ||||||||
| D: Juvenile | 16 | ||||||||
| 16 | 98009 | Classic | Juvenile | 32 | French | No | No | Millat et al. 1999, family 15 | |
| 17 | 90096 | Variant | A: Juvenile | 18 | British | No | Yes | ||
| B: Juvenile | 24 | ||||||||
| 18 | 94139 | Variant | Juvenile | 25 | British | No | No | ||
| 19 | 92114 | Variant | A: Juvenile | 28 | German | No | No | ||
| B: Juvenile | 22 | ||||||||
| 20 | 95070 | Variant | Juvenile | 13 | British | No | No | ||
| 21 | 97089 | Variant | Juvenile | 20 | Dutch | Yes | Yes | ||
| 22 | 96017 | Variant | Adult | 43.5 | French | No | Yes | ||
| 23 | 96157 | Classic | A: Adult | 37 | German | No | No | ||
| B: Adult | 41 | ||||||||
| 24 | 90002 | Variant | Adult without neurological symptoms (simple psychic structure) | 66 | German | Yes | No | Fröhlich et al. 1990 | |
| 25 | 97014 | Classic | Adult without neurological symptoms | 50 | British | No | No | Fensom et al. 1999 | |
| 26 | 96084 | Classic | No neurological symptoms | 7.5 | French | No | Yes | ||
| 27 | 91144 | Variant | No neurological symptoms | 9.0 | French | No | Yes | ||
| 28 | 86065 | Classic | Cholestatic | 1.08 | Tunisian | Yes | No | Vanier et al. 1988, case 39 | |
| 29 | 97114 | Classic | Cholestatic | .25 | Turkish | Yes | Baumkötter et al. 1998 | ||
| 30 | 97119 | Classic | Cholestatic | .5 | French | No | No |
Note.— Families 1–25 are listed in order of decreasing severity of neurological phenotypes. Patients 26–30 could not be classified neurologically (too limited follow-up or early death resulting from liver failure).
Defined by the degree of severity of alterations of intracellular cholesterol processing (Vanier et al. 1991b).
Classification of clinical phenotypes by age at onset of neurological symptoms (except for the cholestatic rapidly fatal form) (Vanier and Suzuki 1996).
DNA and RNA Isolation
Genomic DNA was isolated from skin fibroblasts or from whole blood, according to the protocol of Jeanpierre et al. (1987). Total RNA was extracted from fibroblasts monolayers using the Trizol reagent (Gibco BRL).
Reverse-Transcription and SSCP Analysis
Reverse transcription was performed using the First-Strand cDNA synthesis kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions. NPC1 cDNA was amplified in 18 overlapping fragments, using primers described in table 2. Each PCR was performed in a 20-μl reaction volume containing 0.4 μl of cDNA, 200 μM of each dNTP, 1.5 mM MgCl2, PCR buffer, 1 U of EurobioTaq, 4 pmol of each primer, 0.1 μl α[33P]dATP, and 3,000 Ci/mmol (Amersham Pharmacia Biotech). Two microliters of the radiolabeled PCR product were mixed with 15 μl stop solution (95% formamide, 10 mM NaOH, 0.25% bromophenol blue, and 0.25% xylene cyanol) and denatured for 5 min at 95°C. The sample was loaded on 0.5× MDE gels (Biowhittaker Molecular Applications), with or without 10% glycerol. Electrophoresis was performed overnight in 0.6× TBE running buffer, either at 20 W at 4°C or at 6W at room temperature.
Table 2.
Sequence of Primers Used for RT-PCR
| Primer | Sequence (5′→3′) | cDNACoordinatesa |
| NPC-As | GAG CCC AAC CAG CCG AAC | −51/−34 |
| NPC-Aas | CCG AAC ATC ACA ACA GAG ACT GAC | 234/211 |
| NPC-Bs | CCA TTG CCA AAG GAT GGA TAT G | 148/169 |
| NPC-Bas | GCC TTG TCA TTA CTT GAG GGG G | 515/494 |
| NPC-Cs | CTA CGT CGG ACA GAG TTT TGC C | 438/459 |
| NPC-Cas | GCT ACA TGG TGC TGT GAC CTC ATC | 717/694 |
| NPC-Ds | CAA TGG ACA GGC ACC TTT TAC C | 591/612 |
| NPC-Das | CCG TTT TCT GTA GCA CCA CAC TG | 888/866 |
| NPC-Es | GCT TGG ACG CCA TGT ATG TCA TC | 791/813 |
| NPC-Eas | CCT GAC GAA CAC GCA GTA ATG AAG | 1,100/1,077 |
| NPC-Fs | CAG CAT TTG AGG GCT GCT TG | 986/1,005 |
| NPC-Fas | AAG CGG AGG TCC AAA GGG TAC ATC | 1,305/1,282 |
| NPC-Gs | CAA CCA ATC CAG TTG ACC TCT GG | 1,121/1,143 |
| NPC-Gas | AGC ACG GAA TGG CTG TTC TG | 1,484/1,465 |
| NPC-Hs | CAC GAA CTG CAC CAT TTT GAG TG | 1,428/1,450 |
| NPC-Has | TTG ACA GGG AAG GTA ATC ACA AGG | 1,703/1,680 |
| NPC-Is | TGT GTT GGG AGG CTA TGA TGA TC | 1,635/1,657 |
| NPC-Ias | CCT GCG ACA GCT TTT GAT GTG | 1,941/1,921 |
| NPC-Js | GTG ACA GTG ATG TCT TCA CCG TTG | 1,850/1,873 |
| NPC-Jas | AGG ATG ACA GGA ACA TAC TGG GAG | 2,218/2,195 |
| NPC-Ks | CCA GAG AGA TGA ACG TCT TCA AGG | 2,127/2,150 |
| NPC-Kas | CCC CAA GAG ACT CAC GAA ACA G | 2,352/2,331 |
| NPC-Ls | CGG GAT TGG CAG TCT TCA TTG | 2,291/2,311 |
| NPC-Las | TCC ACC ATG TAG GAG TCA TCT GG | 2,621/2,599 |
| NPC-Ms | AGC ATC GCA GTC CTG AAC AAA G | 2,545/2,566 |
| NPC-Mas | GAT ATT GTC CAC TCG ACA GCA AGA C | 2,886/2,862 |
| NPC-Ns | TTT CGA CTG GGT GAA GCC ACA G | 2,838/2,859 |
| NPC-Nas | GCG TCA ATA AAG TCA GCA GAG GTC | 3,161/3,138 |
| NPC-Os | AAG TGT GGC AAA GGG GGA CAT G | 3,028/3,049 |
| NPC-Oas | ACC GAG GTT GAA GAT AGT GTC GTC | 3,309/3,286 |
| NPC-Ps | ATA GCC AGT AAT GTC ACC GAA ACC | 3,181/3,204 |
| NPC-Pas | TTC ATG CTC ACC GTG AAC GCT C | 3,539/3,518 |
| NPC-Qs | AAC CTG GTG ATG AGC TGT GGC ATC | 3,466/3,489 |
| NPC-Qas | GTA TCG CTC TTC AGT GGC ACA AC | 3,801/3,779 |
| NPC-Rs | CTC CGT GTT CAG TGG AAT CAC AC | 3,588/3,610 |
| NPC-Ras | ACA CAG TTC AGT CAG GAT GCC C | 3,869/3,848 |
Counted from the adenosine residue of the initiation codon.
Mutation Analysis
Sequences of PCR products with aberrant SSCP patterns were determined on cDNA and on genomic DNA using the Thermosequenase cycle sequencing kit, with primers labeled with γ[33P]dATP, 3,000 Ci/mmol (Amersham Pharmacia Biotech). Because of the numerous known polymorphisms of the NPC1 gene (Morris et al. 1999; Yamamoto et al. 1999, 2000), for each new point mutation identified, genomic DNA from the patient and from 95 control samples was amplified using specific primers. PCR products were laid as a dot-blot on a nylon membrane (Hybond-N+, Amersham Pharmacia Biotech). Membranes were hybridized with either wild-type or mutated γ[33P]dATP labeled oligoprobes. Whenever possible (table 1), the mutation was also studied in the parents.
Detection of the P1007A Mutation by Introduction of an NheI Restriction Site
Genomic DNA was amplified using the sense primer 5′-TTGACCCTGCCTGCGTTC-3′ and the antisense primer 5′-CTTTGCCACACTTGGGGCT-3′. The 40 PCR cycles each included steps of 45 s at 92°C for denaturation, at 60°C for annealing, and at 72°C for extension. The 126-bp PCR product was digested for 2 h at 37°C by NheI (Roche Diagnostics) and was analyzed on a 12% polyacrylamide gel. The mutation gave rise to a 105-bp product.
Western Blot
Western blotting was performed as described by Yamamoto et al. (2000). Pellets from confluent fibroblast cultures (in MEM or DMEM, supplemented with 10% fetal calf serum) were homogenized and briefly sonicated on ice in buffer A (50 mM Tris (pH8.0), 150 mM NaCl, 100 μg/ml PMSF, and 1 μg/ml aprotinin). The lysates were centrifuged for 20 min at 100,000 rpm (+4°C). After centrifugation, the supernatants were discarded. The pellets were sonicated again with buffer A containing 0.4% SDS and were centrifuged for 20 min at 13,000 rpm. The protein content of the supernatants was determined by the Coomassie Plus Protein Assay Reagent Kit (Pierce). The lysates (10 μg protein) were subjected to 7.5% SDS/PAGE. After transfer overnight to nitrocellulose membranes (BioRad), NPC1 protein was detected with a polyclonal NPC1 rabbit antibody raised against a peptide corresponding to a 19–amino acid residues (1256–1274) peptide in the C-terminus of human NPC1, used at 1:500 dilution. Final detection was done by chemiluminescence using the ECL detection kit (Amersham Pharmacia Biotech).
Results
Spectrum of NPC1 Mutations
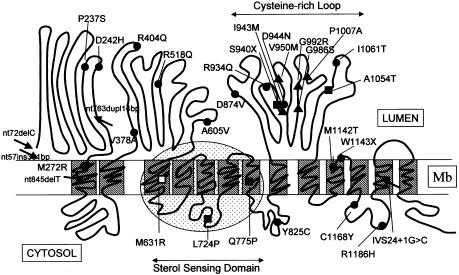
Analysis of cDNA and genomic DNA from 30 patients belonging to the NPC1 complementation group led to the identification of 32 different mutations, 26 of which were novel. A compendium of the mutant alleles is presented in table 3, and their location on a model of the NPC1 protein is represented schematically in figure 1. The largest number (25) of mutant alleles contained point mutations that led to single–amino acid substitutions. None of the novel substitutions was found in the control population of 95 individuals tested. Other abnormalities included two nonsense mutations, two 1-bp deletions, one 381-bp insertion, one 14-bp duplication, and one splice mutation. Mutations were widely scattered and affected all domains of the protein except for the NPC1 domain in the N-terminal end. Three of the five frameshift mutations were in the N-terminal luminal loop, leading to complete or near-complete truncation of the protein, and three missense mutations were located in the putative SSD. Among them, the Q775P allele was found to be homozygous in one family (5) and heterozygous in another (20). It was noteworthy that 11 (i.e., one-third) of the 32 identified mutations were located on the luminal cysteine-rich loop between transmembrane domains 8 and 9. Except for a nonsense mutation, these mutations all resulted in a single–amino acid substitution.Two different changes of the same nucleotide, G2974C (24) and G2974A (27) were observed, both resulting in the G992R substitution. Because of our particular selection of families, the I1061T allele, documented elsewhere as the most common NPC1 mutation (Millat et al. 1999), constituted only 7 of the 60 alleles, but P1007A (Greer et al. 1999) appeared as the second-most-recurrent allele (4/60) in this study.
Table 3.
Mutations in 30 Unrelated Patients with NPC1[Note]
|
Effect on Protein |
||||
| PatientIdentification(Phenotype) | Location | Nucleotide Changea | Amino Acid Change | Affected Domainb |
| 1 98119 (Cl) | Exon 2 | 72delC | Frameshift from codon 24c PTC+33 aa | Stop at the end of signal peptide |
| 72delC or deletion | ||||
| 2 80001 (Cl) | Exon 6 | 763dupl 14 bpc (GCCCCAGCCCCCAC) | Frameshift from codon 255c PTC+59 aa | Stop before TM1 |
| 3 87024 (Cl) | Exon 6 | 845delTc | Frameshift from codon 282cPTC+27 aa | Stop within TM 1 |
| 4 91029 (Cl) | Exon 19 | C2819Ac | S940Xc | Stop within the cysteine-rich luminal loop between TM 8 and 9 |
| 5 81057 (Cl) | Exon 15 | A2324Cc | Q775Pc | Sterol-sensing domain– TM7; residue conserved in SCAP |
| 6 83024 (Cl) | Exon 21 | G3160Ac | A1054Tc | Cysteine-rich luminal loop between TM 8 and 9 |
| 7 83049 (Cl) | Exon 6 | C709Td | P237S | N-terminal loop |
| Exon 8 | G1211A | R404Q | Luminal, between TM 2 and 3 | |
| 8 82052 (Cl) | Exon 12 | T1892G | M631R | TM3 |
| ? | ? | ? | ? | |
| 9 79011 (Cl) | Exon 9 | G1553Ad | R518Q+splicing mutation | Between TM 2 and 3 |
| ? | ? | ? | ? | |
| 10 96115 (Cl) | Exon 14 | T2171C | L724P | Sterol-sensing domain |
| Intron 24 | IVS24+1G→C | Frameshift after codon 1197 PTC+3 aa | Exon 24 skipping+frameshift | |
| 11 82042 (Cl) | Exon 23 | G3557Ae | R1186H | Between TM 11 and 12 |
| ? | ? | ? | ? | |
| 12 95085 (Cl) | Exon 23 | G3503Ac | C1168Yc | Between TM 11 and 12 (cytosolic loop) |
| 13 90104 (Va) | Exon 20 | C3019G | P1007A | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 22 | G3428A | W1143X | Between TM 11 and 12 | |
| 14 93094 (Cl) | Exon 21 | T3182C | I1061T | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 6 | G724C | D242H | N-terminal loop | |
| 15 91048 (Cl) | Exon 21 | T3182Cc,f | I1061Tc | Cysteine-rich luminal loop between TM 8 and 9 |
| 16 98009 (Cl) | Exon 21 | T3182C | I1061T | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 12 | C1814T | A605V | Loop between TM 2 and 3 | |
| 17 90096 (Va) | Exon 21 | T3182C | I1061T | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 20 | C3019Gg | P1007A | Cysteine-rich luminal loop between TM 8 and 9 | |
| 18 94139 (Va) | Exon 21 | T3182C | I1061T | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 20 | C3019Gg | P1007A | Cysteine-rich luminal loop between TM 8 and 9 | |
| 19 92114 (Va) | Exon 21 | T3182C | I1061T | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 20 | C3019Gg | P1007A | Cysteine-rich luminal loop between TM 8 and 9 | |
| 20 95070 (Va) | Exon 20 | G2956A | G986S | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 15 | A2324C | Q775P | Sterol-sensing domain – TM7 | |
| 21 97089 (Va) | Exon 19 | C2829Gc | I943Mc | Cysteine-rich luminal loop between TM 8 and 9 |
| 22 90002 (Va) | Exon 19 | G2848Ac | V950Mc | Cysteine-rich luminal loop between TM 8 and 9 |
| 23 96157 (Cl) | Exon 16 | A2474G | Y825C | Cytosolic loop between TM 7 and 8 |
| ? | ? | ? | ? | |
| 24 96157 (Va) | Exon 20 | G2974Cc | G992Rc | Cysteine-rich luminal loop between TM 8 and 9 |
| 25 97014 (Cl) | Exon 21 | T3182C | I1061T | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 8 | T1133C | V378A | Loop between TM 2 and TM3 | |
| 26 96084 (Cl) | Exon 18 | A2621Tc | D874Vc | Cysteine-rich luminal loop between TM 8 and 9 |
| 27 91144 (Va) | Exon 20 | G2974A | G992R | Cysteine-rich luminal loop between TM 8 and 9 |
| Junction exon 1/exon 2 | 57ins381bp | Frameshift from codon 19 PTC+19 aa | Stop within signal peptide | |
| 28 86065 (Cl) | Exon 19 | G2801Ac,g | R934Qc | Cysteine-rich luminal loop between TM 8 and 9 |
| 29 97114 (Cl) | Exon 6 | T815Gc | M272Rc | TM1 |
| 30 97119 (Cl) | Exon 19 | G2830A | D944N | Cysteine-rich luminal loop between TM 8 and 9 |
| Exon 22 | T3425C | M1142T | TM 10 | |
Note.— PTC = premature stop codon. TM = transmembrane domain. Cl = classic biochemical phenotype. Va = variant biochemical phenotype.
Counted from the adenosine residue of the initiation codon.
The putative spanning membrane domains of the NPC1 protein were determined according to Davies and Ioannou (2000).
Mutation was homozygous in the patient.
Yamamoto et al. 1999.
Carstea et al. 1997.
Millat et al. 1999.
Greer et al. 1999.
Figure 1.
Topology of the mutations identified in 30 patients on an NPC1 protein model and genotype/phenotype correlations. The schematic NPC1 protein model is drawn as proposed by Davis and Ioannou (2000). The hatched areas indicate the putative transmembrane domains. The dotted oval frame delimits the SSD. →, Frameshift mutations, all of them observed in patients with a severe infantile neurological form. ▪, Missense or nonsense mutation definitively associated to a severe infantile neurological form. □, missense mutation likely correlated with a severe infantile neurological form. ▴, Missense mutation definitively correlated with a variant biochemical phenotype. ●, Other missense mutation.
Correlations between Genotypes and the Classic or Variant Biochemical Phenotypes
One aim of our study was to pinpoint possible differences between mutations associated with the classic or the variant biochemical phenotypes (severe or mild impairment of cellular cholesterol trafficking, respectively) on the basis of the observation that the biochemical phenotype was constant within a sibship (Vanier et al. 1991a). In the homozygous state, all frameshift, splice or nonsense mutations and a number of missense mutations (M272R, Q775P, D874V, R934Q, A1054T, I1061T, and C1168Y) led to the classic biochemical phenotype (fig. 1 and table 3). This was also the case for several combinations of two missense mutations (patients 7, 14, 16, 25, and 30) or of L724P associated with a frameshift mutation (patient 10) (table 3).
Among the nine patients with the less common variant phenotype, four were found to carry one P1007A allele, combined either with I1061T (patients 17, 18, and 19) or to a nonsense mutation, W1143X (patient 13). I1061T has definitively been correlated with a classic phenotype when in the homozygous state (Millat et al. 1999), and one P1007A allele thus appeared sufficient to maintain some degree of cholesterol trafficking. These data suggested that the P1007A mutation was a common mutant allele associated with the variant biochemical phenotype.
This hypothesis was confirmed by investigation of genomic DNA in a total population of 55 patients with NPC1, using a simple PCR test based on introduction of an NheI restriction site. The P1007A allele was never observed in the 22 patients with a classic biochemical phenotype but constituted 15/66 alleles (22.7%) in the 33 patients studied who had the variant phenotype (9 from the present mutational study and 24 additional ones).
Four additional mutant alleles could clearly be ascribed to a variant phenotype: I943M (patient 21), V950M (patient 22) and G992R (patient 24) were all found in the homozygous state, and G986S (patient 20) was found in combination with Q775P which is clearly associated with a classic phenotype (patient 5). An intriguing observation is that these four mutations, as well as P1007A, are clustered within a small region of the cysteine-rich luminal loop between transmembrane domains 8 and 9 (fig. 1).
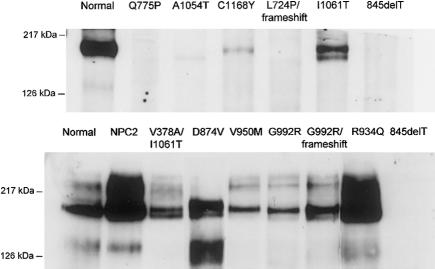
Correlations between Genotypes and Mutant NPC1 Protein Studied by Western Blot
Selected cell lines with missense mutations, most of them homozygous or associated with a frameshift mutation, were studied by immunoblotting (fig. 2). The homozygous 845delT cell line, which gives rise to very early premature termination of the protein, was tested to establish the specificity of the affinity-purified polyclonal antibody, and, as expected, no detectable band was seen in the region of interest. Normal fibroblasts gave a double band of ∼170 and ∼190 kD. Essentially no detectable protein was repeatedly found in three cell lines with missense mutations: homozygous Q775P and L724P, both located in the SSD, but also A1054T in the cysteine-rich region. Furthermore, a clearly diminished amount of the protein was found in cells with the C1168Y mutation. In all other mutations studied, including the classic I1061T, a substantial amount of NPC1 protein could be detected. Definite conclusions about an apparent small reduction of protein in some mutations are difficult to draw, since repeated experiments on different cell pellets could show a variation of at least twofold in levels of expression of the protein, in both normal cells and mutant cells. We observed that the protein was unstable in dilute solution, but variations under strict experimental conditions suggest the influence of yet-undefined factors that regulate expression of NPC1. Interestingly, NPC2 cells carrying the mutation E20X on the HE1 gene (fig. 2) or another mutation (Yamamoto et al. 2000) invariably showed NPC1 protein levels well above those observed in the control cells.
Figure 2.
Western blot of NPC1 protein in cultured fibroblasts from control subjects, 12 patients with NPC1, and 1 patient with NPC2. A C-terminal affinity-purified polyclonal antibody was used. Cells from a patient with a 845delT frameshift mutation served as a negative control. Eight patients with NPC1 had a homozygous missense mutation (lanes with a single label), and three were compound heterozygotes (lanes with a dual label). The NPC2 cell line carried a homozygous E20X mutation of the HE1 gene.
Correlations with the Clinical Phenotype
The three patients with a frameshift mutation and premature stop codon (patients 1, 2, and 3) or a nonsense mutation (patient 4) on both alleles all had an early neurological onset and a short life span. This was also the case for all patients with missense mutations for which immunoblotting revealed a lack of NPC1 protein. Patients 5 and 6 carried a homozygous missense mutation in the SSD or the cysteine-rich luminal loop, respectively. Patient 10, who had a SSD missense mutation associated with a splice mutation leading to exon-24 skipping, had a slightly later neurological onset, but the disease is progressing very rapidly. For patient 8, one allele carried a mutation in the SSD of an amino acid conserved in the patched gene, but the second allele remained unidentified. The significantly reduced amount of NPC1 protein observed in fibroblasts of patient 12, who had the C1168Y mutation, also correlated with the relatively severe late-infantile neurological phenotype.
Conversely, the finding of a near-normal amount of NPC1 protein was of limited predictive value. This pattern was shared by patients with a neurological juvenile onset (patient 15, I1061T), a neurological adult form (patient 22, V950M), or a nonneuronopathic adult form (patient 24, G992R, and patient 25, I1061T/V378A). On the other hand, correlations could clearly be drawn concerning the clinical expression of some missense mutations or combination of missense mutations. The combination of the two most common alleles, I1061T/P1007A, was encountered in three families (17, 18, and 19; five patients) and resulted in a juvenile onset of symptoms and a significantly slower progression of the disease than in homozygous I1061T patients. P1007A combined with a nonsense mutation (patient 13) resulted in a late-infantile neurological form. Homozygous V950M corresponded to an adult onset of neurological symptoms. Study of two adults with splenomegaly and no neurological symptoms (patients 24 and 25) led to the conclusion that G992R and V378A were responsible for this atypical mild phenotype. Interestingly, G992R had only a mild effect on cholesterol trafficking, whereas V378A was associated with severe biochemical alterations, demonstrating that the block in cholesterol transport does not correlate with clinical severity, as discussed elsewhere (Vanier et al. 1991b; Vanier and Suzuki 1998). Future studies should disclose whether correlations would be tighter with the newly discovered, possibly more relevant permease function of the NPC1 protein (Davies et al. 2000).
Patients 26 and 27 are still too young for conclusions to be drawn about their potential neurological subtyping. Mutations found in patients 28–30, initially studied to facilitate prenatal diagnosis, are reported because M272R and M1142T (which affects an amino acid conserved in patched) were the only two missense mutations found in our study in transmembrane domains outside of the SSD. R934Q was reported elsewhere in association with I1061T (Greer et al. 1999). But since the first affected child in those families died early, of liver failure, the complete phenotype associated with these mutations remains unknown.
Discussion
General Distribution of NPC1 Mutations
Although the number of known NPC1 mutations appears to be ∼100, taking into account data presented by seven groups at international meetings (Patterson et al. 2001), few studies have been published except in abstract form. In a survey of 14 Japanese patients with NPC1, with a majority of late-infantile cases, the 14 missense mutations and one in-frame deletion were widely distributed on NPC1 cDNA (Yamamoto et al. 1999, 2000). This was at variance with the initial data on nine patients reported by Carstea et al. (1997), where most mutations were found to affect the carboxy-terminal third of the protein. A later survey of 13 families originating from Canada and Maryland, restricted to the last six NPC1 exons and boundaries, indeed allowed identification of 20/26 mutant alleles (Greer et al. 1999). Greer and colleagues further observed that six of the eight missense mutations found in these patients—including G992W, which is characteristic of Nova Scotian patients—were clustered in a conserved NPC1-specific cysteine-rich loop. More recent topological analysis of the NPC1 protein by Davies and Ioannou (2000) indicated that the most common missense mutation, I1061T (Greer et al. 1999; Millat et al. 1999), initially thought to affect a transmembrane domain, also located to this luminal loop. In the present study, which included mostly white patients but covered a wider clinical spectrum, nearly all domains of the NPC1 protein—except for the leucine-zipper region—were affected. Eleven of the twenty-five identified missense mutations and four frameshift mutations were located in the first two-thirds of the protein, suggesting that the discrepancy between previous results in Japanese versus North American populations might be explained, in part, by differences in phenotypes of the patients. Mutagenesis studies indicated that both the leucine-zipper motif and the putative SSD were particularly critical regions (Watari et al. 1999). Mutations in the SSD have been identified by Yamamoto et al. (2000) (F703S) and in this report (M631R, L724P, and Q775P), but no naturally occurring mutations have been reported so far that affect the NPC1 leucine-zipper domain. In the present study, 14 of the 30 patients showed a proven or almost certain homozygous mutation, which greatly facilitated establishment of genotype/phenotype correlations.
Perinatal Liver Disease Independent of the NPC1 Mutation
It is well known that the neonatal, severe cholestatic form of NPC and neurological forms of the disease may occur in the same sibship (Vanier et al. 1991a); this is further illustrated in this study by families 2, 3, and 5. Most of the published families have shown an association with an infantile neurological form (Patterson et al. 2001), but one of the sibs in the family described by Jaeken et al. (1980) had a neurological juvenile-onset form. We recently found one patient with fetal hydrops/ascites to be homozygous for the I1061T mutation (M. T. Vanier and G. Millat, unpublished data). His affected elder brother, now 24 years old, presented with the expected neurological juvenile form (Millat et al. 1999). Thus, fatal liver disease in a first affected child (patients 28–30) gives no clue as to the possible neurological course in other sibs, unless the mutations correspond to a known neurological subtype. Remarkably, the rapidly fatal neonatal hepatic forms we studied so far all showed severe alterations of cellular cholesterol trafficking (M. T. Vanier, unpublished data).
Missense Mutations in SSD as well as Frameshift and Nonsense Mutations Lead to Absence of Stable NPC1 Protein and Severe Neurological Phenotype
In patient 1, initially described as having “lactosylceramidosis” (Dawson et al. 1971) and later reclassified as having NPC (Wenger et al. 1975; Vanier et al. 1988), a frameshift occurs at the end of the putative signal peptide. Patients in families 2 and 3 (Vanier et al. 1988; Kanoun et al. 1989) also have a very early NPC1 termination caused by a frameshift mutation, whereas those in family 4 have a nonsense mutation. The very severe neurological and biochemical phenotype observed in these families thus is well in line with the nature of the molecular lesion. The Q775P missense molecular lesion found for family 5 is of particular interest, both as the first naturally occurring homozygous mutation described in the NPC1 SSD (amino acids 615–797) and because it affects a SCAP-conserved residue. NPC1 protein (fig. 2) was undetectable both in fibroblasts of this patient and in those of patient 10, in whom another SSD-located substitution, L724P, was associated with a truncation of the C-terminus that should result in abnormal targeting of the protein (Watari et al. 1999). In patient 8, who had a M631R alteration, the second allele remained unidentified, but this patient also had a very severe course of the disease. Absence of detectable levels of NPC1 protein was reported by Yamamoto et al. (2000) in a Japanese patient with a F703S/S813X combination. Our results are in agreement with those of the latter authors, who were the first to point out a correlation between absence or near absence of NPC1 protein and the most severe clinical presentations (Yamamoto et al. 2000). Conversely, SSD mutants created by site-directed mutagenesis (Y643S, Y643C, and P691S), studied after transfection in CT-60 CHO cells, showed the presence (although in reduced amount) of NPC1 protein (Watari et al. 1999). Since we obtained similar results after transfection of Q775P cDNA to mutant FR4 CHO cells (K. Chikh, G. Millat, C. Tomasetto, K. Ohno, and M. T. Vanier, unpublished data), a likely explanation would be that overexpression masks the instability of the native protein. Brain glycolipids, studied in patients 1 (Dawson et al. 1972) and 8 (Vanier 1999), showed striking abnormalities. Taken together, these data clearly show that mutations affecting the putative SSD of the NPC1 protein are particularly deleterious, since they invariably correspond to the most severe neurological form of the disease.
Most missense alterations located to other regions of the NPC1 protein also severely affect cholesterol processing. A majority have a lesser impact on the stability of the protein as seen from immunoblotting, and a varying effect on the clinical picture, as illustrated by C1168Y (late-infantile neurological onset, patient 12), I1061T (juvenile neurological onset; Millat et al. 1999), and V378A, which apparently underlines the nonneurological adult phenotype observed in patient 25.
In spite of the fact that recent data show that NPC1 has an unexpected transmembrane molecular-pump activity and suggest that it is a permease that can transport certain lipids, but apparently not cholesterol, across membranes (Davies et al. 2000), studies in patients definitively highlight a key role of the SSD for the in vivo function of the protein.
Cysteine-Rich Luminal Loop Appears as a Functionally Complex Domain
Greer et al. (1999) were the first to highlight the functional significance of the cysteine-rich domain on the basis of mutational studies in patients. This domain has resemblance to the RING-finger motif regulatory domain of protein kinase C. Watari et al. (2000) further showed that this domain can bind zinc. From the studies of Greer et al. (1999), Yamamoto et al. (2000), and ourselves (Millat et al. 1999 and the present work), no fewer than 16 different missense mutations have already been ascribed to the cysteine-rich domain, which spans amino acids 855 through 1098 (Davies and Ioannou 2000). The first mutation identified, G992W, characterizes patients from Nova Scotia (former Niemann-Pick type D), with a juvenile onset and slowly progressing neurological disease (Greer et al. 1998). Despite the fact that Nova Scotian patients are of Acadian extraction, G992W has not been found so far in French patients. We identified two other base alterations on the same codon, both leading to the G992R substitution. Patient 24, who has isolated splenomegaly (Frölich et al. 1990) and is homozygous for this mutation, still has no neurological signs of NPC at age 66 years (K. Harzer, unpublished data). Cultured fibroblasts from this patient; from patient 27, who has G992R and a frameshift mutation on the other allele; and from patients from Nova Scotia (Byers et al. 1989) all behaved as biochemical “variants,” leading to the conclusion that codon 992 is not essential for cholesterol transport. On the other hand, the most recurrent I1061T mutation, initially described by us (Millat et al. 1999) and by Greer et al. (1999), which constitutes 15%–18% of mutated alleles in countries like the United Kingdom, France, or the United States, is correlated with severe alterations of cholesterol trafficking (the classic phenotype) and a juvenile-onset neurological disease. Immunoblotting studies revealed a quite good level of NPC1 protein in cells homozygous for both the G992R and I1061T mutations. Conversely, the present study has identified the A1054T mutation, for which no NPC1 protein was detectable, and which correlated with a severe infantile neurological onset form (patient 6). Thus, at variance with what was seen for the SSD, consequences of naturally occurring missense mutations affecting the cysteine-rich luminal loop appeared highly variable.
P1007A is a Common Mutated Allele Associated with the Variant Biochemical Phenotype
One patient homozygous for the P1007A mutation, categorized as having the biochemically variant phenotype, was described by Greer et al. (1999). We found four additional patients with the variant phenotype in whom this allele was combined with I1061T or a nonsense mutation and further showed a high prevalence of this allele, 22.7%, in a total population of 33 patients with the variant phenotype. Systematic screening for this mutation thus appears justified in patients with the variant phenotype.
Seven Variant Mutant Alleles So Far Identified Locate to the Cysteine-Rich Luminal Loop
From the study of patients with the P1007A and G992R mutations, we concluded that a single variant allele apparently leads to a variant phenotype, irrespective of the nature of the second allele. Apart from G992W, G992R, and P1007A discussed above, I943M, V950M, G986S (identified in this report) and V889M (Yano et al. 1996; Yamamoto et al. 2000) could all be classified as variant alleles. Although a recent study in Portuguese patients indicated that some variant mutations may affect another domain (Ribeiro et al., in press), those reported so far all located to the cysteine-rich loop, and most of them were clustered within a small region (codons 943–1007) of this loop. The present observation suggests that part of the cysteine-rich domain could be associated with a NPC1 function not tightly associated with cholesterol trafficking. The NPC1 protein seems to be implicated in glycolipid transport (Blanchette-Mackie 2000; Zhang et al. 2001), and, since glycolipid abnormalities have been found in brain of patients with the variant phenotype (Martin et al. 1984; Vanier 1999), more-detailed studies are indicated. Future studies on the three-dimensional structure of the cysteine-luminal loop might also provide new insights on the functional complexity of this domain. Above all, how variant mutations specifically relate to the molecular-pump activity of NPC1 recently described by the group of Ioannou (Davies et al. 2000) is a particularly intriguing question.
Acknowledgments
The authors are grateful to the patients and their families and to all colleagues who, over many years, have provided invaluable clinical information. This research was supported by grants from Vaincre les Maladies Lysosomales, INSERM (U189), and the Japanese Ministries of Education, Science, and Culture and of Health and Welfare, and by an INSERM/JSPS cooperation program. G.M. was the recipient of a fellowship from Vaincre les Maladies Lysosomales. We thank M. Merlin, M. C. Juge, and H. Cornot for expert technical assistance.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genbank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for NPC1 cDNA [accession number AF002020])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Niemann-Pick C [MIM 257220]
References
- Baumkötter J, Freisinger P, Schneider KTM, Harzer K, Vanier MT, Pontz BF (1998) Fetal ascites: a rare presentation of Niemann-Pick disease type C. J Inher Metab Dis Suppl 21:118 [Google Scholar]
- Blanchette-Mackie EJ (2000) Intracellular cholesterol trafficking: role of the NPC1 protein. Biochim Biophys Acta 1486:171–183 [DOI] [PubMed] [Google Scholar]
- Byers DM, Rastogi SR, Cook HW, Palmer FB, Spence MW (1989) Defective activity of acyl-CoA:cholesterol O-acyltransferase in Niemann- Pick type C and type D fibroblasts. Biochem J 262:713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, et al (1997) Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277:228–231 [DOI] [PubMed] [Google Scholar]
- Christomanou H, Vanier MT, Santambrogio P, Arosio P, Kleijer WJ, Harzer K (2000) Deficient ferritin immunoreactivity in tissues from Niemann-Pick type C patients: extension of findings to fetal tissues, H and L ferritin isoforms, but also one case of the rare Niemann-Pick C2 complementation group. Mol Genet Metab 70:196–202 [DOI] [PubMed] [Google Scholar]
- Cruz JC, Chang TY (2000) Fate of endogenously synthesized cholesterol in Niemann-Pick type C1 cells. J Biol Chem 275:41309–41316 [DOI] [PubMed] [Google Scholar]
- Davies JP, Chen FW, Ioannou YA (2000) Transmembrane molecular pump activity of Niemann-Pick C1 protein. Science 290:2295–2298 [DOI] [PubMed] [Google Scholar]
- Davies JP, Ioannou YA (2000) Topological analysis of NPC1 reveals that the orientation of the putative sterol sensing domain is identical to that of HMG CoA and SCAP. J Biol Chem 275:24367–24374 [DOI] [PubMed] [Google Scholar]
- Dawson G (1972) Glycosphingolipid levels in an unusual neurovisceral storage disease characterized by lactosylceramide galactosylhydrolase deficiency: lactosylceramidosis. J Lipid Res 13:207–219 [PubMed] [Google Scholar]
- Dawson G, Matalon R, Stein AO (1971) Lactosylceramidosis: lactosylceramide galactosylhydrolase deficiency and accumulation of lactosylceramide in cultured fibroblasts. J Pediatr 79:423–429 [DOI] [PubMed] [Google Scholar]
- Fensom AH, Grant AR, Steinberg SJ, Ward CP, Lake BD, Logan EC, Hulman G (1999) An adult with a non-neuronopathic form of Niemann-Pick C disease. J Inherit Metab Dis 22:84–86 [DOI] [PubMed] [Google Scholar]
- Fröhlich E, Harzer K, Heller T, Ruhl U (1990) Sonographisch echodichte Milztumoren: Knotige Manifestation eines Morbus Niemann-Pick Typ C. Ultraschall Med 11:119–122 [DOI] [PubMed] [Google Scholar]
- Greer WL, Dobson MJ, Girouard GS, Byers DM, Riddell DC, Neumann PE (1999) Mutations in NPC1 highlight a conserved NPC1-specific cysteine-rich domain. Am J Hum Genet 65:1252–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer WL, Riddell DC, Gillan TL, Girouard GS, Sparrow SM, Byers DM, Dobson MJ, Neumann PE (1998) The Nova Scotia (type D) form of Niemann-Pick disease is caused by a G3097→T transversion in NPC1. Am J Hum Genet 63:52–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins ME, Davies JP, Chen FW, Ioannou YA (1999) Niemann-Pick C1 is a late endosome-resident protein that transiently associates with lysosomes and the trans-Golgi network. Mol Genet Metab 68:1–13 [DOI] [PubMed] [Google Scholar]
- Jaeken J, Proesmans W, Eggermont E, van Hoof F, Den Tandt W, Standaert L, Van Herck G, Corbeel L (1980) Niemann-Pick type C disease and early cholestasis in three brothers. Acta Paediatr Belg 33:43–46 [PubMed] [Google Scholar]
- Jeanpierre M (1987) A rapid method for the purification of DNA from blood. Nucleic Acids Res 15:9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoun N, Trabelsi M, Oueslati A, Damergi R, Ben Dridi MF, Boudhina R, Bennaceur B (1989) Forme infantile précoce de maladie de Niemann-Pick type C. Ann Pediatr (Paris) 36:335–338 [PubMed] [Google Scholar]
- Liscum L, Munn NJ (1999) Intracellular cholesterol transport. Biochim Biophys Acta 1438:19–37 [DOI] [PubMed] [Google Scholar]
- Martin JJ, Lowenthal A, Ceuterick C, Vanier MT (1984) Juvenile dystonic lipidosis (variant of Niemann-Pick disease type C). J Neurol Sci 66:33–45 [DOI] [PubMed] [Google Scholar]
- Millat G, Marçais C, Rafi MA, Yamamoto T, Morris JA, Pentchev PG, Ohno K, Wenger DA, Vanier MT (1999) Niemann-Pick C1 disease: the I1061T substitution is a frequent mutant allele in patients of Western European descent and correlates with a classic juvenile phenotype. Am J Hum Genet 65:1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Zhang D, Coleman KG, Nagle J, Pentchev PG, Carstea ED (1999) The genomic organization and polymorphism analysis of the human Niemann-Pick C1 gene. Biochem Biophys Res Commun 261:493–498 [DOI] [PubMed] [Google Scholar]
- Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P (2000) Identification of HE1 as the second gene of Niemann-Pick C disease. Science 290:2298–2301 [DOI] [PubMed] [Google Scholar]
- Neufeld EB, Wastney M, Patel S, Suresh S, Cooney AM, Dwyer NK, Roff CF, Ohno K, Morris JA, Carstea ED, Incardona JP, Strauss JF, III, Vanier MT, Patterson MC, Brady RO, Pentchev PG, Blanchette-Mackie EJ (1999) The Niemann-Pick C1 protein resides in a vesicular compartment linked to retrograde transport of multiple lysosomal cargo. J Biol Chem 274:9627–9635 [DOI] [PubMed] [Google Scholar]
- Ory DS (2000) Niemann-Pick type C: A disorder of cellular cholesterol trafficking. Biochim Biophys Acta 1529:331–339 [DOI] [PubMed] [Google Scholar]
- Patterson MC, Vanier MT, Suzuki K, Morris JA, Carstea ED, Neufeld EB, Blanchette-Mackie EJ, Pentchev PG (2001) Niemann-Pick disease type C: a lipid trafficking disorder. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 3611–3634 [Google Scholar]
- Pentchev PG, Vanier MT, Suzuki K, Patterson M (1995) Niemann-Pick disease type C: a cellular cholesterol lipidosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 7th ed. McGraw Hill, New York, pp 2625–2639 [Google Scholar]
- Ribeiro I, Marcão A, Amaral O, Sa Miranda MC, Vanier MT, Millat G. Niemann-Pick type C disease: NPC1 mutations associated with severe and mild cellular cholesterol transport alterations. Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Steinberg SJ, Ward CP, Fensom AH (1994) Complementation studies in Niemann-Pick disease type C indicate the existence of a second group. J Med Genet 31:317–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanier MT (1999) Lipid changes in Niemann-Pick disease type C brain: personal experience and review of the literature. Neurochem Res 24:481–489 [DOI] [PubMed] [Google Scholar]
- Vanier MT, Duthel S, Rodriguez-Lafrasse C, Pentchev P, Carstea ED (1996) Genetic heterogeneity in Niemann-Pick C disease: a study using somatic cell hybridization and linkage analysis. Am J Hum Genet 58:118–125 [PMC free article] [PubMed] [Google Scholar]
- Vanier MT, Millat G (2000) Niemann-Pick C disease: insights from studies on mutated NPC1 gene and protein. J Inher Metab Dis Suppl 23:232 [Google Scholar]
- Vanier MT, Millat G, Marçais C, Rafi MA, Yamamoto T, Morris JA, Pentchev PG, Nanba E, Wenger DA (1999) Niemann-Pick C disease: mutational spectrum in NPC1 gene and genotype/phenotype correlations. Am J Hum Genet Suppl 65:A495 [Google Scholar]
- Vanier MT, Pentchev PG, Rodriguez-Lafrasse C, Rousson R (1991a) Type C Niemann-Pick disease: an update. J Inher Metab Dis 14:580–595 [DOI] [PubMed] [Google Scholar]
- Vanier MT, Rodriguez-Lafrasse C, Rousson R, Gazzah N, Juge MC, Pentchev PG, Revol A, Louisot P (1991b) Type C Niemann-Pick disease: spectrum of phenotypic variation in disruption of intracellular LDL-derived cholesterol processing. Biochim Biophys Acta 1096:328–337 [DOI] [PubMed] [Google Scholar]
- Vanier MT, Suzuki K (1996) Niemann-Pick diseases. In: Moser HW (ed) Neurodystrophies and neurolipidoses: handbook of clinical neurology, vol. 66. Elsevier Science, Amsterdam, pp 133–162 [Google Scholar]
- Vanier MT, Suzuki K (1998) Recent advances in elucidating Niemann-Pick C disease. Brain Pathol 8:163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanier MT, Wenger DA, Comly ME, Rousson R, Brady RO, Pentchev PG (1988) Niemann-Pick disease group C: clinical variability and diagnosis based on defective cholesterol esterification: a collaborative study on 70 patients. Clin Genet 33:331–348 [DOI] [PubMed] [Google Scholar]
- Watari H, Blanchette-Mackie EJ, Dwyer NK, Watari M, Burd CG, Patel S, Pentchev PG, Strauss JF III (2000) Determinants of NPC1 expression and action: key promoter regions, posttranscriptional control, and the importance of a “cysteine-rich” loop. Exp Cell Res 259:247–256 [DOI] [PubMed] [Google Scholar]
- Watari H, Blanchette-Mackie EJ, Dwyer NK, Watari M, Neufeld EB, Patel S, Pentchev PG, Strauss JF, III (1999) Mutations in the leucine zipper motif and sterol-sensing domain inactivate the Niemann-Pick C1 glycoprotein. J Biol Chem 274:21861–21866 [DOI] [PubMed] [Google Scholar]
- Wenger DA, Sattler M, Clark C, Tanaka H, Suzuki K, Dawson G (1975) Lactosylceramidosis: normal activity for two lactosylceramide beta-galactosidases. Science 188:1310–1312 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nanba E, Ninomiya H, Higaki K, Taniguchi M, Zhang H, Akaboshi S, Watanabe Y, Takeshima T, Inui K, Okada S, Tanaka A, Sakuragawa N, Millat G, Vanier MT, Morris JA, Pentchev PG, Ohno K (1999) NPC1 gene mutations in Japanese patients with Niemann-Pick disease type C. Hum Genet 105:10–16 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Ninomiya H, Matsumoto M, Nanba E, Ohta Y, Tsutsumi Y, Yamakawa K, Millat G, Vanier MT, Pentchev PG, Ohno K (2000) Genotype-phenotype relationship of Niemann-Pick disease type C: a possible correlation between clinical onsets and levels of NPC1 protein in isolated skin fibroblasts. J Med Genet 37:707–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Taniguchi M, Akaboshi S, Vanier MT, Tai T, Sakuraba H, Ohno K (1996) Accumulation of GM2 ganglioside in Niemann-Pick disease type C fibroblasts. Proc Jpn Acad 72B:214–219 [Google Scholar]
- Zhang M, Dwyer NK, Neufeld EB, Love DC, Cooney A, Comly M, Patel S, Watari H, Strauss JF, III, Pentchev PG, Hanover JA, Blanchette-Mackie EJ (2001) Sterol-modulated glycolipid sorting occurs in Niemann-Pick C1 late endosomes. J Biol Chem 276:3417–3425 [DOI] [PubMed] [Google Scholar]