Abstract
Spondyloepiphyseal dysplasia tarda (SEDL) is a genetically heterogeneous disorder characterized by mild-to-moderate short stature and early-onset osteoarthritis. Both autosomal and X-linked forms have been described. Elsewhere, we have reported the identification of the gene for the X-linked recessive form, which maps to Xp22.2. We now report characterization of an exon-skipping mutation (IVS3+5G→A at the intron 3 splice-donor site) in two unrelated families with SEDL. Using reverse transcriptase (RT)–PCR, we demonstrated that the mutation resulted in elimination of the first 31 codons of the open reading frame. The mutation was not detected in 120 control X chromosomes. Articular cartilage from an adult who had SEDL and carried this mutation contained chondrocytes with abundant Golgi complexes and dilated rough endoplasmic reticulum (ER). RT-PCR experiments using mouse/human cell hybrids revealed that the SEDL gene escapes X inactivation. Homologues of the SEDL gene include a transcribed retropseudogene on chromosome 19, as well as expressed genes in mouse, rat, Drosophila melanogaster Caenorhabditis elegans, and Saccharomyces cerevisiae. The latter homologue, p20, has a putative role in vesicular transport from ER to Golgi complex. These data suggest that SEDL mutations may perturb an intracellular pathway that is important for cartilage homeostasis.
Introduction
Spondyloepiphyseal dysplasia tarda (SEDL) is a rare osteochondrodysplasia that is first evident in affected individuals at 3–12 years of age. Clinical features include short trunk, barrel-shaped chest, and mild-to-moderate short stature. Premature osteoarthritis is common, especially in weight-bearing joints (Taybi and Lachman 1990). The X-linked recessive form (SEDL [MIM 313400]) was first reported by Jacobsen (1939), and its estimated prevalence is 1.7 per million (Wynne-Davies and Gormley 1985). Rare families exhibiting autosomal inheritance patterns have also been described (Pfeiffer et al. 1992; Schantz et al. 1988). Localization of the SEDL gene to Xp was originally reported by Szpiro-Tapia et al. (1988) and was further refined to the ∼2-Mb interval at Xp22, between DXS16 and DXS987, by Heuertz et al. (1995). The critical region was refined to <170 kb, by critical recombination events at DXS16 and AFMa124wc1, in two Australian families with SEDL (Gedeon et al. 1999); subsequently, a highly conserved gene was cloned in silico from a publicly available genomic sequence. In three unrelated Australian families with SEDL, different dinucleotide deletions in this gene were identified that result in frameshifts and generate premature stop codons.
In the present study, we describe a large North American family with SEDL and confirm the location of the disease gene to Xp22.2 in this family by two-point linkage analysis. We demonstrate a mutation at the intron 3 splice-donor site of the SEDL gene in affected individuals and obligate carriers, and we show evidence for abnormal splicing of exon 3 in cell lines from an affected man. We also document the presence of the same mutation in another unrelated patient with SEDL, suggesting that it is a hot spot for mutation within the SEDL gene. Ultrastructural analysis of articular cartilage from an adult SEDL patient carrying this mutation demonstrates unusual chondrocyte histomorphology. Finally, we show that the SEDL gene escapes X inactivation. Its homology to the yeast gene that encodes protein p20 (Sacher et al. 1998), a component of the transport protein particle (TRAPP), leads us to hypothesize that the SEDL gene product has a key role in trafficking chondrocyte proteins from endoplasmic reticulum (ER) to Golgi complexes and therefore is essential for maintaining homeostasis of the cartilage extracellular matrix.
Subjects and Methods
Clinical Summary
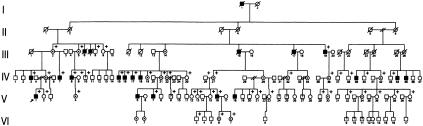
The present study was approved by the Institutional Review Board of Vanderbilt University Medical Center, and informed consent was obtained from all participants. The proband of family 1 (V:1 in fig. 1) was a 19-year-old man who sought medical attention because of chronic lower back pain. The proband's height was 173 cm (25th percentile), and his arm span was 187 cm. His trunk was disproportionately short, and his chest was barrel-shaped. Facial features were unremarkable, and results of the neurologic examination were within normal limits. Radiographs revealed platyspondyly with superior and inferior humping of the vertebral bodies; the pelvis was somewhat narrow, and the femoral necks were short. Family history revealed 21 affected males spread over five generations, a group that constitutes the largest SEDL pedigree ascertained to date (see fig. 1). The surnames in the first generation suggest that the family is of British descent. A maternal first cousin once removed (IV:8 in fig. 1) underwent replacement of the right hip joint at age 31 years, as well as replacement of the left shoulder joint at age 34 years, because of osteoarthritis. Another maternal male relative (IV:15 in fig. 1) underwent hip joint replacement at age 50 years. Five affected men, ages 19–43 years, were examined by one of us (G.E.T.). Heights were 147–173 cm (mean ± SD 159 ± 10 cm), and arm span exceeded height by a mean ± SD of 20 ± 12 cm. Radiographs of an affected man (IV:8 in fig. 1) are shown in figure 2.
Figure 1.
Abbreviated pedigree of a North American family with SEDL. Plus signs (+) indicate genotyped individuals; arrow indicates proband (V:1); question marks (?) indicate questionable clinical status.
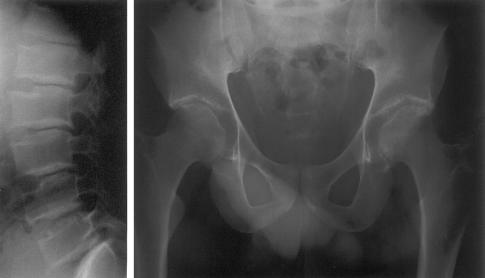
Figure 2.
Radiographs of the lateral spine (left) and pelvis (right) of a 31-year-old man (IV:8 in fig. 1) with SEDL. Note platyspondyly and superior and inferior humping of vertebrae, as well as irregular hip-joint surfaces and short femoral necks.
Family 2 was ascertained at the Universitatskinderklinik in Mainz, Germany. The proband displayed normal linear growth until he was 6 years of age but, since that time, has gradually demonstrated disproportionate short stature. His height at 11.5 years of age was 129 cm (>2 SD below the mean for his age; 50th percentile for 8.5 years), and his arm span exceeded his height by 12 cm. Changes typical of SEDL were also evident on radiographs. He has an unaffected brother and no other affected relatives.
Cell Culture, RNA, and DNA Isolation
Patient lymphoblastoid cell lines were grown and maintained under standard culture conditions. Genomic DNA was extracted from whole blood, cultured cells, or dried buccal swabs, using either the Qiagen blood kit or QIAamp kit (Qiagen). Total RNA was isolated from cultured cells using method of Chomczynski and Sacci (1987) or RNeasy kit (Qiagen).
Linkage Analysis
Fifty-two members of family 1, including 10 of 16 living affected males and 12 obligate female carriers, were genotyped at 18 microsatellite markers spanning the region from DXS1223 to DXS989 at Xp22.2 (Dib et al. 1996; GenBank). Two-point analysis was performed using the MLINK program of LINKAGE, version 5.2 (Lathrop and Lalouel 1984), assuming X-linked recessive inheritance of SEDL with full penetrance and allele frequency of .00001.
Histologic Analysis
Residual articular cartilage was obtained during hip-joint replacement in family member IV:15 (fig. 1). Samples were either frozen or fixed in formalin or buffered glutaraldehyde and were prepared for histologic analysis by routine methods (Brodie et al. 1998).
PCR, RT-PCR, and Sequence Determination
PCR was performed on genomic DNA as described elsewhere (Gedeon et al. 1999), using oligonucleotide primer pairs designed from the unique X-specific sequence that flanked each of the four coding exons on the SEDL gene. The RT-PCR experiments were also performed as described by Gedeon et al. (1999). Genomic DNA was sequenced from both strands, using PCR primers and ThermoSequenase cycle sequencing kit (USB).
X-Inactivation Studies
Mouse/human somatic-cell hybrids that retain either an active or inactive human X chromosome have been described elsewhere (Brown et al. 1997; Carrel et al. 1999). The SEDL primers used were 5′-AGGAGCCATATATTGAAGACCATG-3′ (forward, designated “5′21F”) and 5′-TCCTGAGTATACACCATTGTGG-3′ (reverse, designated “3′52R”). Possible amplification of the pseudogene SEDLP (Gedeon et al. 1999; Gecz et al. 2000) was assessed by digesting RT-PCR products with BsaJ1, which cleaves only the pseudogene amplimer. Conditions for RT-PCR and oligonucleotide primers for MIC2 and A007K03 are as described elsewhere (Carrel et al. 1999).
Linkage of SEDL Phenotype to Xp22 Confirmed
Before the discovery of the SEDL gene, we performed two-point linkage analysis between the SEDL phenotype and 18 microsatellite markers at Xp22.2. Peak LOD scores were obtained at DXS16 and DXS8022 (Z=5.71 and 8.22, respectively, at θ = 0), which were later found to flank the disease locus. However, despite the large size of the North American pedigree, no recombinants were identified that could narrow the candidate region within the 18-cM span from DXS7109 to DXS1226. The critical region was subsequently limited to <170 kb by identifying recombinations in two small Australian pedigrees (Gedeon et al. 1999).
Histologic Analysis of SEDL Cartilage
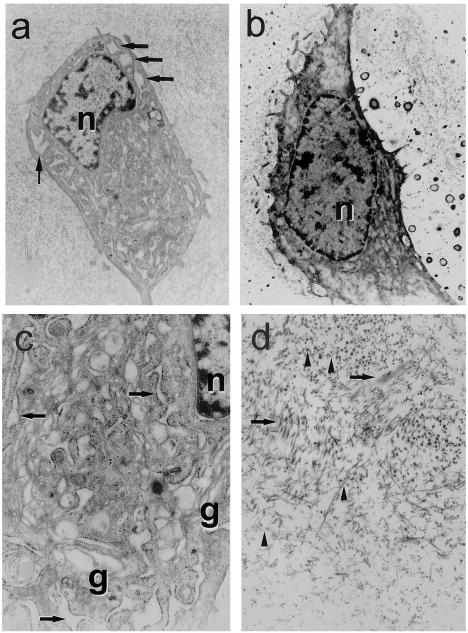
Histologic analysis revealed pathologic changes that are typical of osteoarthritis (Pritzker 1998), including horizontal fibrillation near the articular surface and occasional clustering of chondrocytes in the resting zone (data not shown). Cellularity was unremarkable, and staining with toluidine blue revealed a normal distribution of glycosaminoglycan. Figure 3 shows the ultrastructure of articular cartilage from the hip joint of a family 1 member (individual IV:15). The extracellular matrix contained a normal number of collagen fibrils, some of which appeared short, frayed, or of variable diameter. Chondrocytes contained excessive cytoplasm with abundant Golgi complexes and dilated, rough ER. In contrast, chondrocytes from an age- and sex-matched patient with idiopathic osteoarthritis were mostly apoptotic, with those remaining viable cells demonstrating a normal nucleus:cytoplasm ratio and no organelles of unusual appearance.
Figure 3.
Articular cartilage ultrastructure from the femoral head of a 50-year-old man with SEDL (IV:15 in fig. 1) and a 50-year-old patient with idiopathic osteoarthritis. a, SEDL chondrocyte with dilated rER. Arrows indicate rER, and n indicates nucleus (original magnification, 8,140×). b, Chondrocyte from a 50-year-old man with osteoarthritis (original magnification, 8,140×). Note normal nuclear:cytoplasmic ratio and absence of dilated rER. c, SEDL chondrocyte cytoplasm. The abundant Golgi complexes are indicated by g, and dilated loops of rER are indicated by arrows (original magnification, 23,830×). d, SEDL extracellular matrix. Arrows indicate collagen fibrils that are short and frayed, and arrowheads indicate fibrils of variable diameter (original magnification, 23,830×).
Splice-Donor–Site Mutation in Intron 3
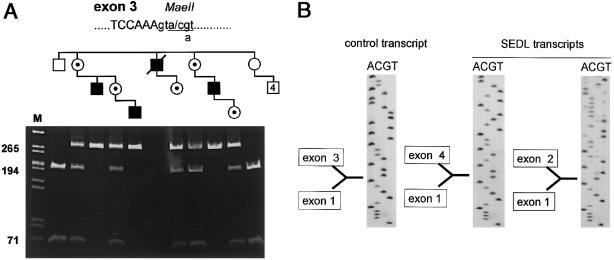
Direct sequencing of gDNA from the proband of each family revealed a G→A transition in the fifth base of the intron 3 splice-donor site. The mutation disrupts a MaeII restriction site, which allowed us to efficiently screen all family members (fig. 4a). The mutation cosegregated with the affected phenotype and in known carriers, with two exceptions. One was a phenotypically normal 10-year-old boy (V:9 in fig. 1), and the other was a man (IV:10 in fig. 1) of normal stature who denied symptoms of osteoarthritis. Despite repeated inquiries, we were unable to obtain radiographs of either individual, and individual IV:10 was unavailable for physical examination. The mutation was not seen in 120 control X chromosomes (CEPH panel) after complete MaeII digestion of gDNA PCR amplimers. The identical mutation was seen in the affected individual and his mother in family 2 (data not shown). No other sequence changes were seen in the coding region or flanking intron sequences in either proband. No mutations in the SEDL gene were detected by genomic sequencing of the control patient with osteoarthritis.
Figure 4.
Splice mutation in SEDL. a, PCR amplification of a fragment of the SEDL gene spanning the exon 3/intron 3 junction. The mutant allele contains a +5G→A transition at the intron 3 splice-donor site, which disrupts a MaeII restriction site. The proband of family 1 is the affected man in the third generation of the pedigree as drawn. b, Sequencing of cDNA from a lymphoblastoid cell line from the proband of family 1. Sequencing reveals two transcripts (each missing exon 3, which contains the translational start site).
cDNA Pattern in SEDL Lymphoblastoid Cells
The predominant SEDL transcript from several normal tissues lacks exon 2 (Gedeon et al. 1999; Gecz et al. 2000). When RT-PCR was performed, using total lymphoblastoid cell RNA from the proband of family 1, two SEDL transcripts were seen (fig. 4b). The longer transcript lacked exon 3 but retained exon 2; the shorter transcript lacked both exon 2 and exon 3. There was no evidence for the use of alternative splice sites (data not shown), and no other consensus sequences for translation initiation (Kozak 1986) are seen in exons 4–6. Because exon 3 contains the translation start site, we hypothesize that no SEDL protein is produced from this mutant allele.
X-Inactivation Studies
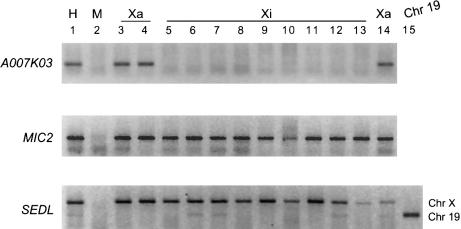
Figure 5 illustrates the expression of SEDL in a series of somatic-cell hybrids that retain either an active or an inactive human X chromosome. Amplification of EST A007K03 (which is subject to inactivation [Carrel et al. 1999]) and MIC2 (which escapes inactivation [Brown et al. 1997]) is shown for comparison. SEDL is expressed in all cell lines tested, indicating that SEDL escapes X inactivation. Identity of the RT-PCR product as the SEDL gene product is further demonstrated by absence of amplification within the parent mouse cell line (lane 2) and insensitivity to digestion by BsaJ1.
Figure 5.
Expression of genes from active or inactive X-containing hybrids. cDNA from mouse/human somatic-cell hybrids that retained either the human active (Xa) or inactive (Xi) X chromosome was amplified with gene-specific primers. A negative image of ethidium bromide–stained products separated by agarose gel electrophoresis is shown. Lanes contain human female cell line (lane 1); mouse tsA1S9az31b cell line (lane 2); t60-12 (lane 3); Aha11aB1 (lane 4); t86-B1maz1b-3a (lane 5); t11-4Aaz5 (lane 6); t75-2maz34-4a (lane 7); t48-1a-Daz4a (lane 8); LT23-1E2Buv5Cl26-7A2 (lane 9); L23-4B (lane 10); t51-S2 (lane 11); tHM-1a (lane 12); tHM-1d (lane 13); A23-1aCl5 (lane 14); genomic DNA from a somatic-cell hybrid cell line retaining human chromosome 19 (lane 15). PCR products in all lanes have been digested with BsaJ1 to distinguish SEDL product from pseudogene SEDLP product, since only the latter is cleaved by BsaJ1. Control transcripts A007K03 (which is subject to inactivation) and MIC2 (which escapes inactivation) demonstrate the inactivation status for each hybrid shown.
Discussion
Effect of IVS3+5G→A Mutation on Splicing
The RT-PCR experiments using lymphoblastoid RNA from the proband in family 1 failed to generate any transcripts with a normal splicing pattern. Some transcripts retained exon 2, which is usually spliced out (Gedeon et al. 1999), but the majority of the transcripts lacked both exon 2 and exon 3. Exon 3 contains the translation start site, and, although there are five additional methionine residues in the open reading frame, none has an associated Kozak consensus sequence for initiation of translation. Therefore, we would not expect any protein to be translated from this mutant allele. In addition to the two families reported here, three additional unrelated families with SEDL have been ascertained that carry this same mutation (Gedeon et al. 2001 [in this issue]). This suggests that the IVS3+5G residue may represent a hypermutable site in the SEDL gene.
The thermodynamic data in the literature are inadequate for direct calculation of the effect of the IVS3+5G→A transition on RNA splicing. However, a rough estimate is attained by using the consensus sequence data compiled by Shapiro and Senapathy (1987). By use of their method, a score of 76.1 is obtained for the wild-type allele, whereas a score of 61.7 is obtained for the mutant allele. For comparison, the human consensus sequence for splice-donor sites (AGgt[a/g]agt) (Zhang 1998) yields a score of 96–100, depending on whether adenine or guanine lies in the third position of the intron. A more convenient way to assess the significance of splice-donor site substitutions is based on the analysis of factor IX splice mutations by Ketterling et al. (1999), where one considers bases −2 to +6 of the splice site. They propose a “5–6” hypothesis, whereby normal splicing is expected to be severely compromised if a donor site matches the consensus sequence at fewer than seven positions. This is exemplified by the IVS3+5G→A transition in SEDL, as well as 12 other such mutations extracted from OMIM, which are listed in table 1. Thus, all thirteen mutations lend further support to Ketterling’s “5–6” hypothesis.
Table 1.
IVS+5G→A Mutations Causing Human Diseases
| Gene | Mutated Sequence | Intron | No. of bp Matched with Consensusa | Reference |
| SEDL | AAgtacat | 3 | 5/8 | Present study |
| β-globin | TTgtgaat | 1 | 5/8 | Lapoumeroulie et al. 1986 |
| COL3A1 | CTgtaaat | 27 | 5/8 | Thakker-Varia et al. 1995 |
| COL1A1 | CTgtaaat | 14 | 5/8 | Bonadio et al. 1990 |
| COL1A2 | GTgtgaat | 33 | 5/8 | Ganguly et al. 1991 |
| HPRT | CTgtaaat | 7 | 5/8 | Gibbs et al. 1989 |
| CYBB | CGgtaaaa | 3 | 5/8 | deBoer et al. 1992 |
| PROS1 | TGgtacat | 10 | 5/8 | Reitsma et al. 1994 |
| WT1 | AAgtgcat | 9 | 5/8 | Bruening et al. 1992 |
| BTK | TGgtaaat | 2 | 6/8 | Duriez et al. 1994 |
| MYBPC3 | ACgtgaat | 7 | 6/8 | Carrier et al. 1997 |
| CYP17 | TGgtgaat | 7 | 6/8 | Yamaguchi et al. 1997 |
| HPRT | ATgtaaat | 8 | 6/8 | Gibbs et al. 1989 |
Splice-donor consensus sequence AGgt(a/g)agt derived from 3,274 exon/intron junctions (Zhang 1998), where capital letters denote last 2 bp of exon.
Two clinically normal male subjects in family 1 (V:9 and IV:10 in fig. 1) were shown to harbor the mutant allele by both haplotype analysis and MaeII restriction analysis of PCR amplimers. Although V:9 may be presymptomatic, IV:10 is a mature adult who states that his height is 183 cm and that he has no skeletal complaints. We were unable to obtain radiographs or a cell sample of either individual and therefore cannot determine whether IV:10 is mildly affected or has normal SEDL transcripts in his tissue.
Significance of SEDL Escaping X Inactivation
The data in figure 5 clearly indicate that the SEDL gene escapes X inactivation. The closest flanking genes identified at Xp22.2 (Rab9 and Cxorf5) also escape X inactivation (Carrel et al. 1999). This clustering further supports a model in which regional mechanisms may control the expression of genes that escape X inactivation (Miller and Willard 1998; Carrel et al. 1999). The majority of the mutations in SEDL patients characterized thus far (Gedeon et al. 1999 and 2001; Mumm et al. 2000; and the present study) are predicted to severely truncate or eliminate the protein product. The observation that SEDL escapes X inactivation suggests that haploinsufficiency at the locus is inadequate to produce any phenotypic changes in female SEDL carriers. Although Whyte et al. (1999) observed subtle radiographic changes in older SEDL carriers, no signs or symptoms of premature osteoarthritis were noted in the women of the family reported here or in those described in the report by Gedeon et al. (1999).
Putative Role for SEDL Gene Product, Sedlin
The SEDL gene is highly conserved, and homologues are found in mouse, rat, Caenorhabditis elegans, Drosophila melanogaster, and Saccharomyces cerevisiae (Gedeon et al. 1999). The yeast homologue, designated p20 or YBR254c, shares 41% identity and 57% amino acid similarity with human SEDL (Sacher et al. 1998). Yeast p20 is a 20-kD hydrophilic protein which is part of a complex of ⩾10 proteins constituting TRAPP. The TRAPP complex has been shown to play a key role in transport of yeast proteins from the ER to the Golgi apparatus (Barrowman et al. 2000). A precedent for human disease in such a transport process has been established by combined deficiency of factors V and VII (Nichols and Ginsburg 1999). This autosomal recessive coagulopathy results from mutations in the ERGIC-53 gene, which encodes a transmembrane glycoprotein–binding component of the ER-Golgi intermediate compartment (ERGIC). Mutations in ERGIC-53 result in impaired secretion of clotting factors V and VIII from megakaryocytes and hepatocytes.
Transient transfection experiments with FLAG- or GFP-tagged SEDL (Gecz et al. 2000) suggested localization of the SEDL protein to perinuclear structures, whereas transfection with truncating mutant transcripts disrupted this localization. Furthermore, the appearance of SEDL chondrocytes from articular cartilage (fig. 3) suggests that there may be disruption in cellular trafficking between rER and Golgi. Since the histopathology of SEDL cartilage has not been described elsewhere, it is uncertain whether the cellular architecture depicted in figure 3 is representative of this genetic disorder. The cellular appearance is dissimilar to the gross dilation of rER that is seen as a result of any of the following: retention of abnormal type II collagen in patients with spondyloepiphyseal dysplasia congenita (Tiller et al. 1995), abnormal type IX collagen in some patients with multiple epiphyseal dysplasia (Spayde et al. 2000), or abnormal COMP in patients with pseudoachondroplasia (Rimoin et al. 1994). Such ultrastructural abnormalities are not seen in fibroblasts of patients with these three disorders, since the abnormal protein is not produced appreciably in fibroblasts. Although the human SEDL gene appears to be expressed ubiquitously, we propose that the tissue-specific nature of the SEDL phenotype may lie in the protein cargo that is transported by the TRAPP system in chondrocytes. Delineation and further study of additional SEDL mutations (Gedeon et al. 2001 [in this issue]) may reveal critical motifs within the SEDL protein structure and afford us a better understanding of its function.
Acknowledgments
We thank the patients and their families for making this study possible. This work was supported by National Institutes of Health grants AR45477 (support to G.E.T.), GM60672 (support to H.F.W. and L.C.), P01HD22657 (support to D.L.R. and W.R.W.), and by the National Health and Medical Research Council of Australia. We also thank Janet Matthews, Betty Mekikian, and Loyda Nolasco, for technical assistance; Huntington Willard, for helpful discussions; and Drs. Andre Edmonds and Stuart Smith, for aid in procurement of cartilage samples.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Web/Genbank/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for SEDL [MIM 313400] and sedlin [MIM 300202])
References
- Barrowman J, Sacher M, Ferro-Novick S (2000) TRAPP stably associates with the Golgi and is required for vesicle docking. EMBO J 19:862–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonadio J, Ramirez F, Barr M (1990) An intron mutation in the human alpha 1(I) collagen gene alters the efficiency of pre-mRNA splicing and is associated with osteogenesis imperfecta type II. J Biol Chem 265:2262–2268 [PubMed] [Google Scholar]
- Brodie SG, Lachman RS, Crandall BF, Fox MA, Rimoin DL, Cohn DH, Wilcox WR (1998) Radiographic and morphologic findings in a previously undescribed type of mesomelic dysplasia resembling atelosteogenesis type II. Am J Med Genet 80:247–251 [PubMed] [Google Scholar]
- Brown CJ, Carrel L, Willard HF (1997) Expression of genes from the human active and inactive X chromosomes. Am J Hum Genet 60:1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruening W, Bardeesy N, Silverman BL, Cohn RA, Machin GA, Aronson AJ, Housman D, Pelletier J (1992) Germline intronic and exonic mutations in the Wilms' tumour gene (WT1) affecting urogenital development. Nat Genet 1:144–148 [DOI] [PubMed] [Google Scholar]
- Carrel L, Cottle AA, Coglin KC, Willard HF (1999) A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci USA 96:14440–14444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier L, Bonne G, Bahrend E, Yu B, Richard P, Niel F, Hainque B, Cruaud C, Gary F, Labeit S, Bouhour JB, Dubourg O, Desnos M, Hagege AA, Trent RJ, Komajda M, Fiszman M, Schwartz K (1997) Organization and sequence of human cardiac myosin binding protein C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ Res 80:427–434 [PubMed] [Google Scholar]
- Chomczynski P, Sacci N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed] [Google Scholar]
- De Boer M, Bolscher BG, Sijmons RH, Scheffer H, Weening RS, Roos D (1992) Prenatal diagnosis in a family with X-linked chronic granulomatous disease with the use of the polymerase chain reaction. Prenatal Diag 12:773–777 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Duriez B, Duquesnoy P, Dastot F, Bougneres P, Amselem S, Goossens M (1994) An exon-skipping mutation in the btk gene of a patient with X-linked agammaglobulinemia and isolated growth hormone deficiency. FEBS Lett 346:165–170 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Baldwin CT, Strobel D, Conway D, Horton W, Prockop DJ (1991) Heterozygous mutation in the G+5 position of intron 33 of the pro-alpha 2(I) gene (COL1A2) that causes aberrant RNA splicing and lethal osteogenesis imperfecta. Use of carbodiimide methods that decrease the extent of DNA sequencing necessary to define an unusual mutation. J Biol Chem 266:12035–12040 [PubMed] [Google Scholar]
- Gecz J, Hillman MA, Gedeon AK, Cox TC, Baker E, Mulley JC (2000) Gene structure and expression study of the SEDL gene for spondyloepiphyseal dysplasia tarda. Genomics 69:242–251 [DOI] [PubMed] [Google Scholar]
- Gedeon AK, Colley A, Jamieson R, Thompson EM, Rogers J, Sillence D, Tiller GE, Mulley JC, Gecz J (1999) Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat Genet 22:400–404 [DOI] [PubMed] [Google Scholar]
- Gedeon AK, Tiller GE, LeMerrer M, Heuertz S, Tranebjaerg L, Chitayat D, Robertson S, Glass I, Savarirayan R, Cole WG, Rimoin DL, Kousseff BG, Ikegawa S, Munnich A, Zabel B, Gecz J, Mulley JC (2001) The molecular basis of X-linked spondyloepiphyseal dysplasia tarda. Am J Hum Genet 68:1386–1397 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Nguyen PN, McBride LJ, Koepf SM, Caskey CT (1989) Identification of mutations leading to the LeschNyhan syndrome by automated direct DNA sequencing of in vitro amplified cDNA. Proc Natl Acad Sci USA 86:1919–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuertz S, Smahi A, Wilkie AOM, LeMerrer M, Maroteaux P, Hors-Cayla MC (1995) Genetic mapping of Xp22.12-p22.31, with a refined localization for spondyloepiphyseal dysplasia (SEDL). Hum Genet 96:407–410 [DOI] [PubMed] [Google Scholar]
- Jacobsen AW (1939) Hereditary osteochondrodystrophia deformans: a family with twenty members affected in five generations. JAMA 113:121–124 [Google Scholar]
- Ketterling RP, Drost JB, Scaringe WA, Liao D-Z, Liu J-Z, Kasper CK, Sommer SS (1999) Reported in vivo splice-site mutations in the factor IX gene: severity of splicing defects and hypothesis for predicting deleterious splice donor mutations. Hum Mutat 13:221–231 [DOI] [PubMed] [Google Scholar]
- Kozak M (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates transcription by eukaryotic ribosomes. Cell 44:283–292 [DOI] [PubMed] [Google Scholar]
- Lapoumeroulie C, Pagnier J, Bank A, Labie D, Krishnamoorthy R (1986) Beta thalassemia due to a novel mutation in IVS 1 sequence donor site consensus sequence creating a restriction site. Biochem Biophys Res Commun 139:709–713 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Miller AP, Willard HF (1998) Chromosomal basis of X chromosome inactivation: identification of a multigene domain in Xp11.21-p11.22 that escapes X inactivation. Proc Natl Acad Sci USA 95:8709–8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm S, Christie PT, Finnegan P, Jones J, Dixon PH, Pannett AA, Harding B, Gottesman GS, Thakker RV, Whyte MP (2000) A five–base pair deletion in the sedlin gene causes spondyloepiphyseal dysplasia tarda in a six-generation Arkansas kindred. J Clin Endocrinol Metab 85:3343–3347 [DOI] [PubMed] [Google Scholar]
- Nichols WD, Ginsburg D (1999) From the ER to the Golgi: insights from the study of combined factors V and VIII deficiency. Am J Hum Genet 64:1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer RA, Suess J, Haagen M (1992) An autosomal recessive variant of spondyloepiphyseal dysplasia in three sibs. Pediatr Radiol 22:83–86 [DOI] [PubMed] [Google Scholar]
- Pritzker KPH (1998) Pathology of osteoarthritis. In: Brandt KD, Doherty M, Lohmander LS (eds) Osteoarthritis. Oxford, New York, pp 50–60 [Google Scholar]
- Reitsma PH, Ploos van Amstel HK, Bertina RM (1994) Three novel mutations in five unrelated subjects with hereditary protein S deficiency type I. J Clin Invest 93:486–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoin DL, Rasmussen IM, Briggs MD, Roughley PJ, Gruber HE, Warman ML, Olsen BR, Hsia YE, Yuen J, Reinker K, Garber AP, Grover J, Lachman RS, Cohn DH (1994) A large family with features of pseudoachondroplasia and multiple epiphyseal dysplasia: exclusion of seven candidate gene loci that encode proteins of the cartilage extracellular matrix. Hum Genet 93:236–242 [DOI] [PubMed] [Google Scholar]
- Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR, Abeliovich H, Ferro-Novick S (1998) TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J 17:2494–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schantz K, Andersen PE, Justesen P (1988) Spondyloepiphyseal dysplasia tarda: report of a family with autosomal dominant transmission. Acta Orthop Scand 59:716–719 [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spayde EC, Joshi AP, Wilcox WR, Briggs M, Cohn DH, Olsen BR (2000) Exon skipping mutation in the COL9A2 gene in a family with multiple epiphyseal dysplasia. Matrix Biol 19:121–128 [DOI] [PubMed] [Google Scholar]
- Szpiro-Tapia S, Sefiani A, Guilloud-Bataille M, Heuertz S, LeMarec B, Frezal J, Maroteaux P, Hors-Cayla MC (1988) Spondyloepiphyseal dysplasia tarda: linkage with genetic markers from the distal short arm of the X chromosome. Hum Genet 81:61–63 [DOI] [PubMed] [Google Scholar]
- Taybi H, Lachman RS (1996) Radiology of syndromes, metabolic disorders, and skeletal dysplasias (4th ed) Mosby, New York, pp 926–927 [Google Scholar]
- Thakker-Varia S, Anderson DW, Kuivaniemi H, Tromp G, Shin HG, van der Rest M, Glorieux FH, Ala-Kokko L, Stolle CA (1995) Aberrant splicing of the type III procollagen mRNA leads to intracellular degradation of the protein in a patient with Ehlers-Danlos type IV. Hum Mutat 6:116–125 [DOI] [PubMed] [Google Scholar]
- Tiller GE, Weis MA, Polumbo PA, Gruber HE, Rimoin DL, Cohn DH, Eyre DR (1995) An RNA-splicing mutation (G+5IVS20) in the gene for type II collagen (COL2A1) in a family with spondyloepiphyseal dysplasia. Am J Hum Genet 56:388–395 [PMC free article] [PubMed] [Google Scholar]
- Whyte MP, Gottesman GS, Eddy MC, McAlister WH (1999) X-linked recessive spondyloepiphyseal dysplasia tarda: clinical and radiographic evolution in a 6-generation kindred and review of the literature. Medicine 78:9–25 [DOI] [PubMed] [Google Scholar]
- Wynne-Davies R, Gormley J (1985) The prevalence of skeletal dysplasias: an estimate of their minimum frequency and the number of patients requiring orthopaedic care. J Bone Joint Surg 67:133–137 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Nakazato M, Miyazato M, Kangawa K, Matsukura S (1997) A 5′-splice site mutation in the cytochrome P450 steroid 17alpha-hydroxylase gene in 17alpha-hydroxylase deficiency. J Clin Endocrinol Metab 82:1934–1938 [DOI] [PubMed] [Google Scholar]
- Zhang MQ (1998) Statistical features of human exons and their flanking regions. Hum Mol Genet 7:919–932 [DOI] [PubMed] [Google Scholar]