Abstract
Human HDL-associated paraoxonase (PON1) hydrolyzes a number of toxic organophosphorous compounds and reduces oxidation of LDLs and HDLs. These properties of PON1 account for its ability to protect against pesticide poisonings and atherosclerosis. PON1 also hydrolyzes a number of lactone and cyclic-carbonate drugs. Among individuals in a population, PON1 levels vary widely. We previously identified three polymorphisms in the PON1 regulatory region that affect expression levels in cultured human hepatocytes. In this study, we determined the genotypes of three regulatory-region polymorphisms for 376 white individuals and examined their effect on plasma-PON1 levels, determined by rates of phenylacetate hydrolysis. The −108 polymorphism had a significant effect on PON1-activity level, whereas the −162 polymorphism had a lesser effect. The −909 polymorphism, which is in linkage disequilibrium with the other sites, appears to have little or no independent effect on PON1-activity level in vivo. Other studies have found that the L55M polymorphism in the PON1-coding region is associated with differences in both PON1-mRNA and PON1-activity levels. The results presented here indicate that the L55M effect of lowered activity is not due to the amino acid change but is, rather, largely due to linkage disequilibrium with the −108 regulatory-region polymorphism. The codon 55 polymorphism marginally appeared to account for 15.3% of the variance in PON1 activity, but this dropped to 5% after adjustments for the effects of the −108 and Q192R polymorphisms were made. The −108C/T polymorphism accounted for 22.8% of the observed variability in PON1-expression levels, which was much greater than that attributable to the other PON1 polymorphisms. We also identified four sequence differences in the 3′ UTR of the PON1 mRNA.
Introduction
Human paraoxonase (PON1 [MIM 168820]) is an HDL-associated enzyme (Kitchen et al. 1973; Don et al. 1975; Mackness et al. 1985). PON1 was first investigated for its ability to hydrolyze organophosphorous (OP) compounds, such as the highly toxic oxon forms of the pesticides parathion (Aldridge 1953; Playfer et al. 1976; Geldmacher-v Mallinckrodt et al. 1979), chlorpyrifos (Furlong et al. 1988, 1989), and diazinon, as well as the nerve agents sarin and soman (Davies et al. 1996). In recent years, PON1 has been found to protect LDL and HDL from oxidation (Watson et al. 1995; Aviram et al. 1998a, 1998b; Mackness et al. 1998b; Cao et al. 1999). The classes of substrates for PON1 that most recently have been identified include a number of drugs and a drug precursor, with important pharmacokinetic implications. Tongou et al. (1998) have shown that the prodrug NM441 is converted to its active form (NM394) by PON1, and Billecke et al. (2000) have shown that PON1 metabolizes lactones and cyclic-carbonate esters, including several of the statin drugs used to control cholesterol levels. The hydrolysis of glucocorticoid γ-lactones and cyclic-carbonate esters by PON1 provides a mechanism to confine the active forms of drugs to their target sites (e.g., lung) by rapidly metabolizing them as they enter the plasma (Biggadike et al. 2000).
Several types of evidence suggest that low levels of PON1 protein raise the risk and severity of both OP poisoning and atherosclerosis. Species with low PON1 activity are more sensitive to OP poisoning than are species with higher PON1 activity (Costa et al. 1987). PON1-null mice on a high-fat diet had larger atherosclerotic lesions than were seen in either wild-type or heterozygous mice (Shih et al. 1998). Additionally, PON1-null mice are 5–10 times more sensitive to cholinesterase inhibition by diazoxon and chlorpyrifos oxon than are wild-type mice (Furlong et al. 1998; Shih et al. 1998; Li et al. 2000); however, intraperitoneal injection of purified PON1 into these mice reconstitutes plasma PON1 and resistance to diazoxon and chlorpyrifos oxon (Li et al. 2000). In humans, wide variation (i.e., ⩾13-fold) in PON1-protein levels has been found among individuals (La Du et al. 1986; Furlong et al. 1989; Davies et al. 1996; Richter and Furlong 1999). Relative to that in controls, PON1 activity has been found to be lower in individuals with either non–insulin-dependent diabetes mellitus (Mackness et al. 1998c; Sakai et al. 1998), familial hypercholesterolemia (Tomas et al. 2000), coronary artery disease (James et al. 2000b), or carotid artery disease (Jarvik et al. 2000).
The PON1-coding region contains two common polymorphisms, a leucine (L) to methionine (M) substitution at codon 55 and a glutamine (Q) to arginine (R) substitution at codon 192 (Adkins et al. 1993; Humbert et al. 1993). The Q192R polymorphism causes substrate-dependent differences in the kinetics of hydrolysis by each PON1192 isoform, such that paraoxon (Adkins et al. 1993; Humbert et al. 1993; Li et al. 2000) and chlorpyrifos oxon (Li et al. 2000) are hydrolyzed in vitro more efficiently by PON1R192, whereas soman and sarin are hydrolyzed more rapidly by PON1Q192 (Davies et al. 1996). The Q192R polymorphism has been reported to affect the enzyme's in vivo ability to hydrolyze oxidized lipids, in both LDL and HDL (Aviram et al. 1998a; Mackness et al. 1998b; Cao et al. 1999). The PON155 polymorphism does not affect the catalytic efficiency of substrate hydrolysis by the enzyme, but the PON1M55 allele is correlated with decreased mRNA and protein levels (Blatter Garin et al. 1997; Leviev et al. 1997; Mackness et al. 1998c; Brophy et al. 2000).
Although the substrate-dependent PON1 Q192R polymorphism has been examined in some detail, the genetic basis for the high interindividual variability in serum PON1 levels has just recently begun to be understood. Our laboratory and two other groups recently identified five polymorphisms in the PON1 regulatory region (Leviev and James 2000; Suehiro et al. 2000; Brophy et al. 2001); these polymorphisms are at −107/−108, −126, −160/−162, −824/−832, and −907/−909, where the base immediately preceding the start codon is numbered as “−1.” The nomenclature differences for four of the polymorphisms are likely due to small variations in the sequences examined by the three different groups. In our laboratory, data generated from cell-culture experiments with a reporter gene indicated that three of the five polymorphisms had a functional effect on PON1 expression (Brophy et al. 2001). In this report, we examine the three PON1 regulatory-region polymorphisms—that is, −108, −162, and −909—to determine their effects on PON1 activity level in a white population sample.
Material and Methods
Samples
Genomic DNA and lithium-heparin–plasma samples were obtained from volunteers who were participants in a project at the Puget Sound Veterans Affairs Health Care System (PSVAHCS) Epidemiology Research and Information Center. The white sample population consisted of control subjects and patients (97.0% male) with varying degrees of carotid artery disease. Individuals with internal stenosis <15% (controls), 15%–80%, or >80% comprised 49.1%, 10.5%, and 40.4% of the population, respectively. No differences in PON1192gene frequencies between the patients and the controls were observed in this group; details of subject selection can be found in the report by Jarvik et al. (2000). Subject DNA was prepared from buffy-coat preparations, by a modification of the procedure of Miller et al. (1988), with Puregene reagents (Gentra). The study was approved by both the University of Washington and the PSVAHCS human-subject review processes. All subjects gave written, informed consent.
Genotyping
All genotyping was conducted by PCR amplification, followed by polymorphism-specific restriction digestion and gel electrophoresis. The Q192R polymorphism was detected by AlwI digestion, and the L55M polymorphism was detected by NlaIII digestion, as described elsewhere (Humbert et al. 1993). The genotype of the −108 polymorphism was determined by PCR amplification with primers GACCGCAAGCCACGCCTTCTGTGCACC and TATATTTAATTGCAGCCGCAGCCCTGCTGGGGCAGCGCCGATTGGCCCGCCGC, with 5% dimethyl sulfoxide (DMSO) and Taq polymerase (Promega) and at an annealing temperature of 63°C, for 25 cycles. The latter primer creates a BstUI site (New England Biolabs) when a C is present at −108. After digestion, the products were analyzed by 3% agarose gel (Sigma). Presence of a −108C allele results in digested bands of 52 bp and 67 bp (instead of an undigested band of 119 bp). The −162A/G polymorphism was PCR amplified with primers GCTATTCTTCAGCAGAGGGT and TGAATCTGTAGCCAGGGCAC, with 5% DMSO and Taq polymerase and at an annealing temperature of 56°C, for 30 cycles. The 1,210-bp PCR product was digested with BstUI and was electrophoresed through 1% agarose. Digested bands (674 bp and 536 bp) indicated the presence of G at −162, whereas absence of digestion indicated the presence of an A allele. The −909G/C polymorphism was PCR amplified with primers AACATGTCACTGTGGCATATATAATGCTC and TATTATAATATATTATATCATTCACAGTAACAGCAGACAGCAGAGAAAAGA, with 5% DMSO and Taq polymerase and at an annealing temperature of 60°C, for 35 cycles. The latter primer removes a second BsmAI site, leaving one BsmAI site when a G is present at −909, resulting in digested bands of 50 bp and 206 bp.
PON1-Activity Assays
Rates of phenylacetate hydrolysis (arylesterase activity) are neutral with respect to the Q192R polymorphism and are in a linear relationship with PON1-expression levels (Furlong et al. 1993; Blatter Garin et al. 1997). Arylesterase activities for individuals with sequence differences in the 3′ UTR were determined as described elsewhere (Kitchen et al. 1973). Arylesterase activities of the population whose regulatory-region polymorphisms were studied were determined as above, except that a SpectraMax Plus (Molecular Devices) plate reader was used to measure rates of hydrolysis. Ten microliters of a 1:40 dilution of human plasma was mixed with 200 μl of substrate (3.26 mM phenylacetate in 9 mM Tris-Cl pH 8 and 0.9 mM CaCl2). The rates of hydrolysis were determined by analysis in a plastic UV-transparent 96-well plate (Costar), at 270 nm for 4 min. The initial, linear rates of hydrolysis were used for rate calculation. Samples were assayed in triplicate. Because of the difference in assay format, a conversion factor was necessary for comparison of the data versus the results of the standard assay, in a 1-cm quartz cuvette. A subset of samples was assayed by both methods, and the arylesterase activity (in U/ml) was calculated; the results were plotted against each other, and a linear-regression line was drawn (R2=.93). To convert the plate-reader results for comparison with standard DU70 spectrophotometer-determined rates, the intercept (18,950 U/ml) was subtracted from the plate-reader assay arylesterase activity, and the result was divided by the slope (2,105.8 U/ml).
Identification of 3′ UTR Sequence Differences
The regions encompassing the poly(A) signal sequences were PCR amplified from genomic DNA preparations from four individuals: one with low plasma PON1 arylesterase activity, one with intermediate plasma PON1 arylesterase activity, and two with high plasma PON1 arylesterase activity. The PCR primers used were GGACATCATGAAGCATCAAAGC and CCTATGTGTCATTGCAACAGG. The PCR products were cloned into plasmid pCRII and then were sequenced by AmpliTaq FS dye terminator cycle sequencing (Perkin-Elmer) and primer M13 (forward).
Statistical Analyses
Statistical analysis was conducted with SPSS 8.0 (SPSS). The χ2 test was used both to test if genotype frequencies deviated from Hardy-Weinberg–equilibrium expectations and to evaluate the significance of the linkage disequilibrium between each polymorphism pair. Arylesterase-activity differences between genotypes were evaluated by either analysis of variance (ANOVA) or t-test, and, of the total variance (vt) in arylesterase activity, the proportion (vg/vt) due to each of the five genetic polymorphisms was estimated two ways: first, the marginal effect that each polymorphism had on unadjusted arylesterase activity was estimated as the genetic variance divided by the total variance; second, the conditional effect of each polymorphism, given the effect of polymorphisms with a greater effect, was sequentially estimated. The PON1−108 polymorphism had the greatest effect; thus, linear regression with two dummy variables was used to adjust arylesterase for the PON1−108 genotypes. The regression residuals were relocated to the arylesterase mean of 100.0742 U/ml, and, of the total variance of each relocated residual genetic variance, the proportion due to each additional polymorphism was computed. The polymoprhism with the greatest effect was then adjusted for, and the new residuals were tested for the effects of the remaining polymorphisms. This process was repeated until all polymorphisms had been considered.
Results
A total of three PON1 regulatory-region polymorphisms (−108C/T, −162A/G, and −909G/C) and two PON1 coding-region polymorphisms (L55M and Q192R) were genotyped in a population of 376 white individuals. The allele frequencies determined, shown in table 1, are consistent with published reports on whites (Cascorbi et al. 1999; James et al. 2000b; Leviev and James 2000). The allele frequencies for both the coding-region and the −162 regulatory-region polymorphisms differed from those of a Japanese population, as expected (Imai et al. 2000; Suehiro et al. 2000), although the −108 allele frequencies did not differ. Genotypes at all five positions do not deviate from Hardy-Weinberg–equilibrium expectations. There is significant linkage disequilibrium across the entire regulatory region (table 2). As described elsewhere (Blatter Garin et al. 1997; Mackness et al. 1998c; Schmidt et al. 1998), the PON155L allele also is in linkage disequilibrium with the PON1192R allele. In addition, we observed that PON155L is in linkage disequilibrium with −108C, −162A, and −909G.
Table 1.
Allele Frequencies of the PON1 Polymorphisms
|
Frequency Reported by |
|||||
| James et al.(2000b) |
|||||
| Positionand Allele | PresentStudy | LevievandJames(2000) | Suehiroet al.(2000) | +CHD | −CHD |
| −909: | |||||
| G | .46 | .41 | … | … | … |
| C | .54 | .59 | … | … | … |
| −162: | |||||
| A | .23 | … | .10 | … | … |
| G | .77 | … | .90 | … | … |
| −108: | |||||
| C | .50 | .46 | .48 | .44 | .51 |
| T | .50 | .54 | .52 | .56 | .49 |
| 55: | |||||
| L | .64 | .65 | .94 | .62 | .60 |
| M | .36 | .35 | .06 | .38 | .40 |
| 192: | |||||
| Q | .73 | .69 | .40 | .67 | .73 |
| R | .27 | .31 | .60 | .33 | .27 |
Table 2.
χ2 P Values for Pairs of PON1 Polymorphisms Demonstrating Linkage Disequilibrium
|
P at |
|||||
| Position | −909 | −162 | −108 | 55 | 192 |
| −909 | … | <.001 | <.001 | <.001 | .004 |
| −162 | … | <.001 | <.001 | .017 | |
| −108 | … | <.001 | .002 | ||
| 55 | … | <.001 | |||
| 192 | … | ||||
Arylesterase activities for each genotype were evaluated by ANOVA to test for evidence of genotype effect on activity level (table 3). Each polymorphism, whether coding or noncoding, is associated with statistically significant variability in arylesterase values. Comparison of individuals with two (fig. 1) or three (table 4) of the regulatory-region polymorphisms associated with high expression versus those with the regulatory-region polymorphisms associated with low expression reveals a twofold difference in activity levels.
Table 3.
Mean ±SD Arylesterase Activity, for Each PON1 Genotype
| Position and Genotype (No. of Cases) | Mean ±SDArylesteraseActivity(U/ml) | Pa |
| −909: | <.001 | |
| GG (79) | 128.8 ± 46.0 | |
| CG (189) | 103.0 ± 37.5 | |
| CC (108) | 73.9 ± 32.9 | |
| −162: | <.001 | |
| AA (18) | 135.9 ± 50.4 | |
| AG (140) | 117.1 ± 41.4 | |
| GG (218) | 86.2 ± 37.2 | |
| −108: | <.001 | |
| CC (94) | 125.8 ± 45.6 | |
| CT (188) | 102.9 ± 37.6 | |
| TT (94) | 68.6 ± 27.9 | |
| 55: | <.001 | |
| LL (147) | 115.0 ± 42.8 | |
| LM (185) | 94.0 ± 39.7 | |
| MM (44) | 75.9 ± 38.3 | |
| 192: | .015 | |
| QQ (195) | 104.1 ± 45.4 | |
| QR (158) | 98.5 ± 40.3 | |
| RR (23) | 77.4 ± 27.4 |
Evaluated by ANOVA.
Figure 1.
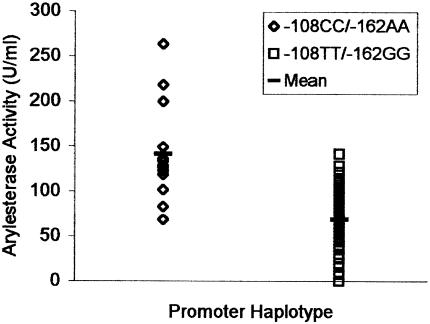
Arylesterase activity in individuals homozygous for both polymorphisms associated with either high or low plasma-PON1 activity. For −108CC/−162AA, N=14 and mean = 140.9 U/ml; for −108TT/−162GG, N=89, mean = 67.5 U/ml, and P<.001 (t-test). (Data available on request from the corresponding author.)
Table 4.
Mean ±SD Arylesterase Activity, for PON1 Regulatory-Region Genotypes
| Position | −909 | −162 | −108 | No. ofCases | Mean ±SDArylesteraseActivity(U/ml) | P |
| −909 | CC | GG | CT | 20 | 96.0 ± 41.5 | .959 |
| CG | GG | CT | 83 | 96.5 ± 35.8 | ||
| −162 | CG | AG | CT | 80 | 109.3 ± 36.9 | .026 |
| CG | GG | CT | 83 | 96.5 ± 35.8 | ||
| −108 | CC | GG | CT | 20 | 96.0 ± 41.5 | .009 |
| CC | GG | TT | 86 | 68.2 ± 28.1 | ||
| All three | GG | AA | CC | 14 | 140.9 ± 52.7 | <.001 |
| CG | AG | CT | 80 | 109.3 ± 36.9 | ||
| CC | GG | TT | 86 | 68.2 ± 28.1 |
Although each polymorphism genotyped in this study is associated with variability in arylesterase activity, the linkage disequilibrium among all of these polymorphisms makes it unclear whether activity differences were due to a specific polymorphism or to linkage disequilibrium with another position. We used two strategies to reduce this confounding effect: first, we subdivided the subjects by genotype at each of the three regulatory-region polymorphisms; second, we examined the residual genetic variance when adjustments were made for sites with greater effects. Each regulatory-region position was examined for PON1-activity differences while the genotype at the other two regulatory-region polymorphism positions was held constant (table 4). The −108 haplotype pair showed a significantly higher PON1-activity level when C was present compared to when T was present. Likewise, the −162A allele had a statistically significant effect, although the arylesterase-activity level differences were less than those for −108 and were not adjusted for linkage disequilibrium with 55 or 192. When the genotypes at 55 and 192 (LM and QQ, respectively) were also held constant, −162 remained significant (P=.025). In contrast, the −909G/C polymorphism appears not to independently affect PON1-activity levels.
Estimated marginal proportions and conditional proportions, vg/vt, of the total variance, vt, in arylesterase activity that are due to each of the five genetic polymorphisms are given in table 5. The greatest effect appears to be that of the PON1−108 site, accounting for 22.8% of the variation in arylesterase activity. The significant linkage disequilibrium between the polymorphisms results in dependence between the estimated marginal vg/vt. Given the effect of the PON1−108 site, 5.7% of the remaining variance is associated with PON1192. Given the PON1−108- and PON1192-site effects, 4.1% of the remaining variance is associated with the PON155 polymorphism. The PON1−162 site accounts for only 1.1% of the variance in PON1−108-, PON1192-, and PON155-adjusted arylesterase activity. The effect of the PON1−909 polymorphism that appeared, for marginal vg/vt, quite great (20.5 %; table 5) is trivial, once the other polymorphisms are accounted for.
Table 5.
Estimated Marginal and Serial Conditional Proportions vg/vt of the Total Variance vt, in Arylesterase Activity, Due to Each of the Five Genetic Polymorphisms
|
Meana |
|||||||
| PON1 Site | vg/vtMarginal | ArylesteraseAdjusted for | vt | 1 | 2 | 3 | vg/vtConditional |
| −108 | .228 | 1,831.31 | 125.80 | 102.94 | 68.61 | .228 | |
| 192 | .022 | 108 | 1,413.22 | 105.88 | 97.50 | 68.53 | .057 |
| 55 | .153 | 108, 192 | 1,330.32 | 109.09 | 95.40 | 89.60 | .041 |
| −162 | .095 | 108, 192, 55 | 1,250.83 | 112.17 | 102.71 | 97.38 | .011 |
| −909 | .205 | 108, 192, 162, 55 | 1,231.25 | 101.16 | 99.09 | 101.01 | .001 |
Means 1, 2, and 3 are those of the three genotypes—pp, pq, and qq—for each site, for the arylesterase trait adjusted for the polymorphisms listed. The p allele for sites −108, 192, 55, 162, and −909, respectively, are C, Q, L, A, and C. The overall arylesterase mean was 100.0742 U/ml.
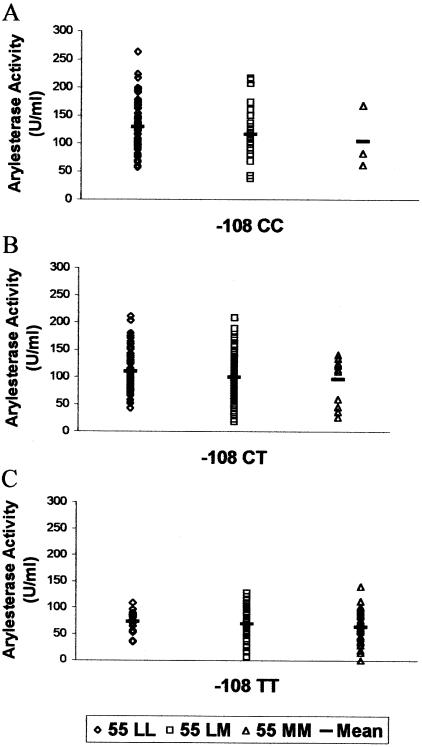
As described elsewhere (Blatter Garin et al. 1997; Mackness et al. 1998c; Brophy et al. 2000), individuals with PON155LL show, on average, a higher level of enzyme activity than do individuals with PON155MM (table 3). The difference in expression level is statistically significant (P<.001) and has been suggested to be due to linkage to other elements, such as regulatory-region polymorphisms (Leviev and James 2000). Our analysis showed that the M allele is indeed in linkage disequilibrium with the regulatory-region polymorphism (i.e., −108T) associated with lower PON1 expression. When the −108 genotype was held constant, arylesterase values shown were not significantly different between individuals with genotype LL and those with genotype MM (fig. 2 and table 6).
Figure 2.
Arylesterase activity in individuals with each 55/−108 haplotype. Means and P values are shown in table 6. The graphs indicate PON1 regulatory-region genotypes of −108CC (A), −108CT (B), and −108TT (C). (Data available on request from the corresponding author.)
Table 6.
Mean ±SD Arylesterase Activity for Each 55/−108 Haplotype
|
Genotype |
||||
| −108 | 55 | No. ofCases | Mean ±SDArylesteraseActivity(U/ml) | P |
| CC | LL | 67 | 129.9 ± 43.2 | .349 |
| CC | LM | 24 | 116.9 ± 50.8 | |
| CC | MM | 3 | 104.4 ± 56.7 | |
| CT | LL | 63 | 110.3 ± 38.6 | .154 |
| CT | LM | 113 | 99.5 ± 36.4 | |
| CT | MM | 12 | 96.2 ± 41.3 | |
| TT | LL | 17 | 73.2 ± 20.4 | .572 |
| TT | LM | 48 | 69.5 ± 28.5 | |
| TT | MM | 29 | 64.5 ± 30.9 | |
The polymorphisms identified in the regulatory region explain part of the variation in PON1-expression levels; however, a significant portion of that variation is still unexplained. We examined the 3′ UTR of PON1 mRNA from four individuals, by sequence analysis. The clones differed in the location of their poly(A) tails, and none of the sequences contained a canonical poly(A) signal of AATAAA; instead, sequences from the clones revealed the use of variants of this sequence (data not shown). Four sequence differences were identified in the 3′ UTR (table 7), but none altered the poly(A) signal sequences. These 3′ sequence differences have not yet been examined for their effects on expression level or for their frequencies in the population.
Table 7.
Examination Results from 3′ UTR of PON1 mRNA
|
Sequence Difference ata |
||||||
| Individual (Allele at 192) | ArylesteraseActivity(U/ml) | No. of Clones | 1290 | 1314 | 1616 | 1647 |
| 1 (Q) | 47 | 6 | A | A | A | C |
| 2 (Q) | 142 | 1 | A | G | A | C |
| 3 | A | A | A | C | ||
| 3 (R) | 103 | 2 | A | A | G | C |
| 3 | A | A | A | C | ||
| 4 (R) | 169 | 1 | G | A | A | C |
| 3 | A | A | A | T | ||
Nucleotide position of mRNA, from initiator ATG.
Discussion
The allele frequencies for PON1 polymorphisms differ among ethnic groups, as shown in table 1. Notably, the only position that does not differ, in allele frequency, between white and Japanese populations, in either the coding or the upstream regulatory region, is −108. This may be a coincidence, or selection pressure(s) may have acted on this polymorphism to maintain specific allele frequencies across different ethnic groups.
The three regulatory and the two coding polymorphisms all show significant linkage disequilibrium to each other, including the four alleles associated with high expression (i.e., −108C, −162A, 55L, and 192Q). The results summarized in table 5 suggest that the PON1−108 polymorphism has the greatest effect on arylesterase activity, followed, in order, by PON1192, PON155, PON1−162, and PON1−909. It is possible that any of the effects listed are, at least in part, due to linkage disequilibrium with other, still unidentified polymorphisms. Our results indicate that the −909G/C polymorphism has little, if any, effect on PON1-activity level but is in linkage disequilibrium with the functional polymorphisms. Interestingly, −108C and −162A, the two regulatory-region alleles that may independently increase PON1 expression (table 4) are in linkage disequilibrium with each other. The −108C allele is also in linkage disequilibrium with the PON1R192 allele, which has been implicated in a lower level of protection against atherosclerosis, although the Q192R polymorphism is associated with activity variance that is independent of the −108C site. It is possible that the −108C allele that increases expression may partly compensate for the lowered protection afforded by the PON1R192 isoform, complicating the apparent relationship between the PON1192 genotype and disease. The L55M polymorphism has been associated with PON1-expression level (Blatter Garin et al. 1997; Leviev et al. 1997; Mackness et al. 1998c; Brophy et al. 2000). The results presented here (fig. 2 and tables 5 and 6) indicate that the PON155 effect is primarily due to linkage disequilibrium of PON155M with the −108T lower-expression variant.
The lack of effect of the −909 polymorphism contrasts with the increased expression from −909G observed in our cell-culture experiments (Brophy et al. 2001). This discrepancy may be due to an inherent difference between a cultured hepatocarcinoma cell line and plasma from normal individuals. Alternatively, sequence-context effects may alter the significance of the −909 polymorphisms; for instance, the −909G allele may have an affect when −162A is present in cis (in vitro data) but not when other polymorphisms are present. We do not have plasma and genotypes from enough individuals of these specific haplotypes to investigate that possibility.
The −108 polymorphism lies within a probable binding site for Sp1, a ubiquitous transcription factor common in TATA-less genes such as PON1. James et al. (2000b) found that the −108T allele was associated with increased risk of coronary artery disease in patients with type 2 diabetes. The −162 polymorphism may be within a possible NF-1 transcription-factor–binding site (Brophy et al. 2001). Interestingly, a recent publication has identified an interleukin-6 (IL-6)–responsive element that shows homology to the sequence including and 3′ of the −162 polymorphism (Ray 2000). IL-6, a proinflammatory cytokine, has been shown to decrease PON1 expression in human HepG2 cells and PON1 activity in the plasma of mice (Van Lenten et al. 2001).
The data shown in figure 1 indicate that possessing efficient PON1 regulatory regions does not alone guarantee a high PON1-activity level. Environmental factors—such as smoking (Nishio and Watanabe 1997; James et al. 2000a; Jarvik et al. 2000) or a high-fat diet (Shih et al. 1996; Hedrick et al. 2000)—may decrease PON1 expression and PON1 activity and may also explain why individuals with efficient regulatory regions have low levels of plasma-PON1 activity. Conversely, consumption of antioxidants—such as those found in pomegranate juice (Aviram et al. 2000), red wine (Hayek et al. 1997), and other alcoholic beverages (when consumed in moderation) (van der Gaag et al. 1999)—may allow an individual with inefficient PON1 regulatory regions to express moderate levels of protein. However, individuals with inefficient regulatory regions appear to be unable to express very high levels of PON1 (fig. 1).
Four sequence differences and variant poly(A) signal sequences were identified in the 3′ UTR of PON1 mRNA. Variant poly(A)-signal sequences are thought to be used inefficiently, resulting in low levels of polyadenylated (stable) mRNA (Sheets et al. 1990; Edwalds-Gilbert et al. 1997). Although the observed sequence differences are not within the variant signal sequences, they may affect the addition of the poly(A) tail or otherwise alter the stability of the PON1 mRNA.
The −108 regulatory-region polymorphism has a significant effect on PON1 expression in humans. Toxicological and epidemiological studies indicate that PON1-expression levels affect the degree of protection that an individual has against pesticide poisoning (Li et al. 1993, 1995, 2000) and heart disease (reviewed in Mackness et al. 1998a; Lusis 2000). Further understanding of PON1 regulatory-region function may reveal approaches for increasing the PON1 expression in at-risk individuals. Because the wide variation in PON1-activity level among individuals cannot be predicted solely by genotype, future studies examining the relationship between PON1 and disease should include determinations of plasma-PON1–activity levels. The determination of PON1 status via the two-dimensional substrate-assay protocol provides an accurate inference of genotype at codon 192, as well as expression levels for individuals (Richter and Furlong 1999; Brophy et al. 2000).
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant ES09883 and by Veteran Affairs Epidemiology Research and Information Center Program award CSP 701S. This study was also supported by National Institutes of Environmental Health Sciences Center grant P30 ES07033, University of Washington Center Grant for Child Environmental Health Risks Research (NIEHS 1 PO1 ES09601/EPA-R826886-01-0), and NIH Training Grant T32 AG 00057-22 (to V.H.B.).
Note added in proof.—
It has recently been brought to our attention that the previously designed primers for genotyping the L55M polymorphism have a mismatch, in the reverse primer, that allows it to bind, but not completely. New primers were created that produce a larger PCR product but still use the same digestion. The new primers, AGAGGATTCAGTCTTTGAGGAAA and CTGCCAGTCCTAGAAAACGTT, were used with Taq polymerase at an annealing temperature of 50°C, for 30 cycles. After digestion, the products were analyzed on a 2% agarose gel (Sigma). Digestion with NlaIII generated either 296-bp and 90-bp fragments, when a 55M allele was present, or an undigested 386-bp fragment, when a 55L allele was present.
Electronic-Database Information
The accession number and URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PON1 [MIM 168820])
References
- Adkins S, Gan KN, Mody M, La Du BN (1993) Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am J Hum Genet 52:598–608 [PMC free article] [PubMed] [Google Scholar]
- Aldridge WN (1953) An enzyme hydrolysing diethyl-p-nitrophenyl phosphate (E-600) and its identity with the A-esterase of mammalian sera. Biochem J 53:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram M, Billecke S, Sorenson R, Bisgaier C, Newton R, Rosenblat M, Erogul J, Hsu C, Dunlop C, La Du B (1998a) Paraoxonase active site required for protection against LDL oxidation involves its free sulfhydryl group and is different from that required for its arylesterase/paraoxonase activities: selective action of human paraoxonase allozymes Q and R. Arterioscler Thromb Vasc Biol 18:1617–1624 [DOI] [PubMed] [Google Scholar]
- Aviram M, Dornfeld L, Rosenblat M, Volkova N, Kaplan M, Coleman R, Hayek T, Presser D, Fuhrman B (2000) Pomegranate juice consumption reduces oxidative stress, atherogenic modifications to LDL, and platelet aggregation: studies in humans and in atherosclerotic apolipoprotein E-deficient mice. Am J Clin Nutr 71:1062–1076 [DOI] [PubMed] [Google Scholar]
- Aviram M, Rosenblat M, Bisgaier CL, Newton RS, Primo-Parmo SL, La Du BN (1998b) Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions: a possible peroxidative role for paraoxonase. J Clin Invest 101:1581–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggadike K, Angell RM, Burgess CM, Farrell RM, Hancock AP, Harker AJ, Irving WR, Ioannou C, Procopiou PA, Shaw RE, Solanke YE, Singh OM, Snowden MA, Stubbs RJ, Walton S, Weston HE (2000) Selective plasma hydrolysis of glucocorticoid γ-lactones and cyclic carbonates by the enzyme paraoxonase: an ideal plasma inactivation mechanism. J Med Chem 43:19–21 [DOI] [PubMed] [Google Scholar]
- Billecke S, Draganov D, Counsell R, Stetson P, Watson C, Hsu C, La Du BN (2000) Human serum paraoxonase (PON1) isozymes Q and R hydrolyze lactones and cyclic carbonate esters. Drug Metab Dispos 28:1335–1342 [PubMed] [Google Scholar]
- Blatter Garin MC, James RW, Dussoix P, Blanche H, Passa P, Froguel P, Ruiz J (1997) Paraoxonase polymorphism Met-Leu54 is associated with modified serum concentrations of the enzyme: a possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J Clin Invest 99:62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy VH, Hastings MD, Clendenning JB, Richter RJ, Jarvik GP, Furlong CE (2001) Polymorphisms in the human paraoxonase (PON1) promoter. Pharmacogenetics 11:77–84 [DOI] [PubMed] [Google Scholar]
- Brophy VH, Jarvik GP, Richter RJ, Rozek LS, Schellenberg GD, Furlong CE (2000) Analysis of paraoxonase (PON1) L55M status requires both genotype and phenotype. Pharmacogenetics 10:453–460 [DOI] [PubMed] [Google Scholar]
- Cao H, Girard-Globa A, Berthezene F, Moulin P (1999) Paraoxonase protection of LDL against peroxidation is independent of its esterase activity towards paraoxon and is unaffected by the Q→R genetic polymorphism. J Lipid Res 40:133–139 [PubMed] [Google Scholar]
- Cascorbi I, Laule M, Mrozikiewicz PM, Mrozikiewicz A, Andel C, Baumann G, Roots I, Stangl K (1999) Mutations in the human paraoxonase 1 gene: frequencies, allelic linkages, and association with coronary artery disease. Pharmacogenetics 9:755–761 [DOI] [PubMed] [Google Scholar]
- Costa LG, Richter RJ, Murphy SD, Omenn GS, Motulsky AG, Furlong CE (1987) Species differences in serum paraoxonase correlate with sensitivity to paraoxon toxicity. In: Costa LG, Galli CL, Murphy SD (eds) Toxicology of pesticides: experimental, clinical and regulatory perspectives. Springer-Verlag, Heidelberg, pp 263–266 [Google Scholar]
- Davies HG, Richter RJ, Keifer M, Broomfield CA, Sowalla J, Furlong CE (1996) The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat Genet 14:334–336 [DOI] [PubMed] [Google Scholar]
- Don MM, Masters CJ, Winzor DJ (1975) Further evidence for the concept of bovine plasma arylesterase as a lipoprotein. Biochem J 151:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res 25:2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong CE, Li WF, Costa LG, Hassett C, Richter RJ, Sundstrom JA, Adler DA, Disteche CM, Omiecinski CJ, Chapline C, Crabb JW, Humbert R (1993) Human and rabbit paraoxonase: purification, cloning, sequencing, mapping and role of the polymorphism in organophosphate detoxication. Chem Biol Interact 87:35–48 [DOI] [PubMed] [Google Scholar]
- Furlong CE, Li WF, Costa LG, Richter RJ, Shih DM, Lusis AJ (1998) Genetically determined susceptibility to organophosphorus insecticides and nerve agents: developing a mouse model for the human PON1 polymorphism. Neurotoxicology 19:645–650 [PubMed] [Google Scholar]
- Furlong CE, Richter RJ, Seidel SL, Costa LG, Motulsky AG (1989) Spectrophotometric assays for the enzymatic hydrolysis of the active metabolites of chlorpyrifos and parathion by plasma paraoxonase/arylesterase. Anal Biochem 180:242–247 [DOI] [PubMed] [Google Scholar]
- Furlong CE, Richter RJ, Seidel SL, Motulsky AG (1988) Role of genetic polymorphism of human plasma paraoxonase/arylesterase in hydrolysis of the insecticide metabolites chlorpyrifos oxon and paraoxon. Am J Hum Genet 43:230–238 [PMC free article] [PubMed] [Google Scholar]
- Geldmacher-v Mallinckrodt M, Hommel G, Dumbach J (1979) On the genetics of the human serum paraoxonase (EC 3.1.1.2). Hum Genet 50:313–326 [DOI] [PubMed] [Google Scholar]
- Hayek T, Fuhrman B, Vaya J, Rosenblat M, Belinky P, Coleman R, Elis A, Aviram M (1997) Reduced progression of atherosclerosis in apolipoprotein E-deficient mice following consumption of red wine, or its polyphenols quercetin or catechin, is associated with reduced susceptibility of LDL to oxidation and aggregation. Arterioscler Thromb Vasc Biol 17:2744–2752 [DOI] [PubMed] [Google Scholar]
- Hedrick CC, Hassan K, Hough GP, Yoo JH, Simzar S, Quinto CR, Kim SM, Dooley A, Langi S, Hama SY, Navab M, Witztum JL, Fogelman AM (2000) Short-term feeding of atherogenic diet to mice results in reduction of HDL and paraoxonase that may be mediated by an immune mechanism. Arterioscler Thromb Vasc Biol 20:1946–1952 [DOI] [PubMed] [Google Scholar]
- Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE (1993) The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet 3:73–76 [DOI] [PubMed] [Google Scholar]
- Imai Y, Morita H, Kurihara H, Sugiyama T, Kato N, Ebihara A, Hamada C, Kurihara Y, Shindo T, Oh-hashi Y, Yazaki Y (2000) Evidence for association between paraoxonase gene polymorphisms and atherosclerotic diseases. Atherosclerosis 149:435–442 [DOI] [PubMed] [Google Scholar]
- James RW, Leviev I, Righetti A (2000a) Smoking is associated with reduced serum paraoxonase activity and concentration in patients with coronary artery disease. Circulation 101:2252–2257 [DOI] [PubMed] [Google Scholar]
- James RW, Leviev I, Ruiz J, Passa P, Froguel P, Garin MC (2000b) Promoter polymorphism T(−107)C of the paraoxonase PON1 gene is a risk factor for coronary heart disease in type 2 diabetic patients. Diabetes 49:1390–1393 [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Rozek LS, Brophy VH, Hatsukami TS, Richter RJ, Schellenberg GD, Furlong CE (2000) Paraoxonase (PON1) phenotype is a better predictor of vascular disease than PON1192 or PON155 genotype. Arterioscler Thromb Vasc Biol 20:2441–2447 [DOI] [PubMed] [Google Scholar]
- Kitchen BJ, Masters CJ, Winzor DJ (1973) Effects of lipid removal on the molecular size and kinetic properties of bovine plasma arylesterase. Biochem J 135:93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Du BN, Adkins S, Bayoumi RA (1986) Analysis of the serum paraoxonase/arylesterase polymorphism in some Sudanese families. Prog Clin Biol Res 214:87–98 [PubMed] [Google Scholar]
- Leviev I, James RW (2000) Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arterioscler Thromb Vasc Biol 20:516–521 [DOI] [PubMed] [Google Scholar]
- Leviev I, Negro F, James RW (1997) Two alleles of the human paraoxonase gene produce different amounts of mRNA: an explanation for differences in serum concentrations of paraoxonase associated with the (Leu-Met54) polymorphism. Arterioscler Thromb Vasc Biol 17:2935–2939 [DOI] [PubMed] [Google Scholar]
- Li WF, Costa LG, Furlong CE (1993) Serum paraoxonase status: a major factor in determining resistance to organophosphates. J Toxicol Environ Health 40:337–346 [DOI] [PubMed] [Google Scholar]
- Li WF, Costa LG, Richter RJ, Hagen T, Shih D, Tward A, Lusis AJ, Furlong CE (2000) Catalytic efficiency determines the in-vivo efficacy of PON1 for detoxifying organophosphorus compounds. Pharmacogenetics 10:767–779 [DOI] [PubMed] [Google Scholar]
- Li WF, Furlong CE, Costa LG (1995) Paraoxonase protects against chlorpyrifos toxicity in mice. Toxicol Lett 76:219–226 [DOI] [PubMed] [Google Scholar]
- Lusis AJ (2000) Atherosclerosis. Nature 407:233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackness B, Durrington PN, Mackness MI (1998a) Human serum paraoxonase. Gen Pharmacol 31:329–336 [DOI] [PubMed] [Google Scholar]
- Mackness B, Mackness MI, Arrol S, Turkie W, Durrington PN (1998b) Effect of the human serum paraoxonase 55 and 192 genetic polymorphisms on the protection by high density lipoprotein against low density lipoprotein oxidative modification. FEBS Lett 423:57–60 [DOI] [PubMed] [Google Scholar]
- Mackness B, Mackness MI, Arrol S, Turkie W, Julier K, Abuasha B, Miller JE, Boulton AJ, Durrington PN (1998c) Serum paraoxonase (PON1) 55 and 192 polymorphism and paraoxonase activity and concentration in non–insulin dependent diabetes mellitus. Atherosclerosis 139:341–349 [DOI] [PubMed] [Google Scholar]
- Mackness MI, Hallam SD, Peard T, Warner S, Walker CH (1985) The separation of sheep and human serum “A”-esterase activity into the lipoprotein fraction by ultracentrifugation. Comp Biochem Physiol B 82:675–677 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio E, Watanabe Y (1997) Cigarette smoke extract inhibits plasma paraoxonase activity by modification of the enzyme's free thiols. Biochem Biophys Res Commun 236:289–293 [DOI] [PubMed] [Google Scholar]
- Playfer JR, Eze LC, Bullen MF, Evans DA (1976) Genetic polymorphism and interethnic variability of plasma paroxonase activity. J Med Genet 13:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A (2000) A SAF binding site in the promoter region of human γ-fibrinogen gene functions as an IL-6 response element. J Immunol 165:3411–3417 [DOI] [PubMed] [Google Scholar]
- Richter RJ, Furlong CE (1999) Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics 9:745–753 [PubMed] [Google Scholar]
- Sakai T, Matsuura B, Onji M (1998) Serum paraoxonase activity and genotype distribution in Japanese patients with diabetes mellitus. Intern Med 37:581–584 [DOI] [PubMed] [Google Scholar]
- Schmidt H, Schmidt R, Niederkorn K, Gradert A, Schumacher M, Watzinger N, Hartung HP, Kostner GM (1998) Paraoxonase PON1 polymorphism Leu-Met54 is associated with carotid atherosclerosis: results of the Austrian Stroke Prevention Study. Stroke 29:2043–2048 [DOI] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP (1990) Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res 18:5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DM, Gu L, Hama S, Xia YR, Navab M, Fogelman AM, Lusis AJ (1996) Genetic-dietary regulation of serum paraoxonase expression and its role in atherogenesis in a mouse model. J Clin Invest 97:1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DM, Gu L, Xia YR, Navab M, Li WF, Hama S, Castellani LW, Furlong CE, Costa LG, Fogelman AM, Lusis AJ (1998) Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 394:284–287 [DOI] [PubMed] [Google Scholar]
- Suehiro T, Nakamura T, Inoue M, Shiinoki T, Ikeda Y, Kumon Y, Shindo M, Tanaka H, Hashimoto K (2000) A polymorphism upstream from the human paraoxonase (PON1) gene and its association with PON1 expression. Atherosclerosis 150:295–298 [DOI] [PubMed] [Google Scholar]
- Tomas M, Senti M, Garcia-Faria F, Vila J, Torrents A, Covas M, Marrugat J (2000) Effect of simvastatin therapy on paraoxonase activity and related lipoproteins in familial hypercholesterolemic patients. Arterioscler Thromb Vasc Biol 20:2113–2119 [DOI] [PubMed] [Google Scholar]
- Tougou K, Nakamura A, Watanabe S, Okuyama Y, Morino A (1998) Paraoxonase has a major role in the hydrolysis of prulifloxacin (NM441), a prodrug of a new antibacterial agent. Drug Metab Dispos 26:355–359 [PubMed] [Google Scholar]
- van der Gaag MS, van Tol A, Scheek LM, James RW, Urgert R, Schaafsma G, Hendriks HF (1999) Daily moderate alcohol consumption increases serum paraoxonase activity; a diet-controlled, randomised intervention study in middle-aged men. Atherosclerosis 147:405–410 [DOI] [PubMed] [Google Scholar]
- Van Lenten BJ, Wagner AC, Navab M, Fogelman AM (2001) Oxidized phospholipids induce changes in hepatic paraoxonase and apoJ but not monocyte chemoattractant protein-1 via interleukin-6. J Biol Chem 276:1923–1929 [DOI] [PubMed] [Google Scholar]
- Watson AD, Berliner JA, Hama SY, La Du BN, Faull KF, Fogelman AM, Navab M (1995) Protective effect of high density lipoprotein associated paraoxonase: inhibition of the biological activity of minimally oxidized low density lipoprotein. J Clin Invest 96:2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]