Abstract
The genomewide screen to search for asthma-susceptibility loci, in the Collaborative Study on the Genetics of Asthma (CSGA), has been conducted in two stages and includes 266 families (199 nuclear and 67 extended pedigrees) from three U.S. populations: African American, European American, and Hispanic. Evidence for linkage with the asthma phenotype was observed for multiple chromosomal regions, through use of several analytical approaches that facilitated the identification of multiple disease loci. Ethnicity-specific analyses, which allowed for different frequencies of asthma-susceptibility genes in each ethnic population, provided the strongest evidence for linkage at 6p21 in the European American population, at 11q21 in the African American population, and at 1p32 in the Hispanic population. Both the conditional analysis and the affected-sib-pair two-locus analysis provided further evidence for linkage, at 5q31, 8p23, 12q22, and 15q13. Several of these regions have been observed in other genomewide screens and linkage or association studies, for asthma and related phenotypes. These results were used to develop a conceptual model to delineate asthma-susceptibility loci and their genetic interactions, which provides a promising basis for initiation of fine-mapping studies and, ultimately, for gene identification.
Introduction
Asthma (MIM 600807) is a common respiratory disease characterized by variable airflow obstruction, inflammation of the airways, and bronchial hyperresponsiveness (BHR). Although family studies, twin studies, adoption studies, and segregation analyses have provided evidence for a genetic component in the etiology of asthma, the mode of inheritance is complex and not yet fully understood (Ober and Moffatt 2000). It is likely that several genes, each with moderate-to-major effects, act together with environmental exposures to determine an individual’s overall risk of development of asthma.
Genetic analytic techniques that permit simultaneous consideration of susceptibility genes in multiple chromosomal regions may enhance the mapping of genes controlling risk to common multigenic diseases. Conditional analysis incorporates linkage evidence from one chromosomal region while assessing evidence for linkage at a second region (or multiple regions), by weighting families according to their evidence for linkage at the first region (Kong and Cox 1997; Cox et al. 1999). A two-locus identity-by-descent (IBD) method, which simultaneously evaluates IBD sharing at two unlinked marker loci, represents a similar approach (Cordell et al. 1995, 2000). When two genes influence an individual’s susceptibility to a disease, affected sib pairs may show a predictable relationship in their IBD scores, although quite different genetic models can generate similar profiles of IBD sharing. Once reproducible evidence for interaction has been identified, the likelihood that the regions actually contain susceptibility loci is increased.
The Collaborative Study on the Genetics of Asthma (CSGA) is a multicenter collaborative study supported by the National Heart, Lung and Blood Institute of the NIH (NHLBI/NIH), whose purpose is to identify important loci that contribute to the development of asthma and asthma-associated phenotypes. We have previously reported the results of an initial genomewide screen of the first 140 families (CSGA 1997) and have now collected and genotyped more individuals in the first 140 families as well as in 126 new families. All 266 families were ascertained through sib pairs with asthma at the CSGA centers in the United States. Three hundred twenty-three polymorphic markers, evenly distributed across the genome, were genotyped in the 266 families. Because it is possible that different genes, as well as environmental exposures, influence asthma and asthma-associated phenotypes in individuals of different ethnic backgrounds, three ethnic groups (African Americans, European Americans, and Hispanics) were ascertained in this study.
We report here the results of a systematic genomewide search for asthma-susceptibility genes in the total sample and in each of the three ethnic groups, using multipoint analyses based on allele sharing among affected relatives, to determine evidence for linkage. In addition, we conducted a systematic search for gene-gene interactions, using conditional analysis and the affected-sib-pair two-locus approach. Conditional linkage analysis was performed for the four regions identified in the initial genomewide screen (one region from the entire population and one from each of the three ethnic groups), by assignment of weights to families, on the basis of their nonparametric linkage (NPL) scores at these loci. In the affected-sib-pair two-locus analysis, the observed joint IBD sharing for two marker loci among affected siblings was maximized over several two-locus models. LOD scores were calculated by comparing the likelihood of the observed IBD to that expected under the null hypothesis of no linkage. On the basis of these analyses, we propose a model for loci contributing to asthma susceptibility and for their potential interactions.
Subjects and Methods
Family Ascertainment
The ascertainment scheme has been reported in detail elsewhere (CSGA 1997). In brief, families were ascertained at the four CSGA centers: (1) Johns Hopkins University (Baltimore), with additional recruitment of families at Howard University (Washington, D.C.); (2) The University of Chicago; (3) the University of Minnesota; and (4) the University of Maryland, with additional recruitment of families at the University of New Mexico. Families were ascertained through two siblings with asthma and then were expanded to include other affected relatives, by extending the family either through relatives with asthma or through one unaffected relative. The siblings with asthma in each family met the following criteria: (1) age ⩾6 years; (2) either (a) bronchial hyperresponsiveness, defined by a fall in baseline forced expiratory volume at one second (FEV1) ⩾20% at ⩽25 mg/ml methacholine or (b) bronchodilator reversibility, defined as a ⩾15% increase in baseline FEV1 after inhalation of a bronchodilator (albuterol), in subjects whose FEV1 was reduced; (3) at least two symptoms (cough, wheeze, and dyspnea) consistent with asthma; (4) fewer than three pack years of cigarette exposure; and (5) physician’s diagnosis of asthma and no conflicting pulmonary diagnoses.
All subjects were evaluated by standardized protocols. Baseline spirometry was performed according to American Thoracic Society (ATS) criteria (ATS 1991, 1995). Methacholine was administrated in doubling concentrations (0.15 mg/ml to 10.0 mg/ml, then 25 mg/ml) at 5-min intervals, inhaled through a DeVilbiss #646 nebulizer attached to a solenoid trigger, activated on inspiration for 0.6 s, for each of five breaths (Chai et al. 1975; Chatham et al. 1982). The PC20 FEV1 was defined as the concentration of methacholine that results in a 20% fall in FEV1. Subjects with a PC20 FEV1 ⩽25 mg/ml methacholine were considered to demonstrate BHR. Airway reversibility to a β2 agonist was assessed by administration of albuterol by either a metered dose inhaler or a hand-held nebulizer. Spirometry was repeated 15 min after this treatment.
The standard ATS respiratory questionnaire was modified to include questions about the frequency, severity, and duration of symptoms of both asthma and atopy, to identify conflicting diagnoses, and to assess current and prior asthma therapy. Relatives of probands were classified as having asthma if they reported a past or current history of at least two of the three respiratory symptoms (cough, wheeze, or dyspnea), had a physician's diagnosis of asthma, and demonstrated either BHR or bronchodilator reversibility.
Genotyping
DNA was extracted from whole blood, by standard methods, at each of the four centers and was sent to the Mammalian Genotyping Service in Marshfield, WI, for genotyping. DNA samples were genotyped for 323 polymorphic autosomal markers. The order of and distances between the markers were estimated by CRIMAP (∼5% of these markers were genotyped at individual CSGA centers) (Lander and Green 1987). The resulting maps were consistent with the Marshfield map, and there was no region in which the intermarker distance was >20 cM.
Analytical Methods
Estimates of allele sharing were based on the marker-allele frequencies estimated from the pedigree founders for each ethnic group. However, the impact of marker-allele frequencies on LOD scores was negligible in this study because the parents were genotyped in the majority of the families. Linkage results were very similar, whether the marker-allele frequencies were estimated from each ethnic group or from the overall sample.
Evidence for linkage over the entire genome was obtained by nonparametric multipoint linkage analysis, using the “S(all)” statistic of the computer program GENEHUNTER-PLUS (Kruglyak et al. 1996; Kong and Cox 1997). One-parameter allele-sharing–model LOD scores were calculated based on the distribution of test statistics under the null hypothesis and conditional on the data using the ASM computer program (Kong and Cox 1997), for each population separately and for the three populations combined. For the initial genomewide screen, equal weights were assigned to each family. LOD scores from this analysis can be converted to a χ2 statistic by multiplying by 4.6, which is approximately a χ2 distribution with a mixture of degrees of freedom of 1 and 0, under the null hypothesis of no linkage (Faraway 1993). P values were calculated as .5×[1-(1-P1)(1-P1)], where P1 is the P value of χ2 with 1 df.
Conditional analyses were performed four times, by conditioning on each of four regions identified in the genomewide screen (the region with the highest LOD score, from the total sample as well as from each of the three ethnic populations) and then searching the rest of the genome for evidence of additional loci. In the conditional analyses, the weighting of each family was based on their evidence for linkage at these regions. Two weighting schemes were used to model a positive and a negative relationship between loci on different chromosomes. A positive relationship (gene-gene interaction) was modeled by assigning a weight of 0 to families with linkage scores that either equaled 0 or were negative and a weight of 1 to families with linkage scores that were positive (weight0-1). This positive relationship may be due to various underlying biological models, such as a multiplicative model or positive epistasis. A negative relationship (heterogeneity gene-gene interaction) was modeled by assigning a weight of 1 to families with linkage scores that were negative and a weight of 0 to families with linkage scores that either equaled 0 or were positive (weight1-0).
The empirical significance of the LOD scores from the conditional analyses was estimated from computer simulations. The original linkage score at a specific location for each family was obtained by GENEHUNTER-PLUS, on the basis of the actual data. A fixed proportion representing the actual distribution of linkage scores, by family, at the region that is being conditioned on was used. Weights were then randomly assigned to each family so that the number of families was the same as that included in the actual analysis, and the LOD score was then calculated. This procedure was repeated 5,000 times, and the empirical significance of the observed LOD score was estimated as the proportion of the replicates exceeding the observed conditional LOD score.
The two-locus analyses (joint analysis of two loci simultaneously) were performed by the computer program TWOLOC (Farrell 1997), which uses the output from the program VITESSE (O’Connell and Weeks 1995). These analyses were limited to the markers that had elevated LOD scores in the conditional analyses. The joint probability of affected sib pairs sharing allele i IBD at locus 1 and allele j IBD at locus 2 (zij) was maximized, with the genetic restrictions taken into account, by a quasi-Newtonian algorithm (Cordell et al. 1995). The maximum LOD score (MLS) was then calculated by comparing the maximum likelihood to the likelihood under the null hypothesis of no involvement at either locus. Three two-locus MLS values were calculated—for the general, multiplicative, and heterogeneity models. In the general two-locus model, zij varied freely within the ranges that are genetically valid. In the multiplicative and heterogeneity two-locus models, the maximization procedure was restricted so that the joint IBD sharing was allowed to vary only in a way consistent with the specific model. Relative values of LOD-score statistics for different models can be used to identify that model which best describes the relationship between the different loci. The general model will always fit at least as well as any other model, since all other models are nested within the general model. LOD scores under the multiplicative model equal the sum of the single-locus LOD scores (Cordell et al. 1995).
Although, through use of asymptotic theory (Cox and Hinkley 1974), it may be possible to calculate the distribution of the two-locus LOD score, it would be complicated because of the nonstandard genetic restrictions on this maximization procedure (Cordell et al. 1995). Therefore, computer simulation was performed to evaluate the significance of the observed two-locus LOD scores. Two independent markers were simulated as being unlinked to a dominant disease locus in the 266 families, by means of FASTSLINK (Weeks et al. 1990; Statgen Software Web site). The disease-gene frequency was assumed to be .05, with penetrances of 90% and 1% for the gene carriers and gene noncarriers, respectively. These parameters are consistent with a population prevalence of 10% for asthma and a 10% phenocopy rate. The exact pedigree structure and availability of DNA samples in these CSGA families were used in this simulation. The number of alleles for each marker, as well as the marker-allele frequencies, also reflected the actual markers genotyped. Five thousand replicates were generated, and each replicate was analyzed by TWOLOC (Farrell 1997). The proportion of the 5,000 replicates with a LOD score (under general, heterogeneity, and multiplicative models) exceeding the observed values from analyses of the actual data was used as a nominal P value.
Results
Family Characteristics
The clinical characteristics of all family members with asthma, in each ethnic group, are listed in table 1. Our sample represents the total number of families for whom data has been collected by the CSGA and is a much larger sample than that reported for the set of families in the original study (CSGA 1997). There are no significant differences, in these clinical characteristics, between the first and second set of families (data not shown).
Table 1.
Clinical Characteristics of Family Members with Asthma
|
Mean ± SD (n) |
|||
| Clinical Characteristic | African American | European American | Hispanic |
| Age at exam (years) | 19±12 (350) | 24±15 (444) | 15±11 (91) |
| Age at first symptom (years) | 7±8 (344) | 9±10 (434) | 5±5 (89) |
| Sex ratio (M:F) | 168:18 | 206:23 | 50:4 |
| FEV1 (% predicted)a | 85±18 (350) | 90±17 (444) | 92±19 (91) |
| FEV1/forced vital capacity | 82±10 (350) | 80±10 (444) | 79±10 (91) |
| PC20 FEV1(mg/ml methacholine) | 4.5±5.9 (252) | 5.2±6.5 (394) | 4.8±5.7 (79) |
| Bronchodilator reversibility (%) | 28±13.3 (97) | 33.4±23.3 (50) | 38.9±17.3 (11) |
| ⩾1 positive skin test (%) | 74±44 (345) | 77±42 (438) | 92±27 (91) |
| Mean log of immunoglobulin E (IgE) (IU/ml) | 2.35±.66 (346) | 2.14±.68 (439) | 2.49±.55 (86) |
| Geometric mean IgE concentration | 225 (346) | 139 (439) | 306 (86) |
Because of physical differences in upper-body size, % predicted values are adjusted for age, sex, height, and ethnicity, using the ATS Standards (ATS 1991, 1995). It is important to note that % predicted values are given for comparison purposes only. Actual FEV1 measurements during the methacholine challenge were used to determine whether a subject had bronchial hyperresponsiveness, one of the criteria for asthma.
Genomewide Screen
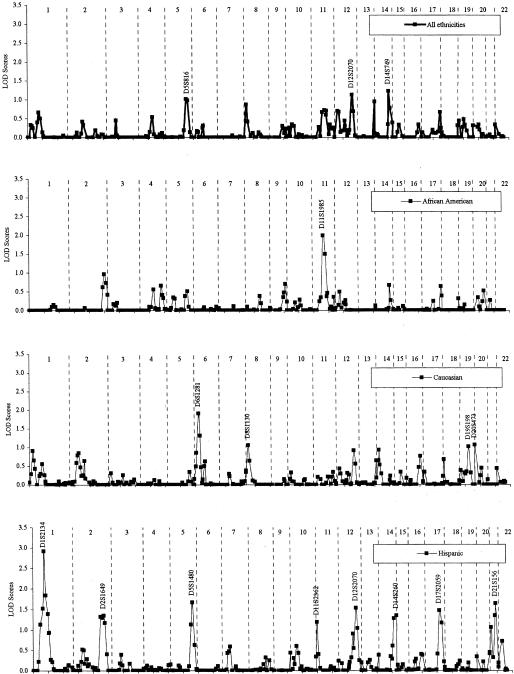
The results of the genomewide screen in the 266 families are presented in figure 1. Multiple regions across the genome showed some evidence for linkage in the total sample, although no chromosomal region had a LOD score >2.0. The highest LOD score was at 14q32 (LOD 1.23, P=.017). Two other regions (5q31 and 12q22) had LOD scores >1.0, whereas seven other regions (1p, 4q, 8p, 11q, 12p, 13q, and 17q) had LOD scores >0.5.
Figure 1.
Results of genomewide screen for asthma-susceptibility loci. Allele-sharing multipoint LOD scores are shown, and the marker identification is given for LOD scores >1. Results for the total CSGA data set are shown in the top graph, followed by the results for each ethnic group.
Analyses were also performed separately for each ethnic group, to investigate genetic differences between the groups. In the 107 African American families, evidence for linkage was found at 11q21, with a LOD score of 2.00 (P=.002) at marker D11S1985, with no other regions having a LOD score ⩾1. In the 129 European American families, evidence for linkage was found at 6p21, with a LOD score of 1.91 (P=.003) at marker D6S1281. LOD scores ⩾1 were found at 8p23, with a LOD score of 1.06 at marker D8S1130; at 19q13, with a LOD score of 1.02 at marker D19S198; and on 20p13, with a LOD score of 1.07 at marker D20S473. In the 30 Hispanic families, the highest LOD score over the whole genome was found on 1p32, with a value of 2.92 (P=.0002) at marker D1S2134. Seven other regions had LOD scores >1 (2q, 5q, 11p, 12q, 14q, 17q, and 21q).
Conditional Analysis
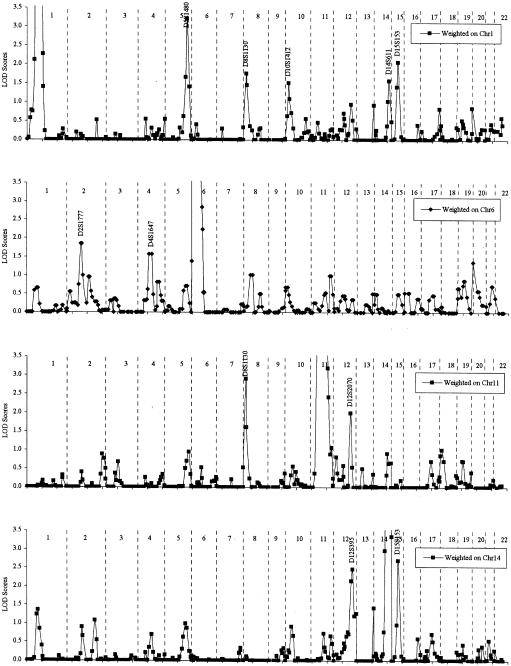
The results of the genomewide conditional analyses in these 266 families are presented in figure 2. Conditional analyses were performed by weighting the families on the basis of the linkage results at four regions—1p32, 6p21, 11q21, and 14q32 (the regions with the strongest evidence for linkage in each ethnic group and in the total sample)—and under the assumption of a positive relationship between loci (i.e., weight0-1). LOD scores that were significantly increased, compared with those in the unconditional analysis, were observed at only four loci: the LOD score at 5q31 (D5S1480) increased from 0.98 to 3.21 when conditioned on the evidence for linkage results at 1p32 (D1S2134); the LOD score at 8p23 (D8S1130) increased from 0.87 to 2.90 when conditioned on linkage results at 11q21 (D11S2002); the LOD scores at 15q13 (D15S153) increased from 0.33 to 2.70 and 2.06, when conditioned on linkage results at 14q32 (D14S749) and 1p32 (D1S2134), respectively; and the LOD scores at 12q22 (D12S2070) increased from 1.13 to 2.15 and 2.00, when conditioned on linkage results at 14q32 (D14S749) and 11q21 (D11S2002), respectively. A similar conditional analysis was performed under the assumption of heterogeneity, but no LOD scores significantly increased over the original results were observed (data not shown).
Figure 2.
Results of conditional analyses for asthma-susceptibility loci, for the total CSGA data set. The genomewide conditional analysis was performed four times, conditional on the evidence for linkage to each of the chromosomes 1, 6, 11, and 14. Allele-sharing multipoint LOD scores are shown, and marker identification is given at the peak LOD-score values.
Computer simulation was used to evaluate empirically the significance of these increased LOD scores obtained from this conditional analysis of the 266 families. The increased LOD scores at four of these chromosomal regions were statistically significant (table 2). In the 5,000 replicates of the conditional analyses, a LOD score ⩾3.21 at 5q31 was observed only once, leading to an empirical P value of .0002. Similarly, no conditional LOD score >2.90 at 8p23 was observed among 5,000 replicates, which leads to an empirical P value <.0002. Evidence for linkage at 15q13 was observed in conditional analyses of both chromosomes 1p32 (P=.0016) and 14q32 (P<.0002).
Table 2.
Summary of Conditional Linkage Analyses of 266 Families
|
MLS |
||||
| Region andConditional Marker | Region andMarker Detected | Primary | Conditional | Empirical P Valuea |
| 1p32 at D1S2134 | 5q31 at D5S1480 | .98 | 3.21 | .0002 |
| 15q13 at D15S153 | .33 | 2.06 | .0016 | |
| 11q21 at D11s2002 | 8p23 at D8S1130 | .87 | 2.90 | <.0002 |
| 12q22 at D12S2070 | 1.13 | 2.00 | .04 | |
| 14q32 at D14S749 | 12q22 at D12S2070 | 1.13 | 2.15 | .02 |
| 15q13 at D15S153 | .33 | 2.70 | <.0002 | |
For conditional analyses, determined from simulations of 5,000 replicates. Conditional analysis was performed for 6p21, but no regions with increased evidence for linkage were detected.
Two-Locus Analyses
Results of the affected-sib-pair two-locus IBD-sharing analyses, as well as those of the single-locus IBD-sharing analysis for the regions identified in the conditional analyses, are presented in table 3. These results can be summarized as follows:
-
1.
There was evidence for linkage in several chromosomal regions when the interactive two-locus analysis was used. Specifically, the two-locus LOD score was 3.23 (P=.001) for markers D14S749 and D12S2070 and 2.88 (P=.003) for markers D11S2002 and D12S2070. The evidence for linkage at the second locus, conditional on linkage at the first locus, is determined by the difference between the general and locus 1 LOD scores; for example, the resulting LOD scores for chromosome 12, conditional on linkage at chromosomes 11 and 14, are 2.88-0.05=2.83 and 3.23-1.84=1.39, respectively.
-
2.
LOD scores under the heterogeneity model were always less than or equal to the LOD scores under the multiplicative model, similar to the results found in the conditional analysis, in which significant linkage results were observed only under a multiplicative model.
-
3.
The two-locus LOD score under the general model for markers D11S2002 and D12S2070 (LOD score 2.88) was higher than the LOD score under either the heterogeneity or the multiplicative model (LOD score 0.94 or 0.98, respectively). This may reflect a complex interaction between genes in these two regions, as could be expected to occur for common diseases, such as asthma, which are likely to have more than two genes—in addition to environmental factors—involved in control of susceptibility. The interaction of the two trait genes may differ among families and across populations, especially if the background effects of other genes and environmental factors vary.
Table 3.
Summary of Two-Locus IBD Models Using Affected Sib Pairs from 266 Families
|
LOD Score (Empirical P Valuea) |
|||||
| Single-Locus |
Two-Locus |
||||
| Markers(Locus 1/Locus 2) | Locus 1 | Locus 2 | Heterogeneity | Multiplicative | General |
| D1S2134/D5S1480 | 1.24 (.02) | .53 (.08) | 1.67 (.02) | 1.77 (.01) | 2.11 (.01) |
| D1S2134/D15S153 | 1.24 (.02) | .05 (.40) | 1.26 (.04) | 1.29 (.04) | 1.68 (.03) |
| D11S2002/D8S1130 | .05 (.40) | .28 (.17) | .33 (.31) | .33 (.31) | .34 (.37) |
| D11S2002/D12S2070 | .05 (.40) | .93 (.03) | .94 (.08) | .98 (.07) | 2.88 (.003) |
| D14S749/D12S2070 | 1.84 (.004) | .93 (.03) | 2.59 (.003) | 2.73 (.001) | 3.23 (.001) |
| D14S749/D15S153 | 1.84 (.004) | .05 (.40) | 1.87 (.01) | 1.89 (.01) | 1.98 (.01) |
Determined from simulation of 5,000 replicates.
Discussion
Genomewide screens for complex disorders often fail to identify chromosomal regions with evidence for linkage compelling enough to justify follow-up studies. The genomewide screen in our 266 families from three ethnic populations provided preliminary evidence for linkage at 14q32, 5q31, and 12q22, with LOD scores of 1–2 (figure 1). Genomewide analysis was performed separately for each of the three ethnic groups, and the strongest evidence for linkage was seen at 6p21 in European Americans, at 11q21 in African Americans, and at 1p32 in Hispanics. These results appear to reflect an inherent mismatch between the genetic complexity of these traits and the rather simplistic genetic analyses usually performed for primary linkage studies. Although we routinely characterize complex disorders as arising from the actions and interactions of many genetic and nongenetic factors, primary analyses generally do not consider the effects from multiple susceptibility factors simultaneously. When multiple loci contribute simultaneously to a trait but only a single susceptibility locus is considered in analysis, the magnitude of the loss of power to detect linkage depends on the underlying genetic model (Knapp et al. 1994). There are, however, clear examples, in animal studies, of loci showing little or no major effects in a primary analysis but, nevertheless, contributing substantially to the genetics of the trait, when analyzed for gene-gene and/or gene-environment interactions (Leiter et al. 1998; Clark 2000; Kuida and Beier 2000).
Results of our primary analysis of the complete CSGA sample is, perhaps, typical of results of genome screens for complex disorders. Although some chromosomal regions reported in previous studies of asthma also provided nominally significant results in the present study (e.g., 12q, 5q, 11q) (Marsh et al. 1994; Barnes et al. 1996; Daniels et al. 1996; Doull et al. 1996; Ober et al. 1998; Bleecker et al. 1999; Wjst et al. 1999), these were not the regions with the highest LOD scores in the current genomewide analyses. Moreover, only a subset of the regions showing the strongest evidence for linkage in the preliminary CSGA (1997) data showed a comparable effect in the total CSGA sample. The differences in magnitude of the LOD scores were due to both the addition of new families and the expansion of a portion of the original families to include additional relatives. The challenge is now to prioritize regions for follow-up fine-mapping studies—and to optimize gene identification, by the development of testable models for interactions between chromosomal regions likely to contain asthma-susceptibility loci.
We have utilized the results from the conditional and two-locus analyses to develop a model for gene-gene interaction. Three observations from the results of the conditional analyses are noteworthy. First, increased evidence for linkage at several regions was repeatedly observed when a positive relationship between the two loci was assumed (table 2). For example, conditional analyses based on evidence of linkage at either 11q21 or 14q32 resulted in significantly increased evidence of linkage at 12q22; the analyses conditional on linkage at either 1p32 or 14q32 resulted in significantly increased evidence for linkage at 15q13; and the analyses conditional on linkage at either 1p32 or 11q21 resulted in significantly increased evidence for linkage at 8p23. Second, increased evidence for linkage at several regions was observed in multiple ethnic groups. Although the evidence for linkage at 5q31 was nominally significant in the Hispanic population in the genomewide screen, increased evidence for linkage (LOD score 3.21) at this region was observed in all three populations after conditioning on linkage at 1p32: the LOD scores increased from 0.98 to 3.21 in the entire group and to 2.1, 1.33, and 0.77 at 5q31 in Hispanics, African Americans, and European Americans, respectively. Third, several regions identified through use of conditional analyses are consistent with those identified in previously published studies. For asthma and related phenotypes, linkage to 5q has been found in several different populations, including the Amish (Marsh et al. 1994), the Dutch (Meyers et al. 1994; Postma et al. 1995), and the Hutterites (Ober et al. 1998). Although originally described for atopy only, linkage to 11q was one of the first linkages reported for atopy or asthma (Cookson et al. 1989). Evidence for linkage at 12q has also been observed in several studies (Barnes et al. 1996; Ober et al. 1998; Wjst et al. 1999). Many of these reported regions are broad, and it is not clear whether there may be multiple susceptibility genes or whether different genes may be detected in different studies.
Affected-sib-pair two-locus analysis, an independent analytical approach complementing conditional analysis, provides further evidence of linkage in several of the regions seen in the conditional analyses (table 4). For example, the estimated joint IBD sharing at 12q22 and 14q32 and at 12q22 and 11q21 were both significantly higher than expected among affected sib pairs, similar to the results at 12q22 after conditioning on linkage at either 14q32 or 11q21. The joint IBD sharing at 1p34 and 5q31 was significantly higher than expected among affected sib pairs, similar to the results seen for 5q31 after conditioning on linkage at 1p32. It is worth noting that, because of computational limitations, nominal P values, which do not consider multiple tests, were reported in the two-locus analyses. Caution should be used in interpreting the level of significance.
Table 4.
Summary of Evidence for Linkage to Chromosomes 1, 5, 11, 12, and 14
|
LOD scores (Empirical P Value) |
||||
| Single Locus |
||||
| Markers (Locus 1/Locus 2) | Locus 1 | Locus 2 | Two Locus | Conditionala |
| D1S2134/D5S1480 | 1.24 | .53 | 2.11 (.01) | 3.21 (.0002) |
| D11S2002/D12S2070 | .05 | .93 | 2.88 (.003) | 2.00 (.04) |
| D14S749/D12S2070 | 1.84 | .93 | 3.23 (.001) | 2.15 (.02) |
Summary of conditional analyses from table 2.
By the nature of the analyses, subsets of pedigrees are used in the conditional analysis, whereas the complete set of pedigrees is used in the two-locus analysis. The pros and cons of the use of a subset versus use of the complete set of families are dependent on the true genetic mechanisms, which remain unknown. However, it is encouraging that similar results were obtained from the two different analytical approaches.
On the basis of the results both of the primary linkage studies in the complete CSGA sample and of subsequent conditional and two-locus analyses, we suggest a model (fig. 3) summarizing the chromosomal regions with evidence for linkage and the possible gene-gene interaction. This model should be tested further, either through primary linkage analysis or through conditional or two-locus analysis of different data sets (Goring et al. 2000). This model may be useful in the setting of priorities for follow-up studies, as well as for the narrowing of regions in fine-mapping studies. For example, the relatively modest LOD score for the 14q region, as well as this region's novel location, would have given it uncertain priority for follow-up studies. However, two different regions showed significantly increased evidence for linkage conditional on evidence for linkage at 14q32, and each of those regions was independently identified in analyses with other loci. This increases the likelihood that 14q32 contains an asthma-susceptibility locus. Although it is not simple to assess the overall significance of the conditional and two-locus analyses, both the consistency of these results across chromosomal regions in our different study populations and their consistency with previously reported asthma linkage studies reinforce the likelihood that these regions contain asthma-susceptibility loci.
Figure 3.
Summary of conditional analysis of 266 CSGA families: conditional on chromosome 11q21, a positive relationship was observed with loci on 12q22 and 8p23; conditional on 14q32, a positive relationship was observed for 12q22 and 15q13; and, conditional on 1p32, a positive relationship was observed for 8p23, 15q13, and 5q31.
Acknowledgments
This study was supported by the following NIH grants: HL49612 (to Johns Hopkins University), HL49596 and MR1 RR00055 (to the University of Chicago), HL49602 (to the University of Maryland; now Wake Forest University), HL49609 and MO1 RR00400 (to the University of Minnesota), HL58977 (to Wake Forest University), HV-48141 (to the Mammalian Genotyping Service), and HRRC 93-196 (to the University of New Mexico). We are indebted to our patients and to the families who have participated in this study. In addition to the authors, other coinvestigators and key support staff for the CSGA include the following: Johns Hopkins University—Alkis Togias, Shau Ku Huang, Linda Freidhoff, Marion Stockton, Eva Ehrlich, and Beverly Plunkett; University of Chicago—Raoul Wolf, Heidi Gidley, Rhonda Peterson, Stephanie Willadsen, Patrick Klimczyck, and Anya Tsalenko; University of Maryland (and University of New Mexico)—Shirley Murphy, Jonathon Samet, O Colin Stine, Lilly Zheng, Adam Boldt, Betsy Rechtsteiner, and Shannon Gierczak; University of Minnesota—William Oetting, Marcia Brott, Duaine Jackola, Andreas Rosenberg, Edward Corazalla, Sandra Leikam, and Lisa Daniels; and Wake Forest University (Data Coordinating Center)—June Pierce and Allison Florance. We wish to acknowledge the Observational and Safety Monitoring Board—William Busse (chair), Ellen Wijsman, and Jeffrey Chamberlain; and National Heart, Lung, and Blood Institute staff—Susan Banks-Schlegel. The Chair of the Steering Committee is rotated between the principal investigators: Carole Ober (current), Terri Beaty, Eugene Bleecker, and Malcolm Blumenthal.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for asthma [MIM 600807])
- Statgen software, http://watson.hgen.pitt.edu/register/soft_doc.html (for FASTSLINK software)
References
- American Thoracic Society (1991) Lung function testing: selection of reference values and interpretive strategies. Am Rev Respir Dis 144:1202–1218 [DOI] [PubMed] [Google Scholar]
- ——— (1995) Standardization of spirometry. 1994 update. Am J Respir Crit Care Med 152:1107–1136 [DOI] [PubMed] [Google Scholar]
- Barnes KC, Neely JD, Duffy DL, Freidhoff LR, Breazeale DR, Schou C, Naidu RP, Levett PN, Renault B, Kucherlapati R, Lozzino S, Ehrlich E, Beaty TH, Marsh DG (1996) Linkage of asthma and total serum IgE concentration to markers on chromosome 12q: evidence from Afro-Caribbean and Caucasian populations. Genomics 37:41–50 [DOI] [PubMed] [Google Scholar]
- Bleecker ER, Postma DS, Howard TD, Koppelman GH, Meijer GG, Xu J, Stine OC, Meyers DA (1999) Genome screen for susceptibility loci for bronchial hyperresponsiveness in a genetically homogeneous Dutch population. Am J Respir Crit Care Med 159:A645 [Google Scholar]
- Chai H, Farr RS, Froehlich LA, Mathison DA, McLean JA, Rosenthal RR, Sheffer AL, Spector SL, Townley RG (1975) Standardization of bronchial inhalation challenge procedures. J Allergy Clin Immunol 56:323–327 [DOI] [PubMed] [Google Scholar]
- Chatham M, Bleecker ER, Norman P, Smith PL, Mason P (1982) A screening test for airways reactivity—an abbreviated methacholine inhalation challenge. Chest 82:15–18 [DOI] [PubMed] [Google Scholar]
- Clark AG (2000) Limits to prediction of phenotype from knowledge of genotypes. In: Clegg MT, Hecht MK, MacIntyre RJ (eds) Evolutionary biology: limits to knowledge in evolutionary genetics, vol 32. Plenum Press, New York, pp 205–224 [Google Scholar]
- Collaborative Study on the Genetics of Asthma (1997) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 15:389–397 [DOI] [PubMed] [Google Scholar]
- Cookson WO, Sharp PA, Faux JA, Hopkin JM (1989) Linkage between immunoglobulin E responses underlying asthma and rhinitis and chromosome 11q. Lancet 8650:1292–1295 [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Todd TA, Bennett ST, Kawaguchi Y, Farrall M (1995) Two locus maximum LOD score analysis of a multifactorial trait: joint consideration of IDDM2 and IDDM4 with IDDM1 in type I diabetes. Am J Hum Genet 57:920–934 [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ, Wedig GC, Jacobs KB, Elston RC (2000) Multilocus linkage tests based on affected relative pairs. Am J Hum Genet 66:1273–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- Cox DR, Hinkley DV (1974) Asymptotic theory. In: Theoretical statistics. Chapman & Hall, London, pp 279-363 [Google Scholar]
- Daniels SE, Bhattacharrya S, James A, Leaves NI, Young A, Hill MR, Faux JA, Ryan GF, LeSouef PN, Lathrop GM, Musk AW, Cookson WO (1996) A genome-wide search for quantitative loci underlying asthma. Nature 383:247–250 [DOI] [PubMed] [Google Scholar]
- Doull IJM, Lawrence S, Watson M, Begishvili T, Beasley RW, Lampe F, Holgate ST, Morton NE (1996) Allelic association of gene markers on chromosomes 5q and 11q with atopy and bronchial hyperresponsiveness. Am J Respir Crit Care Med 153:1280–1284 [DOI] [PubMed] [Google Scholar]
- Faraway JJ (1993) Distribution of the admixture test for the detection of linkage under heterogeneity. Genet Epidemiol 10:75–83 [DOI] [PubMed] [Google Scholar]
- Farrell M (1997) Affected sib pair linkage tests for multiple linked susceptibility genes. Genet Epidemiol 14:103–115 [DOI] [PubMed] [Google Scholar]
- Goring HHH, Terwilliger JD, Blangero J (2000) Genome scans for quantitative trait loci using variance components linkage analysis: upward bias in heritability estimates attributable to individual quantitative trait loci at lod score peaks. Am J Hum Genet Suppl 67:218 [Google Scholar]
- Knapp M, Seuchter SA, Baur MP (1994) Two-locus disease models with two marker loci: the power of affected-sib-pair tests. Am J Hum Genet 55:1030–1041 [PMC free article] [PubMed] [Google Scholar]
- Kong A, and Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kuida S, Beier DR (2000) Genetic localization of interacting modifiers affecting severity in a murine model of polycystic kidney disease. Genome Res 10:49–54 [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter EH, Reifsnyder PC, Flurkey K, Partke H-J, Junger E, Herberg L (1998) Non-insulin dependent diabetes genes in mice: deleterious synergism by both parental genomes contributes to diabetogenic thresholds. Diabetes 47:1287–1295 [DOI] [PubMed] [Google Scholar]
- Marsh DG, Neely JD, Breazeale DR, Ghosh B, Friedhoff LR, Ehrlich-Kautzy E, Schou C, Krishnaswamy G, Beaty TH (1994) Linkage analysis of IL-4 and other chromosome 5q31.1 markers and total serum IgE concentrations. Science 264:1152–1156 [DOI] [PubMed] [Google Scholar]
- Meyers DA, Postma DS, Panhuysen CIM, Xu J, Amelung PJ, Levitt RC, Bleecker ER (1994) Evidence for a locus regulating total serum IgE levels mapping to chromosome 5. Genomics 23:464–470 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- Ober C, Cox NJ, Abney M, DiRienzo A, Lander ES, Changyaleket B, Gidley H, Kurtz B, Lee J, Nance M, Petterson A, Prescott J, Richardson A, Schlenker E, Summerhill E, Willadsen S, Parry R (1998) Genome-wide search for asthma susceptibility loci in a founder population. Hum Mol Genet 7:1393–1398 [DOI] [PubMed] [Google Scholar]
- Ober C, Moffatt MF (2000) Contributing factors to the pathobiology: the genetics of asthma. Clin Chest Med 21:245–261 [DOI] [PubMed] [Google Scholar]
- Postma DS, Bleecker ER, Amelung PJ, Holroyd KJ, Panhuysen CIM, Meyers DA, Levitt RC (1995) Genetic susceptibility to asthma: bronchial hyperresponsiveness coinherited with a major gene for atopy. N Engl J Med 333:894–900 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop GM (1990) SLINK: a general simulation program for linkage analysis. Am J Hum Genet Suppl 47:A204 [Google Scholar]
- Wjst M, Fischer G, Immervoll T, Jung M, Saar K, Rueschendorf F, Reis A, et al (1999) A genome-wide search for linkage to asthma German Asthma Genetics Group. Genomics 58:1–8 [DOI] [PubMed] [Google Scholar]