Abstract
The sequence in the first hypervariable segment (HVS-I) of the control region has been used as a source of evolutionary information in most phylogenetic analyses of mtDNA. Population genetic inference would benefit from a better understanding of the variation in the mtDNA coding region, but, thus far, complete mtDNA sequences have been rare. We determined the nucleotide sequence in the coding region of mtDNA from 121 Finns, by conformation-sensitive gel electrophoresis and subsequent sequencing and by direct sequencing of the D loop. Furthermore, 71 sequences from our previous reports were included, so that the samples represented all the mtDNA haplogroups present in the Finnish population. We found a total of 297 variable sites in the coding region, which allowed the compilation of unambiguous phylogenetic networks. The D loop harbored 104 variable sites, and, in most cases, these could be localized within the coding-region networks, without discrepancies. Interestingly, many homoplasies were detected in the coding region. Nucleotide variation in the rRNA and tRNA genes was 6%, and that in the third nucleotide positions of structural genes amounted to 22% of that in the HVS-I. The complete networks enabled the relationships between the mtDNA haplogroups to be analyzed. Phylogenetic networks based on the entire coding-region sequence in mtDNA provide a rich source for further population genetic studies, and complete sequences make it easier to differentiate between disease-causing mutations and rare polymorphisms.
Introduction
Most phylogenetic analyses of mtDNA have been based on sequence variation in the first hypervariable segment (HVS-I) of the control region (Richards et al. 1998; Macaulay et al. 1999). Both high mutation rate and variation in the substitution rate among sites make the evolution of HVS-I complex, however, and make phylogenetic analyses subject to errors. Parallel mutations at some sites and lack of information at other sites may cause the analyses to fail and to lead to networks either low in resolution or with an incorrect topology.
RFLPs in the coding region have been used to define mtDNA haplogroups (Torroni et al. 1996). Those observed in European populations can be subsumed within four mtDNA haplogroup clusters: HV, UK, TJ, and WIX. The most accurate phylogenetic networks for European mtDNA have been constructed by use of sequence data from HVS-I (Richards et al. 1998), augmented with data from RFLP analyses of the coding region (Macaulay et al. 1999) and from HVS-II sequences (Helgason et al. 2000). The use of HVS-I sequence data has led to inconsistent definition of the haplogroup subclusters, however, since some of the haplogroups have been divided into inappropriately small subgroups whereas it has not been possible to subclassify others at all.
The substitution rate varies between regions in the mtDNA (Pesole et al. 1999), so that the hypervariable segments HVS-I and HVS-II, for example, evolve more rapidly than the coding region (Sigurðardóttir et al. 2000). Population genetic inference would benefit from a better understanding of the variation in the mtDNA coding region, but, thus far, complete mtDNA sequences covering this region have been rare. We have found conformation-sensitive gel electrophoresis (CSGE) to be a highly sensitive and specific method for the screening of mutations and polymorphisms in mtDNA (Finnilä et al. 2000). Our recent data have shown that complete coding-region sequences can be arranged into unambiguous networks (Finnilä et al. 2000, and in press; Finnilä and Majamaa 2001), and, in the present article, we report on construction of accurate phylogenetic networks that are based on 192 complete mtDNA sequences from the Finns. The mtDNA of the Finnish population has shown both high homogeneity in its variation and a clear European pattern (Vilkki et al. 1988; Sajantila et al. 1996; Torroni et al. 1996; Richards et al. 1998; Macaulay et al. 1999), and therefore the networks reveal the variation in European mtDNAs and, in fact, a major part of the maternal genealogy of humans.
Subjects, Material, and Methods
Subjects and Samples
A total of 480 blood samples from healthy donors were obtained from Finnish Red Cross offices in the capitals of the provinces of northern Ostrobothnia, central Ostrobothnia, Kainuu, and northern Savo. It was required that the donors and their mothers should be free of diabetes mellitus, sensorineural hearing impairment, and neurological ailments and that the mothers should have been born in the same province. After this information was obtained, the samples were made anonymous. The research protocol was approved by the Ethics Committees of the Medical Faculty at the University of Oulu and by the Finnish Red Cross. Total DNA was isolated by use of a QIAamp Blood Kit (Qiagen), and the detection of restriction-fragment polymorphisms was used to define the haplogroups (table 1).
Table 1.
Samples in the Study Population
|
No. of Samples |
|||
| Haplogroup Definitiona | Total | Sequenced | |
| H: −7025AluI, −10394DdeI | 188 | 31 | |
| V: −4577NlaIII, −10394DdeI | 27 | 27 | |
| U: +12305DdeI,b −10394DdeI | 134 | 31 | |
| K: +12305DdeI,b +10394DdeI | 12 | 12 | |
| T: +15606AluI, −10394DdeI | 12 | 11 | |
| J: −13704MvaI, +10394DdeI | 26 | 17 | |
| I: −1715DdeI, +8251AvaII, +10394DdeI | 15 | 13 | |
| W: +8251AvaII, −10394DdeI | 46 | 37 | |
| X: −1715DdeI, −10394DdeI | 7 | 4 | |
| Z: +10397AluI, +10394DdeI) | 10 | 9 | |
| Other (not defined) | 3 | 0 | |
| Total | 480 | 192 | |
A plus sign (+) denotes gain of a restriction site; a minus sign (−) denotes loss of restriction site.
Created by use of a mismatched oligonucleotide (Finnilä et al. 2000).
CSGE
CSGE was performed as described elsewhere (Finnilä et al. 2000), the mtDNA coding region and part of the D loop (nucleotides 328–16090) being amplified in 64 partially overlapping fragments. The template DNA was amplified in a total volume of 50 μl, by PCR, in 30 cycles of denaturation at 94°C for 1 min, annealing at a primer-specific temperature for 1 min, and extension at 72°C for 1 min, with a final extension at 72°C for 10 min. The quality of the amplified fragment was estimated visually on a 1.5% agarose gel, and a suitable amount of the PCR product, usually 3–10 μl, was then taken for heteroduplex formation. Each amplified fragment was mixed with the corresponding fragment amplified on a control template with a known sequence (Finnilä et al. 2000). The amplified fragments were denatured at 95°C for 5 min, and the heteroduplexes were subsequently allowed to anneal at 68°C for 30 min. Heteroduplex formation was also allowed to occur autogenously, in order to detect possible heteroplasmic mutations.
The polyacrylamide gel was prepared as described elsewhere (Finnilä et al. 2000) and was preelectrophoresed for 30 min, and the heteroduplex samples were electrophoresed through it, at a constant voltage of 400 V overnight at room temperature. After electrophoresis, the gel was stained in 150 μg of ethidium bromide/liter, for 5 min, on the glass plate, then was destained in water, and finally transferred to a UV transluminator and was photographed (Grab-IT Annotating Grabber 2.04.7; UVP).
Sequencing
Those PCR fragments from within the coding region that showed differential mobility in CSGE were analyzed by automated sequencing (ABI PRISM™ 377 Sequencer using Dye Terminator Cycle Sequencing Ready Kit; Perkin Elmer) after treatment with exonuclease I and shrimp alkaline phosphatase (Werle et al. 1994). The primers used for sequencing the coding region were the same as those used in the amplification reactions for CSGE. The D loop was amplified in a fragment spanning nucleotides 15975–725, and the sequence between nucleotides 16024 and 400 was determined by use of forward primers with their 5′ nucleotides at positions 15975 and 16449, respectively. The sequences in some samples were also determined by use of a reverse primer with the 3′ nucleotide at position 16555. The sequence of the 3′ end of the D loop was determined by CSGE analysis of a fragment spanning nucleotides 328 and 725.
Evolutionary Analysis of Sequences
The phylogenetic network based on the coding-region sequence was constructed by use of a reduced-median algorithm (Bandelt et al. 1995) as implemented in the Network 2.0d program (available at the Life Sciences and Engineering Technology Solutions website). Position 10398 was down-weighted in the analysis, but otherwise the weights of the nucleotide positions were equal. The present data set included 121 coding-region sequences, and an additional 71 sequences were obtained from previous reports (table 1) (Finnilä et al. 2000, and in press; Finnilä and Majamaa 2001). The network based on the nucleotide variation in the D loop was constructed separately, in order to show all the details in the data. All hypervariable sites were down-weighted in the initial construction of the network, and these positions were included in the network afterward.
Estimation of Nucleotide Variation among mtDNA Sequences
The average number of sites differing between a gene region in mtDNA and a putative root, ρGR, was calculated over all the haplotypes for the HVS-I sequences, for the third nucleotides in the structural genes, for the tRNAs, and for the rRNAs. ρGR=Σ(nj/n)ρj, where nj is the number of samples in the jth haplotype, n is the total number of haplotypes, and ρj is the number of mutations observed between the jth haplotype and the root. The lengths of the genes were calculated on the basis of the Cambridge reference sequence (CRS) (Anderson et al. 1981), and this reference was also used for the identification of nucleotide positions.
Results
Sequence Variation in mtDNA from 192 Finns
The data set included all the haplogroups that have been detected in the Finnish population (Meinilä et al., in press). The mtDNA sequence was determined in 121 healthy Finns belonging to haplogroups H, V, W, I, X, or Z, by use of CSGE and direct sequencing. Previously reported sequences from 71 Finns belonging to haplogroups U, K, T, and J were also included in these analyses (Finnilä et al. 2000, and in press; Finnilä and Majamaa 2001). Furthermore, HVS-II sequences were determined for these additional 71 samples.
A total of 297 segregating sites were detected in the coding region characterizing 101 haplotypes, and the number of segregating sites in the entire mtDNA was 413. After the exclusion of the hypervariable sites 303 and 16519, a total of 134 haplotypes could be detected. Mean pairwise sequence difference was 21.2 within the coding region and 10.5 within the D loop. Calculations of the average numbers of nucleotide differences in the HVS-I, tRNA genes and rRNA genes, and in the third nucleotide positions in the structural genes suggested that the variation among the tRNA and rRNA genes is quite low, being <10% of that observed in the HVS-I (table 2). The variation in the third nucleotide positions in the structural genes was also lower than that in HVS-I, being ∼22%.
Table 2.
Nucleotide Variation in Different Genomic Regions of mtDNA, Relative to HVS-I[Note]
| Gene or Genomic Region | No. of Nucleotides | ρGR | Relative Rate |
| HVS-I | 377 | 3.292 | 1 |
| Structural genes, third nucleotides | 3,795 | 7.151 | .21579 |
| rRNA | 2,513 | 1.474 | .0672 |
| tRNA | 1,512 | .776 | .0588 |
Topology of Phylogenetic Networks Based on mtDNA Variation in the Coding Region
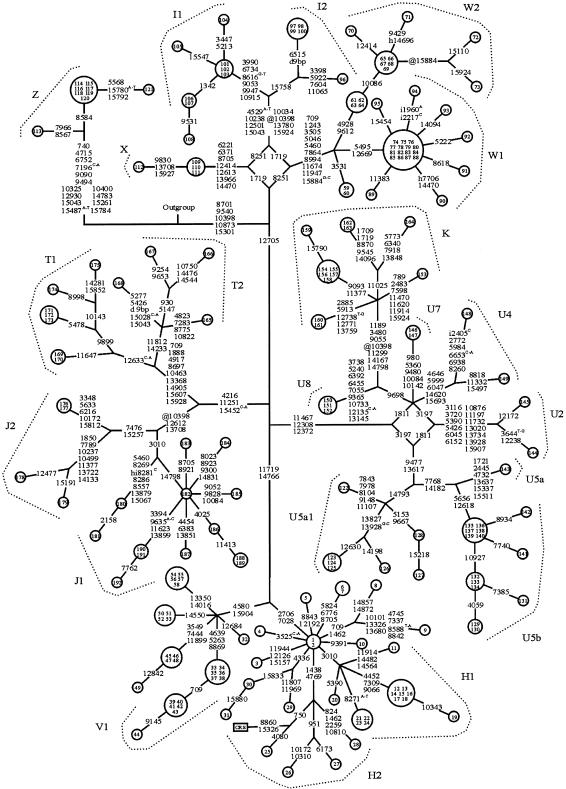
A reduced-median network was constructed by making use of the sequence variation in the coding region and by placing an African mtDNA sequence (Ingman et al. 2000) as the outgroup (fig. 1). Haplogroups H and V formed a cluster that had the 14766C polymorphism in common. The haplogroup H network was highly starlike, and at least two subclusters emerged; the first subcluster, H1, was determined by 3010G→A and encompassed 45% of the samples in haplogroup H, and the second subcluster, H2, was determined by 1438A→G and 4769A→G and encompassed 13% of the samples. The CRS could be placed in subcluster H2 (fig. 1), on the assumption that the following departures from the published sequence were errors: 3106delC, 3423G→T, 4985G→A, 9559G→C, 11335T→C, 13702G→C, 14199G→T, 14272G→C, 14365G→C, 14368G→C and 14766T→C. The nearest neighbor in the network differed from the CRS by 4080T→C, 8860A→G, and 15326A→G. Our findings support the proposed revision of the CRS (Andrews et al. 1999)—with the exception of the status of positions 8860 and 15326, which could not be supported or refuted.
Figure 1.
Phylogenetic network based on variation in the coding sequence in mtDNA from 192 Finnish subjects. The outgroup is mtDNA from an African individual (Ingman et al. 2000; GenBank accession number AF346980). Numbers inside the nodes denote samples. Unless marked otherwise, the polymorphic variants, shown on the lines connecting the nodes, are transitions. Superscripts indicate transversions or inserted nucleotides. Position 10398 was down-weighted in the analysis, but otherwise the weights of the nucleotide positions were equal. A reticulation due to variation at position 15884 within haplogroup W was resolved by assuming a back mutation in samples 72 and 73. The following substitutions were common to all samples: 3423G→T, 4985G→A, 9559G→C, 11335T→C, 13702G→C, 14199G→T, 14272G→C, 14365G→C, 14368G→C, and 3106delC. i = insertion; d 9bp = 9-bp deletion in the CO II-tRNALys intergenic region; @ = back mutation; h = heteroplasmic mutation.
Haplogroup V was determined by polymorphisms 4580G→A and 15904C→T. The network was highly starlike, and at least subcluster V1 could be identified. This subcluster was determined by polymorphisms 4639T→C, 5263C→T, and 8869A→G and encompassed 44% of the samples belonging to haplogroup V. Furthermore, three equally frequent subclusters were present within haplogroup V, but comparison with 129 HVS-I sequences (Torroni et al. 1998) did not reveal similar haplotypes, except for those within subcluster V1. The three other subclusters may therefore include haplotypes specific to the Finns and therefore were not named.
Haplogroup cluster WIX (fig. 1) harbored an unresolved reticulation composed of 1719G→A and 8251G→A, which, however, specifically determined the individual haplogroups. Haplogroup W could be divided into two subclusters. Subcluster W1 was highly starlike and was determined by polymorphisms 5495T→C and 12669C→T. The subcluster formed a large “pincushion and needles” structure, in which 15 samples had an identical genotype and 7 diverged from this by one or two polymorphisms. Subcluster W2 was determined by the polymorphisms 4928T→C and 9612G→A and encompassed 35% of the samples in haplogroup W. Interestingly, the heteroplasmic mutations 7706G→A in COX II and 14696A→G in tRNAGlu were confidently identified in haplogroup W.
Haplogroup I consisted of two subclusters; subcluster I1 was determined by 3990C→T, 6734G→A, 8616G→T, 9053G→A, 9947G→A, and 10915T→C and included 62% of the samples, and subcluster I2 was characterized by the coding-region polymorphism 15758A→G and included 38% of the samples. Only four samples belonged to haplogroup X. The nine samples belonging to haplogroup Z clearly formed an outlier group in the material used in the present study, with 18 unique polymorphisms in the coding region (fig. 1).
Haplogroups U and K (Finnilä et al. 2000, and in press) and haplogroups T and J (Finnilä and Majamaa 2001) were accommodated in the network. The polymorphisms in the coding region (fig. 1) yielded a topology that showed distinct clusters of haplogroups HV, UK, TJ, and WIX and that had a good concordance with the European-mtDNA tree based on variation in the HVS-I (Richards et al. 1998). Specific clusters of polymorphisms served to determine all the haplogroups, and, furthermore, a major division could be made on the basis of the status at position 12705, so that haplogroups H, V, U, K, T, and J harbored 12705C whereas haplogroups I, W, X, and Z harbored 12705T.
Comparison of Networks Based on Variation in the Coding Region and the D Loop
Since the previous networks obtained for Europeans rely on the HVS-I, the coding-region network was compared with that based on the variation in the D loop (fig. 2). Networks were first constructed for each haplogroup separately, and then they were combined to show the general topology. Reticulations were resolved whenever possible by assuming seven back mutations in the less frequent haplotypes. Subclusters could be confidently identified in the D-loop network also, with only a few exceptions: subcluster J2 was divided in two, and subclusters W1 and W2 were partly overlapping, but otherwise there was a good concordance between the subclusters detected in the network based on the coding-region sequence and those detected in the network based on the D-loop sequence.
Figure 2.
Phylogenetic network of mtDNA, based on variation in the D-loop sequence. The outgroup is mtDNA from an African individual (Ingman et al. 2000; GenBank accession number AF346980). Fast-evolving sites 303, 311, and 16519 were not included in the network. i = insertion; d = deletion; @ = back mutation. The superscripts indicate transversions or inserted or deleted nucleotides. For further information, see the legend to figure 1.
Homoplasies in the Coding Region
The total number of homoplasic positions among the 192 mtDNAs was 70 (tables 3 and 4). In the coding region, 21 positions were homoplasic; 7 of these were nonsynonymous, 10 were synonymous, and 6 were other mutations. Homoplasic sites constituted 0.14% of the sites in the coding region and 4.4% of those in the D loop, suggesting a rate ratio of 1:31.
Table 3.
Parallel Mutations Detected in the Coding Region of mtDNA in 192 Finnish Samples
|
Haplogroupa |
||||||||||||
| Position | Gene | Amino AcidSubstitution | H | V | U | K | T | J | W | I | X | Z |
| 709 | 12S rRNA | … | + | + | − | − | + | − | + | − | − | − |
| 1462 | 12S rRNA | … | 2 | − | − | − | − | − | − | − | − | − |
| 1719 | 12S rRNA | … | − | − | − | + | − | − | − | + | + | − |
| 3010 | 16S rRNA | … | + | − | − | − | − | + | − | − | − | − |
| 5390 | ND2 | … | + | − | + | − | − | − | − | − | − | − |
| 5426 | ND2 | … | − | − | + | − | + | − | − | − | − | − |
| 5460 | ND2 | Ala→Thr | − | − | − | − | − | + | + | − | − | − |
| 8269 | Noncoding | del 9 bpb | − | − | − | − | + | − | − | + | − | − |
| 8705 | ATPase6 | Met→Thr | + | − | − | − | − | + | − | − | + | − |
| 10084 | ND3 | Ile→Thr | − | − | + | − | − | + | − | − | − | − |
| 10172 | ND3 | Synonymous | + | − | − | − | − | + | − | − | − | − |
| 10398 | ND3 | Thr→Ala | − | − | − | + | − | + | − | + | − | + |
| 11377 | ND4 | Synonymous | − | − | − | + | − | + | − | − | − | − |
| 11914 | ND4 | Synonymous | + | − | − | + | − | − | − | − | − | − |
| 12414 | ND5 | Synonymous | − | − | − | − | − | − | + | − | + | − |
| 13708 | ND5 | Ala→Thr | − | − | − | − | − | + | − | − | + | − |
| 13928c | ND5 | Ser→Thr/Asn | − | − | 2 | − | − | − | − | − | − | − |
| 14470 | ND6 | Synonymous | − | − | − | − | − | − | + | − | + | − |
| 14798 | Cytochrome b | Phe→Leu | − | − | − | + | − | + | − | − | − | − |
| 15043 | Cytochrome b | Synonymous | − | − | − | − | + | − | − | + | − | + |
| 15924 | tRNAThr | … | − | − | − | + | − | − | + | + | − | − |
A plus sign (+) denotes presence of the mutation; a minus sign (−) denotes absence of the mutation; a numeral denotes the number of repetitions of the mutation.
9-bp deletion in the intergenic region COX II and tRNALys.
A transversion (Ser→Thr) was observed in subcluster U5, and a transition (Ser→Asn) was observed in subcluster U2.
Table 4.
Parallel Mutations Detected in the D Loop of mtDNA in 192 Finnish Samples[Note]
|
Haplogroupa |
|||||||||||
| Position | Nucleotide Change | H | V | U | K | T | J | W | I | X | Z |
| 16051 | A→G | − | − | 2 | − | − | − | − | − | − | − |
| 16093 | T→C | − | − | + | + | − | − | − | − | − | − |
| 16129 | G→A | − | − | + | − | − | − | − | + | − | + |
| 16145 | G→A | − | − | − | − | − | + | − | + | − | − |
| 16146 | A→G | − | − | + | − | + | − | − | − | − | − |
| 16172 | T→C | − | − | − | − | + | + | − | + | − | − |
| 16182 | A→C | − | − | − | − | 2 | − | − | − | − | − |
| 16183 | A→C | − | − | + | − | 2 | − | − | − | + | − |
| 16186 | C→T | − | − | − | − | + | 2 | − | − | − | − |
| 16189 | T→C | − | − | 4 | − | 2 | + | − | − | + | − |
| 16192 | C→T | − | − | 3 | − | − | + | − | − | − | − |
| 16223b | C→T | − | − | + | − | + | − | + | + | + | + |
| 16224 | T→C | − | − | − | + | − | − | − | − | − | + |
| 16256 | C→T | − | − | + | − | + | − | − | − | − | − |
| 16261 | C→T | + | − | − | − | − | + | − | − | − | − |
| 16274 | G→A | + | − | − | − | − | + | − | − | − | − |
| 16278 | C→T | − | − | − | − | − | + | − | − | + | − |
| 16292 | C→T | − | − | − | − | + | − | + | − | − | − |
| 16294 | C→T | − | − | 2 | − | + | − | − | − | − | − |
| 16298 | T→C | − | + | − | − | + | − | − | − | − | + |
| 16304 | T→C | + | − | − | − | 2 | − | − | − | − | − |
| 16311 | T→C | − | − | + | + | − | + | − | + | − | − |
| 16362 | T→C | − | − | 2 | − | − | − | − | − | − | − |
| 16558 | G→A | − | 2 | − | − | − | − | − | + | + | − |
| 73c | A→G | + | − | + | + | + | + | + | + | + | + |
| 93 | A→G | − | 3 | − | − | − | − | − | − | − | − |
| 143 | G→A | − | − | − | − | − | − | + | + | − | − |
| 146 | T→C | + | − | + | + | − | − | − | − | − | − |
| 150 | C→T | − | − | + | − | − | 2 | − | − | − | − |
| 152 | T→C | + | − | + | + | + | + | − | + | − | + |
| 185 | G→A | − | − | − | − | − | 2 | − | − | − | − |
| 188 | A→G | − | − | − | − | − | 3 | − | − | − | − |
| 189 | A→G | − | + | − | − | − | + | + | − | − | − |
| 195 | T→C | − | + | + | 2 | 2 | 2 | 2 | − | + | − |
| 199 | T→C | − | − | − | − | + | − | − | + | − | − |
| 207 | G→A | − | + | − | − | − | − | + | + | − | − |
| 217 | T→C | − | − | 2 | − | − | − | − | − | − | − |
| 227 | A→G | − | 2 | − | − | − | − | + | − | + | − |
| 228 | G→A | − | − | − | − | − | + | − | − | − | − |
| 248 | delA | − | − | + | − | − | − | − | − | − | + |
| 295 | C→T | − | − | − | − | − | + | − | + | − | − |
| 311 | C→T | 2 | + | − | − | + | + | − | + | − | − |
| 322 | G→A | − | − | − | − | − | − | + | + | − | − |
| 462 | C→T | − | + | − | − | − | + | − | − | − | − |
| 489 | T→C | − | − | − | − | − | + | − | − | − | + |
| 497 | C→T | − | − | − | + | − | − | − | − | − | − |
| 498 | C→T | − | − | − | + | − | − | − | − | − | − |
| 514 | insCA | 2 | − | 2 | 2 | + | + | − | − | − | − |
| 568 | insCC | − | + | + | − | − | − | − | − | − | − |
Note.— The following mutational events were not included in the table: 16166A→G in haplogroup H, 16166A→C in haplogroup I, 16166delA in subcluster U4, 16129G→C in subcluster U2, 16183A→G in haplogroup V, 456delC and 456C→T in haplogroup H, 514insCACA in haplogroup H, 514delCA in subcluster U5, 568insC in subcluster U5, 568insCCCC, 568insCCCCC in haplogroup I. Hypervariable positions 303 and 16519 were also excluded.
Notation is as in table 3.
Has shared ancestry in haplogroups I, W, X, and Z but occurs as a parallel mutation in haplogroups T and U.
Has shared ancestry in haplogroups U, K, T, J, I, W, X, and Z but occurs as a parallel mutation in haplogroup H.
Most of the homoplasies occurred in different haplogroups. Only two (9.5%) of the sites in the coding region harbored homoplasies within a haplogroup, whereas in the D loop the corresponding figure was 19 sites (39%). Some of the homoplasies in the D loop occurred at sites that have been used to determine haplogroups; for example, 16223C→T is a mutation that, because of their shared ancestry, occurred in all the samples in haplogroups I, W, X, and Z but was also present in some samples in haplogroups T and U. Similarly, 16298T→C was found in all the samples in haplogroups V and Z and in some samples in haplogroup T, and 16292C→T was found in all but one of the samples in haplogroup W and in one sample in haplogroup T.
Discussion
Topology of mtDNA Phylogenetic Networks
Complete sequences of the human mtDNA have just begun to emerge (Finnilä et al. 2000; Ingman et al. 2000; Elson et al. 2001). In the present article, we have determined the mtDNA sequence for 121 Finns, and, after complementing our recent data (Finnilä and Majamaa 2001; Finnilä et al., in press), we were able to construct a phylogenetic network based on complete mtDNA sequences for 192 Finns, making this the largest set of human mtDNA sequences reported so far. Samples from all the mtDNA haplogroups present in this population were included. Certain nuclear markers (Cavalli-Sforza et al. 1994) and markers on the Y chromosome have suggested that the Finns are outliers in Europe (Sajantila et al. 1996; Zerjal et al. 1997), but the number of mtDNA lineages shared between the Finns and other Europeans is high, suggesting that the Finnish samples represent European lineages (Vilkki et al. 1988; Sajantila et al. 1996; Torroni et al. 1996; Richards et al. 1998; Macaulay et al. 1999). Therefore, the present networks should be useful references for studies on mtDNA population genetics among European populations.
Previous phylogenetic networks for mtDNA have been based on the HVS-I sequence, and only selected polymorphisms in the coding region have been used to construct them (Richards et al. 1998; Macaulay et al. 1999; Helgason et al. 2000). This approach has some shortcomings, owing to the fact that (a) the mutation rate in the HVS-I is several times higher than that in the coding region (Pesole et al. 1999) and (b) the HVS-I sequence is highly variable. We found that, in most haplogroups, the topology of networks based on the coding-region sequence differed from that of networks based on the D-loop sequence. In spite of these differences, however, the polymorphisms in the D loop could, in most cases, be unambiguously placed in networks based on the coding-region sequence. The differences in network topology were due to the high frequency of homoplasies in the D loop, so that a single-nucleotide polymorphism may not be useful for determining a haplogroup. The polymorphisms 16223C→T, 16292C→T, and 16298T→C have been used for this purpose, although they occur as homoplasies in other haplogroups as well. The complete network enabled us to determine the relationships between the haplogroups. Interestingly, each haplogroup was defined by many polymorphisms in the coding region, whereas only a few substitutions in the D loop appeared to be important in this respect.
Solution to Reticulations in the Coding Sequence Network
The network based on the coding-region sequence was unambiguous, although it involved two unresolved reticulations—one in the root of subclusters U2 and U5 and a group composed of subclusters U4, U7, and U8 and haplogroup K and the other in the root of haplogroups I, W, and X. Knowledge on the complete mtDNA sequence enables us to propose a solution to the reticulation within haplogroup UK. The average number of sites differing between a haplotype and the most recent common ancestor was 7.7 in subcluster U5 but was 16.5 in subcluster U2 and 11.6 in the remaining haplotypes (P<.001 for difference; Kruskal-Wallis test). If we assume that mtDNA coding region evolves in a roughly clocklike manner, this difference suggests that subcluster U5 has evolved more recently than its counterparts in the reticulation. Therefore, we suggest that the most recent common ancestor of haplogroups U and K gained, first, 1811A→G, leading to the generation of haplogroup K and subclusters U4, U7 and U8, and, subsequently, 3197T→C, leading to the generation of subcluster U2. At a later point in evolution, the most recent common ancestor gained 3197T→C as a parallel mutation, leading to the generation of subcluster U5. The alternate route of evolution would require a parallel mutation at position 1811.
Homoplasies in the Coding Region
In the present study, we have obtained the first known estimate for the number of homoplasies in the coding region. Previously, only occasional polymorphisms had been found to be homoplasic in the coding region, including the polymorphisms at positions 1719 and 10398 and the loss of a RsaI site at position 3337 (Macaulay et al. 1999). We found the first two polymorphisms in haplogroups K, I, and X and in haplogroups K, J, I and Z, respectively, whereas the third was not a member of any of the networks. The loss of a RsaI site at 3337 suggests a mutation in one of the nucleotides between 3337 and 3340, but none of our networks harbored a polymorphism at any of these positions. Polymorphisms neighboring the homoplasies provide a means for evaluating the possibility of the recombination that has been suggested to take place in mtDNA (Awadalla et al. 1999; Eyre-Walker et al. 1999). The homoplasies identified among the 192 mtDNAs did not share neighboring polymorphism, indicating that they had arisen as solitary mutations rather than as a consequence of the recombination of a larger fragment. The coding region and the D loop differed in the frequency of homoplasies, which was 31-fold higher in the latter. This difference may imply either a higher mutation rate or more-relaxed selection in the D loop. The number of homoplasies in HVS-I was lower than that estimated on the basis of a larger European set of lineages (Richards et al. 1998; Macaulay et al. 1999), probably because of the wider population employed in the latter studies, a difference that suggests that we have underestimated the frequency of coding-region homoplasies. Indeed, 13 homoplasies have been recently discovered in the coding region of 64 European samples (Elson et al. 2001), but only 5 of them were shared with the samples in the present study. The two data sets on homoplasies also differ in the distribution of the homoplasic sites in the genome, since the latter data set includes 12 synonymous polymorphisms and only 1 nonsynonymous polymorphism (Elson et al. 2001).
Coding-Region Sequences as a Basis for mtDNA Genomics
A start for mitochondrial genomics was announced recently (Hedges 2000). The number of complete mtDNA sequences will inevitably grow in the near future, since they provide a rich source of information for studies in population genetics and in mtDNA disease genetics. In the present study, we have employed the sequence data for calculation of certain basic variables showing that the nucleotide variation was lowest in the ribosomal RNA and transfer RNA genes. The phylogenetic networks constructed represent all the haplogroups present in the Finnish population, and, consequently, they should be applicable to studies dealing with other European populations. Moreover, the networks confirm the branching structure of the mtDNA genealogy. Such networks enable further population genetic studies to be performed by use of single-nucleotide polymorphisms and, in mtDNA disease genetics, are required for distinguishing between rare polymorphisms and disease-causing mutations.
Acknowledgments
The expert technical assistance of Mss. Irma Vuoti, Kaisu Korhonen, and Anja Heikkinen is acknowledged. This work was supported, in part, by grants from the Medical Research Council of the Academy of Finland, the Sigrid Juselius Foundation, the Neurology Foundation, the Maire Taponen Foundation, and the Finnish Medical Foundation.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Life Sciences and Engineering Technology Solutions, http://www.fluxus-engineering.com
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for outgroup mtDNA from an African individual [accession number AF346980])
References
- Anderson S, Bankier AT, Barrell BG, deBruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 [DOI] [PubMed] [Google Scholar]
- Awadalla P, Eyre-Walker A, Smith JM (1999) Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science 286:2524–2525 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Elson JL, Andrews RM, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (2001) Analysis of European mtDNAs for recombination. Am J Hum Genet 68:145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Smith NH, Smith JM (1999) How clonal are human mitochondria? Proc R Soc Lond B Biol Sci 266:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S, Hassinen IE, Ala-Kokko L, Majamaa K (2000) Phylogenetic network of the mtDNA haplogroup U in northern Finland based on sequence analysis of the complete coding region by conformation-sensitive gel electrophoresis. Am J Hum Genet 66:1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S, Hassinen IE, Majamaa K. Phylogenetic analysis of mitochondrial DNA in patients with an occipital stroke: evaluation of mutations by using sequence data on the entire coding region. Mutat Res (in press) [DOI] [PubMed] [Google Scholar]
- Finnilä S, Majamaa K (2001) Phylogenetic analysis of mtDNA haplogroup TJ in a Finnish population. J Hum Genet 46:64–69 [DOI] [PubMed] [Google Scholar]
- Hedges SB (2000) Human evolution: a start for population genomics. Nature 408:652–653 [DOI] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir, Gulcher JR, Wars R, Stefánsson K (2000) mtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet 66:999–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Jaksch M, Bezold R, Mertens S, Aholt S, Paprotta A, Gerbitz KD (1997) Population genetics and disease susceptibility: characterization of central European haplogroups by mtDNA gene mutations, correlation with D loop variants and association with disease. Hum Mol Genet 6:1835–1846 [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinilä M, Finnilä S, Majamaa K. Evidence for mtDNA admixture between the Finns and the Saami. Hum Hered (in press) [DOI] [PubMed] [Google Scholar]
- Pesole G, Gissi C, De Chirico A, Saccone C (1999) Nucleotide substitution rate of mammalian mitochondrial genomes. J Mol Evol 48:427–434 [DOI] [PubMed] [Google Scholar]
- Richards MB, Macaulay VA, Bandelt HJ, Sykes BC (1998) Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet 62:241–260 [DOI] [PubMed] [Google Scholar]
- Sajantila A, Salem AH, Savolainen P, Bauer K, Gierig C, Pääbo S (1996) Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc Natl Acad Sci USA 93:12035–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurðardóttir S, Helgason A, Gulcher JR, Stefansson K, Donnelly P (2000) The mutation rate in the human mtDNA control region. Am J Hum Genet 66:1599–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt HJ, D́Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savontaus ML, Bonné-Tamir B, Scozzari R (1998) mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet 62:1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilkki J, Savontaus ML, Nikoskelainen EK (1988) Human mitochondrial DNA types in Finland. Hum Genet 80:317–321 [DOI] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt HJ (1997) Mitochondrial footprints of human expansions in Africa. Am J Hum Genet 61:691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werle E, Schneider C, Renner M, Volker M, Fiehn W (1994) Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res 22:4354–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhovel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183 [PMC free article] [PubMed] [Google Scholar]