Abstract
In 1997, a locus for benign familial infantile convulsions (BFIC) was mapped to chromosome 19q. Further data suggested that this locus is not involved in all families with BFIC. In the present report, we studied eight Italian families and mapped a novel BFIC locus within a 0.7-cM interval of chromosome 2q24, between markers D2S399 and D2S2330. A maximum multipoint HLOD score of 6.29 was obtained under the hypothesis of genetic heterogeneity. Furthermore, the clustering of chromosome 2q24–linked families in southern Italy may indicate a recent founder effect. In our series, 40% of the families are linked to neither chromosome 19q or 2q loci, suggesting that at least three loci are involved in BFIC. This finding is consistent with other autosomal dominant idiopathic epilepsies in which different genes were found to be implicated.
Benign familial infantile convulsions (BFIC [MIM 601764]) is an epileptic phenotype that recently has been recognized as a distinct syndrome segregating as an autosomal dominant trait (Vigevano et al. 1992). Clinical features of BFIC include partial seizures, occurring in clusters lasting over a 2–4-d period, between the ages of 4 and 11 mo. Seizures are characterized by psychomotor arrest, slow deviation of the head and eyes to one side, and asynchronous limb jerks. Ictal EEG shows discharges arising from central-occipital regions and then spreading over the entire brain. Interictal EEG is normal, and no metabolic disorders or brain lesions are observed (Vigevano et al. 1992). Clinically, this disorder is quite homogeneous, and age at onset, features, frequency, and the benign course of partial seizures are very similar among patients from different families.
In 1997, a locus for BFIC was mapped on chromosome 19q, between markers D19S49 and D19S245, by a study of five families of Italian descent (Guipponi et al. 1997). In the same year, Szepetowski et al. (1997) described a related autosomal dominant phenotype (ICCA [MIM 602066] showing benign infantile convulsions associated with paroxysmal choreoathetosis. Mapping of the gene to chromosome 16p demonstrated that BFIC and ICCA are not allelic to the same gene.
Recently, we have further investigated the role of the chromosome 19q BFIC locus by studying a new set of Italian families affected with BFIC. Since no evidence of linkage was found, we have hypothesized genetic heterogeneity (Gennaro et al. 1999).
To identify new BFIC loci, we have performed a genomewide search by studying a large Italian kindred, as described by Giordano et al. (1999) (fig. 1). Since seizures disappear after age 1 year, leaving no clinical sign of the disease, and diagnosis in adult individuals can be established only on the basis of anamnestic data, we have focused our analysis on those family branches segregating the disease to individuals for whom complete clinical documentation was available (fig. 1; family 1, generations III and IV, ages 18 mo to 25 years).
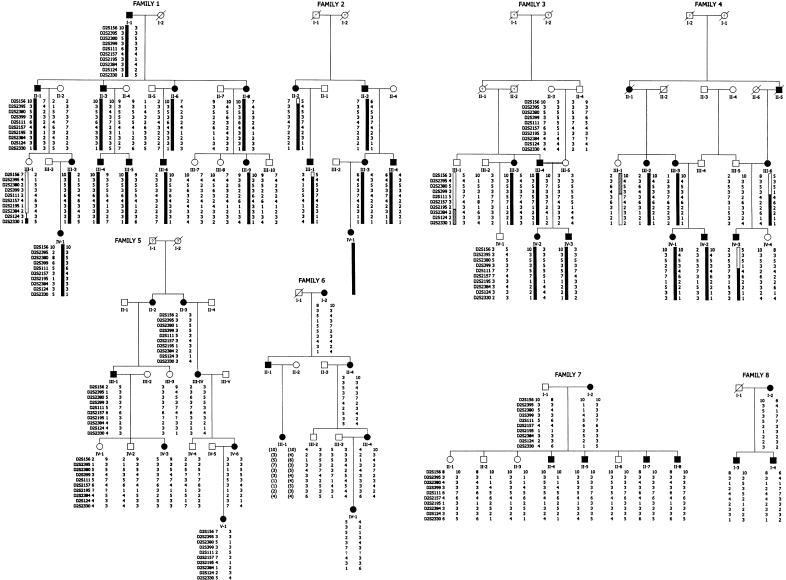
Figure 1.
Pedigrees and chromosome 2q24 haplotypes in eight Italian families affected with BFIC. Families 1–4 are linked, whereas families 5–8 are unlinked. Blackened symbols indicate affected subjects, and unblackened symbols indicate individuals with no history of seizures. Question marks (?) within genetic symbols indicate undetermined status. Black vertical bars represent segregation of the disease chromosome within linked families. Hatched bars indicate the presence of a potential disease chromosome in absence of parental genotypes. In individual III-1 of family 6, haplotypes (in brackets) were arbitrarily assigned because of the lack of parental information. Families 1, 5, 6, 7, and 8 are, respectively, families 7, 5, 4, 6, and 2 in the Gennaro et al. (1999) report.
Three hundred eighty-eight fluorescence-labeled markers of the ABI Prism linkage-mapping set, version 2 (PE Biosystems), were typed by PCR, and genotypes were analyzed by GENESCAN 2.0 and GENOTYPER 3.0 software on a 377 ABI Prism genetic analyzer. Parametric two-point linkage analysis was performed using the LINKAGE 5.1 package (Lathrop et al. 1985) in the FASTLINK 4.0 implementation (Cottingham et al. 1993). LOD scores were calculated under the assumption of a fully penetrant dominant trait with a prevalence of .001 and a phenocopy rate of .05. Equifrequent marker alleles were used throughout the analysis.
Preliminary evidence of linkage was found for marker D2S2330 (maximum pairwise LOD score 1.67 at θ=.1; data not shown). Therefore, we typed additional markers from the Généthon linkage map (Dib et al. 1996) and performed multipoint parametric linkage analysis using the ALLEGRO 1.1 software (Gudbjarrtsson et al. 2000). A maximum multipoint LOD score of 3.78 was obtained between markers D2S111 and D2S2384 (table 1). Furthermore, we have extended our analysis to seven additional families affected with BFIC (families 5, 6, 7, and 8 were described in Gennaro et al. (1999); families 2, 3, and 4 are previously unpublished) (fig. 1).
Table 1.
Family-Specific Multipoint LOD Scores for Markers on Chromosomes 2q and 19q[Note]
|
Multipoint LOD for Family |
|||||||||
| Marker | Distance from Previous Marker (cM) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| D2S156 | … | 3.7721 | −1.3126 | 1.2314 | −1.7773 | −2.5455 | −.6492 | −1.4839 | −.4768 |
| D2S2395 | 1.3 | 3.7820 | 1.0968 | 1.2351 | −1.7173 | −2.7421 | −1.2267 | −1.4972 | −.4768 |
| D2S2380 | .1 | 3.7828 | 1.1050 | 1.2354 | −1.4664 | −2.7668 | −1.3870 | −1.4972 | −.4768 |
| D2S399 | 1.9 | 3.7832 | 1.1339 | 1.2387 | −.7206 | −1.5462 | −1.3813 | −1.4972 | −.4768 |
| D2S111 | .2 | 3.7836 | 1.1343 | 1.2417 | 2.2855 | −1.5364 | −1.3904 | −1.4972 | −.4768 |
| D2S2157 | .1 | 3.7836 | 1.1343 | 1.2428 | 2.3079 | −1.5236 | −1.3905 | −1.4972 | −.4768 |
| D2S2195 | .1 | 3.7836 | 1.1343 | 1.2428 | 2.3106 | −1.5055 | −1.3905 | −1.4972 | −.4768 |
| D2S2384 | .1 | 3.7836 | 1.1343 | 1.1271 | 2.3113 | −1.4953 | −1.3906 | −1.4972 | −.4768 |
| D2S124 | .1 | 3.4821 | 1.1343 | 1.1094 | 2.3114 | −1.4873 | −1.2597 | −1.4972 | −.4768 |
| D2S2330 | .1 | −7.5200 | 1.1325 | 1.1021 | 2.3113 | −.1796 | −1.2058 | −1.4972 | −.4768 |
| D19S414 | … | −7.0732 | −.3647 | −.5417 | −1.1035 | −2.5564 | 1.4508 | −1.4972 | −.4768 |
| D19S868 | 2.7 | −7.8131 | −.3265 | −.6857 | −3.4402 | −3.1953 | 1.4530 | −1.4972 | −.4768 |
| D19S416 | 2.2 | −7.7989 | −.3488 | −.6863 | −2.9683 | −3.1155 | 1.4341 | −1.4972 | −.4768 |
| D19S425 | .6 | −7.8131 | −.2764 | −.6607 | −1.6626 | −3.1103 | 1.4265 | −1.4972 | −.4768 |
Note.— Genetic distances were deduced from the Généthon linkage map (Dib et al. 1996).
Linkage to chromosome 2q was consistent for families 2, 3, and 4 and was excluded for families 5, 6, 7, and 8 (table 1). A maximum cumulative multipoint LOD score of 3.60 was obtained for marker D2S2195. When locus heterogeneity was allowed, the multipoint HLOD score rose to 6.29 with 47% of linked families (data not shown).
To exclude the involvement of the chromosome 19q BFIC locus, we have typed families 2, 3, and 4 with markers D19S414, D19S868, D19S416, and D19S425 (genotypes not shown). Parametric multipoint linkage analysis provided further evidence that the chromosome 19q is not a major locus for BFIC (table 1). So far, in our sample, only family 7 shows evidence (weak evidence) of linkage to chromosome 19q.
To further investigate genetic heterogeneity, we have performed the admixture test implemented in the HOMOG program (Ott 1991) by using chromosome 2q multipoint LOD scores. Significant evidence for genetic heterogeneity was obtained (table 2). No evidence for three-locus genetic heterogeneity was found when chromosome 19 data were added to the analysis (HOMOG3R), because of the lack of linkage to chromosome 19q locus in our family sample.
Table 2.
HOMOG Analysis of Multipoint Data
| Hypothesesa | df | χ2 | LikelihoodRatio |
| H2 vs. H1 | 1 | 10.35 | 176.73 |
| H1 vs. H0 | 1 | 18.64 | 1.12×104 |
| H2 vs. H0 | 2 | 28.99 | 1.97×106 |
H0 = no linkage; H1 = linkage and homogeneity; H2 = linkage and heterogeneity.
The analysis of haplotypes showed recombinations in an ancestral chromosome (individual III-6, family 4), between markers D2S399 and D2S111, and a key recombination, between marker D2S2384 and D2S2330, in family 1, individual III-1 (fig. 1). The BFIC locus therefore must lie within a 0.7-cM interval on chromosome 2q24. Moreover, a key recombination further narrowing the critical region to 0.4 cM could have occurred in individual III-1, family 3, between markers D2S2195 and D2S2384. However, since parental genotypes are not available and the heterozygosity state of flanking marker(s) could not be determined, this crossover can not be precisely localized (fig. 1).
In our series, about half of the families should be associated with the chromosome 2q locus, and other genes are likely involved in the etiology of BFIC. Analysis of the geographical origins of our BFIC families (as far back as the 19th century) indicates that families are clustered in definite areas, suggesting possible founder effects (fig. 2); families linked to chromosome 2q24 are clustered in southern Italy, and unlinked families are clustered in northeastern Italy. To date, no evidence for a shared ancestral haplotype was found in our family sample (fig. 1). It has been shown, however, that linkage disequilibrium may extend to only a few kb from mutated alleles, depending on the age of mutations and the demographic history of populations (Kruglyak 1999). The development of a physical map of the region and the identification of a several closely spaced markers will therefore be necessary to detect linkage disequilibrium. Therefore, we have performed a detailed analysis of clinical data, to identify possible clinical subsets underlying genetic heterogeneity. No significant phenotypic differences have been found among families. Our data fit well the emerging scenario of autosomal dominant idiopathic epilepsies.
Figure 2.
Geographical distribution of BFIC families. Linked families are indicated with underlined numbers.
In autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE [MIM 600513, 603204, and 605375]), two neuronal acetylcholine receptor subunit genes—CHRNA4 on chromosome 20q (Steinlein et al. 1995) and CHRNB2 on chromosome 1q (De Fusco et al. 2000)—were found to be mutated, and a third locus has been mapped to chromosome 15q (Phillips et al. 1998). In benign familial neonatal convulsions (BFNC [MIM 121200 and 121201]) mutations in two different neuronal voltage-gated potassium channel genes (KCNQ2 and KCNQ3) were identified (Charlier et al. 1998; Singh et al. 1998). Similarly, two neuronal voltage-gated sodium channel subunit genes, SCN1B and SCN1A, were associated to generalized idiopathic epilepsy and febrile convulsions in GEFS+ families (GEFS+ [MIM 604236 and 604233]; Wallace et al. 1998; Escayg et al. 2000).
The involvement of neuronal ion-channel subunits and genetic heterogeneity appear so far the most representative traits of autosomal dominant idiopathic epilepsies. In BFIC, the presence of features common to the above-mentioned epilepsies—such as the benign outcome of the seizures and the autosomal dominant pattern of inheritance—suggests that ion-channel genes could be also involved. Genetic heterogeneity would also support this hypothesis, since ion-channel genes are highly redundant in the human genome. According to this assumption, the 0.7-cM critical region on chromosome 2q could harbor four potential candidate genes for BFIC: SCN1A, SCN2A, SCN3A, and KCNJ3.
SCN1A, SCN2A, and SCN3A are highly expressed in brain and form a sodium-channel α-subunit gene cluster mapped on chromosome 2q23-24. Since SCN1A recently was demonstrated to be associated with generalized epilepsy and febrile convulsions (Escayg et al. 2000), the detection of mutations in families affected with BFIC would demonstrate that GEFS+ and BFIC are allelic to the same gene. Interestingly, a similar association is found on chromosome 19q, where the BFIC locus was mapped to the same chromosomal region harboring the GEFS+ gene, SCN1B (Wallace et al. 1998). A recent study, however, failed to detect mutations at SCN1B in families linked to the chromosome 19 BFIC locus (Moulard et al. 2000). In contrast, SCN2A and SCN3A have not yet been associated with human phenotypes.
KCNJ3 encodes a G-protein-coupled potassium inwardly rectifying channel that is thought to be involved in receptor-mediated inhibitory postsynaptic potentials (Kubo et al. 1993; Chen and Yu 1994). KCNJ3 is a potential candidate for BFIC for at least two reasons: first, the reduction of inward K+ current in neurons, caused by mutations in voltage-gated potassium channels, was associated with BFNC, a syndrome that is closely related to BFIC (Charlier et al. 1998; Singh et al. 1998); second, a mouse model of epilepsy—weaver—is associated with mutations in the related gene KCNJ6 (Patil et al. 1995). In brain, KCNJ3 channels are functionally activated through the formation of heterotetrameric complexes with KCNJ6 (Duprat at al. 1995). Thus, we can assume that KCNJ3 mutations affecting the same complex may underlie seizures in humans too.
In summary, we have identify a second locus for BFIC on chromosome 2q24 and have provided evidence for genetic heterogeneity, suggesting the presence of different genes involved in the etiology of BFIC. The identification of the mutated gene will either provide further evidence of the primary role of neuronal ion-channel genes in the etiology of idiopathic epilepsy or will lead to the identification of new epileptogenic mechanisms.
Acknowledgments
This work is supported by Telethon Italy (grant 213bi to F.Z.). We thank the family which kindly consented to the study. Cell lines are stored at the DNA bank of the Italian League against Epilepsy at National Neurological Institute “C. Besta” and at the Galliera Genetic Bank (supported by Telethon Italy, grant C42).
Electronic-Database Information
- Généthon, http://www.genethon.fr
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BFIC [MIM 601764], ICCA [MIM 602066], ADNFLE [MIM 600513, 603204, 605375]), BFNC [MIM 121200, 121201], GEFS+ [MIM 604236, 604233])
References
- Charlier C, Singh NA, Ryan SG, Lewis TB, Reus BE, Leach RJ, Leppert M (1998) A pore mutation in a novel KQT-like potassium channel gene in an idiopathic epilepsy family. Nat Genet 18:53–55 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yu L (1994) Differential regulation by cAMP-dependent protein kinase and protein kinase C of the μ-opioid receptor coupling to a G protein-activated K+ channel. J Biol Chem 269:7839–7842 [PubMed] [Google Scholar]
- Cottingham RW, Idury RM, Shaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- De Fusco M, Becchetti A, Patrignani A, Annesi G, Gambardella A, Quattrone A, Ballabio A, Wanke E, Casari G (2000) The nicotinic receptor β2 subunit is mutant in nocturnal frontal lobe epilepsy. Nat Genet 26:275–276 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome on 5.264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Duprat F, Lesage F, Guillemare E, Fink M, Hugnot JP, Bigay J, Lazdunski M, Romey G, Barhanin J (1995) Heterologous multimeric assembly is essential for K+ channel activity of neuronal and cardiac G-protein-activated inward rectifiers. Biochem Biophys Res Commun 212:657–663 [DOI] [PubMed] [Google Scholar]
- Escayg A, MacDonald BT, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A, Meisler MH (2000) Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+. Nat Genet 24:343–345 [DOI] [PubMed] [Google Scholar]
- Gennaro E, Malacarne M, Carbone I, Riggio MC, Bianchi A, Bonanni P, Boniver C, Dalla Bernardina B, De Marco P, Giordano L, Guerrini R, Santorum E, Sebastianelli R, Vecchi M, Veggiotti P, Vigevano F, Dagna Bricarelli F, Zara F (1999) No evidence of a major locus for benign familial infantile convulsions on chromosome 19q12-q13.1. Epilepsia 40:1799–1803 [DOI] [PubMed] [Google Scholar]
- Giordano L, Accorsi P, Valseriati D, Tiberti A, Menegati E, Zara F, Vignoli , Vigevano F (1999) Benign infantile familial convulsions: natural history of a case and clinical characteristic of a large Italian family. Neuropediatrics 30:99–101 [DOI] [PubMed] [Google Scholar]
- Gudbjarrtsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Guipponi M, Rivier F, Vigevano F, Beck C, Crespel A, Echenne B, Lucchini P, Sebastianelli R, Baldy-Moulinier M, Malafosse A (1997) Linkage mapping of benign familial infantile convulsions (BFIC) to chromosome 19q. Hum Mol Genet 6:473–477 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 [DOI] [PubMed] [Google Scholar]
- Kubo Y, Baldwin TJ, Jan YN, Jan LY (1993) Primary structure and functional expression of a mouse G protein-coupled muscarinic potassium channel. Nature 362:127–133 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1985) Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 37:482–498 [PMC free article] [PubMed] [Google Scholar]
- Moulard B, Buresi C, Malafosse A (2000) Study of the voltage-gated sodium channel beta 1 subunit gene (SCN1B) in the benign familial infantile convulsions syndrome (BFIC). Hum Mut 16:139–142 [DOI] [PubMed] [Google Scholar]
- Ott J (1991) Analysis of human genetic linkage, 2d ed. John Hopkins University Press, Baltimore [Google Scholar]
- Patil N, Cox DR, Bhat D, Faham M, Myers RM, Peterson AS (1995) A potassium channel mutation in weaver mice implicates membrane excitability in granule cell differentiation. Nat Genet 11:126–129 [DOI] [PubMed] [Google Scholar]
- Phillips HA, Scheffer IE, Crossland KM, Bhatia KP, Fish DR, Marsden CD, Howell SJL, Stephenson BP, Tolmie J, Plazzi G, Eeg-Olofsson O, Singh R, Lopes-Cendes I, Andermann E, Andermann F, Berkovic SF, Mulley JC (1998) Autosomal dominant nocturnal frontal-lobe epilepsy: genetic heterogeneity and evidence for a second locus at 15q24. Am J Hum Genet 63:1108–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NA, Charlier C, Stauffer D, DuPont BR, Leach RJ, Melis R, Ronen GM, Bjerre I, Quattlebaum T, Murphy JV, McHarg ML, Gagnon D, Rosales T, Peiffer A, Anderson E, Leppert M (1998) A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat Genet 18:25–29 [DOI] [PubMed] [Google Scholar]
- Steinlein OK, Mulley JC, Propping P, Wallace RH, Phillips HA, Sutherland GR, Scheffer IE, Berkovic SF (1995) A missense mutation in the neuronal nicotinic acetylcoline receptor a4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet 11:201–203 [DOI] [PubMed] [Google Scholar]
- Szepetowski P, Rochette J, Berquin P, Piussan C, Lathrop GM, Monaco AP (1997) Familial infantile convulsions and paroxysmal choreoathetosis: a new neurological syndrome linked to the pericentromeric region of human chromosome 16. Am J Hum Genet 61:889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigevano F, Fusco L, Di Capua M, Ricci S, Sebastianelli R, Lucchini P (1992) Benign infantile familial convulsions. Eur J Pediatr 151:608–612 [DOI] [PubMed] [Google Scholar]
- Wallace RH, Wang DW, Singh R, Scheffer IE, George AL Jr, Phillips HA, Saar K, Reis A, Johnson EW, Sutherland GR, Berkovic SF, Mulley JC (1998) Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel β1 subunit gene SCN1B. Nat Genet 19:366–370 [DOI] [PubMed] [Google Scholar]