Abstract
To date, functional genomic studies have been confined to either cell-based assays or germline mutations, using transgenic or knockout animals. However, these approaches are often unable either to recapitulate complex biologic phenotypes, such as tumor metastasis, or to identify the specific genes and functional pathways that produce serious diseases in adult animals. Although the transcription factor NF-κB transactivates many metastasis-related genes in cells, the precise genes and functional-pathways through which NF-κB regulates metastasis in tumor-bearing hosts are poorly understood. Here, we show that the systemic delivery of plasmid-based ribozymes targeting NF-κB in adult, tumor-bearing mice suppressed NF-κB expression in metastatic melanoma cells, as well as in normal cell types, and significantly reduced metastatic spread. Plasmid-based ribozymes suppressed target-gene expression with sequence specificity not achievable by using synthetic oligonucleotide-based approaches. NF-κB seemed to regulate tumor metastasis through invasion-related, rather than angiogenesis-, cell-cycle- or apoptosis-related pathways in tumor-bearing mice. Furthermore, ribozymes targeting either of the NF-κB-regulated genes, integrin β3 or PECAM-1 (a ligand-receptor pair linked to cell adhesion), reduced tumor metastasis at a level comparable to NF-κB. These studies demonstrate the utility of gene targeting by means of systemic, plasmid-based ribozymes to dissect out the functional genomics of complex biologic phenotypes, including tumor metastasis.
NF-κB, a proximal regulator of gene expression, seems to play an important role in tumor progression (1–3). However, the primary target genes and functional pathways through which it mediates tumor progression are unclear, because NF-κB has been shown to regulate tumor cell apoptosis (4), cycling (5), and adhesion (6), as well as tumor angiogenesis (7). Recent results from in vitro studies suggest that blocking NF-κB gene expression in tumors expressing wild-type p53 (such as B16-F10 melanoma cells (8) may actually increase tumor growth (9), further complicating the role of NF-κB in tumor progression. In addition, NF-κB has been shown to transactivate a number of genes that produce significant antimetastatic effects (including interleukin-2, interleukin-12, and granulocyte macrophage-colony stimulating factor; ref. 1). Thus, the overall function of NF-κB in metastasis, as well as the critical pathway(s) through which it actually controls metastatic spread in tumor-bearing hosts, remains unclear. The use of targeted disruption of selected genes in embryonic stem cells to generate knockout mice is commonly attempted to identify gene function in animals. However, knockout technology can disrupt important developmental pathways, produce in utero lethality, and has a limited ability to recapitulate complex phenotypes such as tumor metastasis. Conversely, systemic, ribozyme-based gene targeting in adult mice does not interfere with development, obviates in utero lethality, and should permit the assessment of the targeted gene's function in virtually any biologic setting. Moreover, gene targeting via efficient, long-expressing DNA plasmids encoding ribozymes should circumvent the major limitations (nonspecific activity, short duration of action and expense) associated with the use of synthetic phosphorothioate oligodeoxynucleotides (10).
We used systemic administration of cationic liposome: DNA complexes (CLDC) to deliver and express plasmid-based hammerhead ribozymes targeting a variety of putative tumor-progression genes directly in tumor-bearing animals. We targeted NF-κB to assess its overall role in the metastatic phenotype and the critical functional pathway (apoptosis, cell cycle, angiogenesis, or invasion) through which NF-κB regulates metastasis. Furthermore, we used CLDC-based ribozyme targeting to identify NF-κB-regulated genes that mediate the effects of NF-κB on metastasis in tumor-bearing hosts. Systemic delivery of plasmid-based ribozymes targeting NF-κB-p65 into adult mice blocked NF-κB expression in metastatic tumor cells, as well as in vascular endothelial cells, a critical normal cell type that regulates both tumor angiogenesis and tumor invasion (11). Conversely, p65-knockout mice die in utero (12), thereby precluding their use to evaluate phenotypes manifested primarily in adult life, such as tumor metastasis. The systemic, plasmid-based approach for expressing ribozymes was only recently made possible through the development of improved cationic liposome formulations and an Epstein–Barr virus (EBV)-based expression plasmid that can express delivered genes at therapeutic levels for prolonged periods in immunocompetent mice. Specifically, i.v.-injected CLDC can now transfect the majority of both lung vascular endothelial cells (13) and melanoma cells metastatic to lung (8). In addition, we used an EBV-based expression plasmid containing both the Epstein–Barr virus-encoded nuclear antigen (EBNA)-1 cDNA and the EBV family of repeats sequence. This plasmid substantially prolongs the expression of delivered genes at therapeutic levels in adult animals (14). Although the utility of ribozyme targeting to identify gene function in cells is well established (15, 16), the development of a systemic ribozyme-based approach to identify gene function in animals has been lacking.
Materials and Methods
Plasmid Construction.
The plasmid backbone pVector used for plasmid construction has been described (14). Construction and preparation of control plasmids containing the luciferase and murine angiostatin cDNAs was also described (8). DNA encoding various ribozymes was inserted into the Pst I-Not I site of the plasmid's multiple-cloning site. The sequences of the various ribozyme DNAs used for cloning were p65-R, 3′-CTCCACAAAGCAGGAGTGCCTGAGTAGTCAAAGTG-5′, targeting the GUA sequence at position 740 of murine p65; p50-R, 3′-GTTTACAAAGCAGGAGTGCCTGAGTAGTCAGTAAA-5′, targeting the GUU sequence at position 328 of murine p50; anti-β3 ribozyme, 3′-CTACCCAAAGCAGGAGTGCCTGAGTAGTCAGGTCC-5′, targeting the GUC sequence at position 11 of murine β3; anti-PECAM-1 ribozyme, 3′-TTTCACCAAAGCAGGAGTGCCTGAGTAGTCTTCTCT-5′, targeting the GUC sequence at position 1,247 of murine PECAM-1; and anti-FLK-1 ribozyme, 3′-TTTGAACAAAGCAGGAGTGCCTGAGTAGTCAATTTA-5′, targeting the GUC sequence at position 942 of murine FLK-1. B16-F10 cells were grown and CLDC using the liposome DOTMA was prepared as described (8).
Stable transformants were generated by cotransfection of 2 × 105 B16-F10 cells with 2 μg of p65-R and 0.2 μg of a neomycin-carrying plasmid and complexed with DOTIM:cholesterol in a 1:4 ratio. Cells were plated on 60-mm dishes in RPMI medium 1640/5% FBS, cultured at 37°C; after 48 h, the cells were resuspended in G418 (800 μg/ml) selection media. The media was changed every 3 days, and after 3 weeks, a few growing colonies were isolated and grown in RPMI medium 1640/5% FBS. The cells were resuspended 24 h later in G418 selection media, and the presence of ribozyme DNA was confirmed by PCR analysis after DNA extraction.
Murine Studies.
Individual C57Bl6 (Simonsen Laboratories, Gilroy, CA) 8-week-old female mice in groups of eight were injected i.v. on day 0 with 25,000 B16-F10 cells and on day 7 with 200 μl of CLDC (i.v.) containing 650 nmol of pure DOTMA MLV complexed to 25 μg of p-angiostatin, p-65-R, or p-LUC. Mice were killed 30 days later and processed as described (8). For reverse transcription (RT)-PCR and immunohistochemistry, CLDC containing the same control or ribozyme sequences were injected again 1 day before killing. For in vivo nested RT-PCR, total RNA was isolated from homogenizing tumor-bearing lungs and purified by Ultraspec RNA Isolation System (Biotecx Laboratories, Houston). Five micrograms of RNA was used as the starting material for reverse transcriptase (Superscript II, GIBCO/BRL). The cDNA product (500 ng) of the RT-RNA reaction was used in the PCR reaction under the following conditions: 95°C for 5 min (95°C for 45 sec, 55°C for 30 sec, 72°C for 45 sec) for a total of 30 cycles, and 72°C for 10 min. The sequences of the vector-specific primer pair used in the PCR reaction were nest primer I, 5′-CCGTGGCGGTAGGGTATGTGTCTGAAAATG-3′, and nest primer II, 5′-CAGATCGCAGCAATCGCGCCC CTATCTTG-3′. For antitumor studies, the number of melanotic tumor nodules was counted under a dissecting microscope by an individual blinded to the identity of the groups. The total tumor count for each mouse included tumor nodules <2 mm and ≥2 mm. The potential statistical significance of differences between various groups was assessed by using an unpaired, two-sided Student's t test.
Immunohistochemical Analysis.
Immunohistochemical staining of NFκB-p65 was performed by using a peroxidase-conjugated avidin-biotin technique with DAKO Universal Autostaining System as described (14). For monoclonal antibody against p65, microwave antigen retrieval was used and blocked with universal blocking solution (Zymed). Mouse monoclonal or rabbit polyclonal IgG antibody against a recombinant protein corresponding to amino acids 1–286 mapping at the amino terminus of p65 (Santa Cruz Biotechnology) diluted at a concentration of 0.5 μg/ml was incubated overnight at 4°C.
Western Blotting.
Ten micrograms of nuclear proteins were electrophoresed onto four SDS/15% PAGE gels and transferred onto nitrocellulose membrane (Hybond, Amersham Pharmacia). The membrane was blocked with 10% (wt/vol) nonfat dry milk in Tris-buffered saline [TBST; 10 mM Tris⋅HCl, 0.15 M NaCl, 0.05% (wt/vol) Tween 20], incubated with primary antibodies, goat anti-p65 diluted 1:1,000 (vol/vol) in 10% milk/TBST (Santa Cruz Biotechnology), and mouse anti-actin diluted 1:8,000 (vol/vol) in 10% milk/TBST (Chemicon) for 1 hr, washed with TBST (3 × 15 min), incubated with specific horseradish peroxidase-conjugated secondary antibody diluted 1:3,000 (vol/vol) in 10% milk/TBST (Santa Cruz Biotechnology) for 45 min, and washed with TBST (3 × 15 min). Immunoreactive bands were visualized with an enhanced chemiluminescence kit (ECL, Amersham Pharmacia) and exposed with BioMax ML film (Kodak).
Nuclear Extracts Preparation and Gel Shift (Supershift) Assays.
Stable cell lines of B16-pLUC and B16-p65-R were grown in RPMI medium 1640/5% FBS supplemented with 800 μg/ml of neomycin (GIBCO/BRL) till confluency and harvested into a buffer containing 10 mM Hepes, pH 7.9/1.5 mM MgCl2/10 mM KCl/0.1% Nonidet P-40/1× protease inhibitor mixture (Roche Molecular Biochemicals). Nuclear extracts were prepared as described (17). Briefly, the nuclear pellet was resuspended in a buffer containing 20 mM Hepes (pH 7.9), 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% glycerol, and 1× protease inhibitor mixture, centrifuged, and the supernatant (nuclear extract) was collected. The protein concentration was determined by using Bradford protein assay reagent (Bio-Rad). All binding reactions were carried out on ice unless otherwise indicated. Five micrograms of protein from each nuclear extract were used in each binding reaction in a total volume of 20 μl containing 20 mM Hepes (pH 7.9), 2.5 mM MgCl2, 1 mM DTT, 1 μg poly[d(I-C)] (Roche Molecular Biochemicals), and 25,000 cpm of end-labeled double-stranded consensus oligo (5′-CAGTTGAGGGGACTCCAGGCC-3′). End labeling was carried out by using T4 polynucleotide kinase (Promega) with [γ32P]ATP and purified with a nucleotide removal kit (Qiagen, Chatsworth, CA). In the competition assay, 200-fold molar excess of cold double-stranded consensus oligo was added. In supershift assays, 2 μg of either anti-p65 IgG (Santa Cruz Biotechnology) or normal rabbit IgG was incubated with nuclear extracts for 1 h on ice before the addition of labeled oligo. Samples were loaded onto a 4.5% polyacrylamide gel (29:1) and electrophoresed with a buffer containing 50 mM Tris (pH 8.5), 0.38 M glycine, 2 mM EDTA, and 0.5 mM β-mercaptoethanol at 200 V for 2 h at 4°C. The gel then was dried and exposed to Kodak BioMax film at −70°C.
Functional Studies.
Mitotic and apoptotic figures were counted on 4 μm hematoxylin and eosin stained slides with a conventional light microscope. Actual counts of mitotic and apoptotic figures were made from the 10 largest nodules. Apoptotic bodies and mitotic figures were counted according to morphologic criteria described (18, 19). The apoptotic and mitotic rates were calculated based on the degree of tumor cellularity and expressed as the number of apoptotic or mitotic figures per 1,000 cells. In situ detection of cleaved, apoptotic DNA fragments (TUNEL) was performed by using the TdT-FragEL Detection Kit (Oncogene Science) according to the manufacturer's protocol. The frequency of labeled cells was calculated by counting at least 1,000 cells in areas with the highest number of TdT-labeled nuclei. Matrigel assay and Boyden chamber analysis were performed as described (20).
Results and Discussion
Hammerhead ribozymes were designed complementary to sequences containing cleavage sites in the p65 and p50 subunits of murine NF-κB (21, 22), and the 35-bp ribozyme sequences were inserted into an HCMV-IE1-driven expression plasmid containing both the EBNA-1 cDNA and the EBV family of repeats (14). The resulting plasmids, p65-R and p50-R, respectively, expressed the corresponding ribozyme sequences in metastatic tumors, after CLDC-based i.v. injection into tumor-bearing mice, as determined by RT-PCR (Fig. 1A, lanes 3–6). The effects of CLDC-based injection of p65-R on both target-gene expression and tumor metastasis then were assessed in C57Bl6 mice bearing syngeneic B16-F10 melanoma tumors.
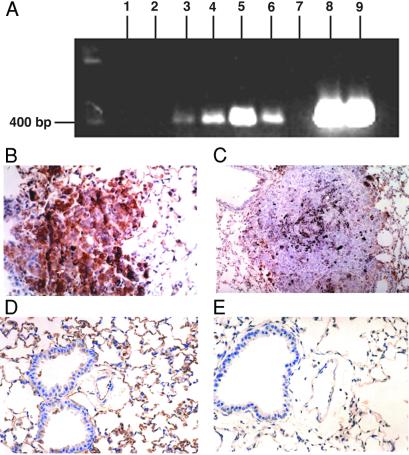
Figure 1.
(A) RT-PCR analysis of anti-NF-κB ribozyme expression in tumor-bearing mice. Lanes 1 and 2, metastatic lung tumors from untreated mice; lanes 3 and 4, tumors from CLDC-p65-R-treated mice; lanes 5 and 6, tumors from CLDC-p50-R-treated mice; lane 7, distilled water; lanes 8 and 9, p65-R and p50-R plasmid DNA, respectively. (B) Photomicrograph (40× ocular) of p65 immunostaining of lung from the pVector-treated control demonstrates pulmonary parenchyma with a centrally placed metastatic melanoma tumor containing black, granular melanin pigment within the cytoplasm. Tumor shows golden-brown immunopositive staining for p65. (C) Photomicrograph (20× ocular) of p65 immunostaining of lung from the p65-R-treated group demonstrates a metastatic melanoma tumor in which immunostaining for p65 is essentially negative. This tumor also demonstrates some black melanin pigment similar to A. (D) Photomicrograph (40× ocular) of p65 immunostaining of lung from pVector-treated (control) group. Respiratory epithelium shows staining of some cells. Vascular endothelium is strongly positive. Alveolar epithelium also demonstrates moderate to intense reactivity of the majority of cells. Vascular endothelial cells and alveolar epithelial cells were identified by using classic microscopic criteria. We have previously shown that endothelial cells are positive for immunoreactivity using antifactor VIII antibodies and negative using anti-cytokeratin antibodies AE1/AE3, with a reverse expression pattern for alveolar lining cells (14). (E) Photomicrograph (40× ocular) of p65 immunostaining of lung from the p65-R-treated group. Respiratory (pseudostratified, ciliated) epithelium, stromal cells, and vascular endothelium are largely negative for p65 immunostaining. Alveolar lining cells show minimal, focal reactivity of low intensity (golden-brown).
Cationic liposomes complexed to either p65-R, or to the same expression plasmid lacking the 35-bp ribozyme insert, pVector, were injected i.v. into mice bearing metastatic B16-F10 tumors. Injection (i.v.) of p65-R significantly reduced the levels of p65 protein in both metastatic tumor cells and in three different normal lung cell types. Specifically, immunohistochemistry for p65 revealed significant reductions in p65-immunoreactivity in (i) metastatic B16-F10 lung tumors (P < 0.005), (ii) vascular endothelial cells (P < 0.0005), (iii) alveolar epithelial cells (P < 0.05), and (iv) bronchial lining cells (P < 0.05) from mice i.v.-injected with CLDC-p65-R, when compared with those treated with CLDC-pVector (Fig. 1 B–E, and Table 1). Furthermore, despite the fact that some cells retained p65-immunoreactivity in the p-65R-treated group (Table 1), the intensity of p65-immunoreactivity was significantly reduced in all cell types from mice injected with CLDC-p65-R (Fig. 1 C and E), when compared with those from mice treated with CLDC-pVector (Fig. 1 B and D). p65 staining did not differ significantly between CLDC-pVector-treated mice, CLDC-pLUC (the same plasmid as pVector, but containing the luciferase cDNA)-treated mice, or tumor-bearing mice not treated with CLDC (data not shown). The ability of i.v., plasmid-based ribozymes to reduce NF-κB expression most significantly in metastatic B16-F10 tumor cells and in lung vascular endothelial cells is consistent with prior observations showing that these are the two cell types most efficiently transfected in the lung after i.v. injection of CLDC (8, 13). Immunohistochemical determination of p50 immunoreactivity served as an internal control, as the expression of intratumoral p50 protein did not differ significantly between the lungs of mice treated with CLDC-p65-R (76.0 + 5.3%) vs. CLDC-pVector (86.2 + 3.3%; P > 0.1). The use of systemic, plasmid-based ribozymes allowed down-regulation of target-gene expression in critical normal cells as well as in tumor cells, thereby permitting a more comprehensive assessment of gene function in vivo than does the inoculation of stably transfected tumor cell lines, which alters target gene expression only in tumor cells.
Table 1.
p65 positive cells in metastatic tumor cells and in normal lung cell types from CLDC-p65-R and CLDC-pVector-treated mice, as determined by immunohistochemical analysis for p65
| Cell type | p65- R-treated, % | pVector-treated, % | P value |
|---|---|---|---|
| Lung tumor cells | 12.9 ± 3.2 | 43.9 ± 8.1 | P < 0.005 |
| Vascular endothelial cells | 12.2 ± 1.5 | 52.5 ± 4.5 | P < 0.0005 |
| Alveolar lining cells | 61.0 ± 9.3 | 83.1 ± 3.4 | P < 0.05 |
| Bronchial lining cells | 29.4 ± 8.8 | 46.3 ± 8.4 | P < 0.05 |
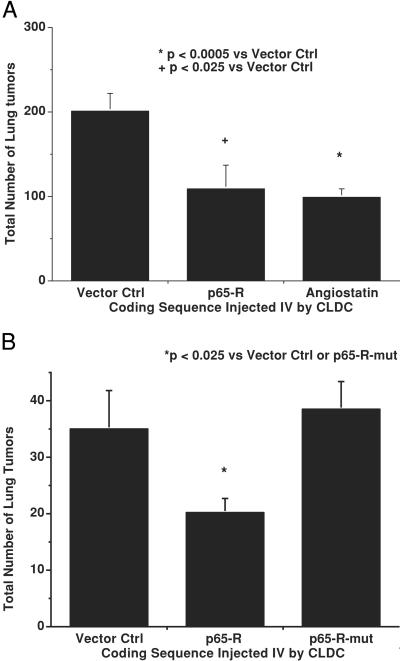
We then assessed whether CLDC-mediated systemic delivery of plasmids encoding hammerhead ribozymes targeting NF-κB could alter the metastatic spread of B16-F10 melanoma cells in syngeneic C57Bl6 mice. CLDC-based i.v. delivery of plasmid p65-R, which reduced p65 expression in the lung, also significantly reduced the metastatic spread of B16 melanoma tumors in mice. Specifically, a single injection of CLDC containing p65-R, 7 days after i.v. injection of 25,000 B16-F10 cells, reduced the total number of lung metastases (P < 0.025) when compared with tumor-bearing mice treated with CLDC containing pVector (Fig. 2A). The anti-p65 ribozyme was as effective as CLDC-based delivery of the murine angiostatin gene, whose overexpression produces significant antimetastatic effects against B16-F10 (8).
Figure 2.
Antimetastatic activity of anti-NF-κB ribozymes. (A) Comparison of antitumor activity of CLDC containing pVector, p65-R, and angiostatin cDNA. (B) Effects of i.v. treatment with CLDC containing pVector, p65-R, and p65-R-mut on number of metastatic B16 lung tumors.
Previously, synthetic oligonucleotide-based antisense constructs have been shown to produce significant nonspecific effects in biologic systems, and absolute sequence specificity is not attainable by using phosphorotioate-linked oligomers (10). Without such sequence specificity, the gene functions identified can, in reality, represent physiologic responses to nonantisense effects. Therefore, we compared the antimetastatic activity produced by systemic delivery of CLDC containing p65-R to that produced by CLDC containing p65-R-mut, a mutated anti-p65 ribozyme plasmid that differed from p65-R by a single base substitution in its catalytic core, required for cleavage activity (23). This single bp substitution abolished the antimetastatic activity of the anti-p65 ribozyme, reducing its activity to that associated with CLDC-pVector (Fig. 2B). CLDC-based delivery of p50-R also significantly reduced (P < 0.025) the metastatic spread of B16-F10 (22 ± 7 lung metastases) when compared with p50-R-mut, its single-base mutant counterpart (55 ± 16 lung metastases). This result is in contrast to studies performed with continuous infusions of ribozyme oligonucleotides, in which the mutant ribozyme retained antimetastatic effects (24). Thus, systemic plasmid-based ribozyme activity seemed to be highly sequence-specific in mice.
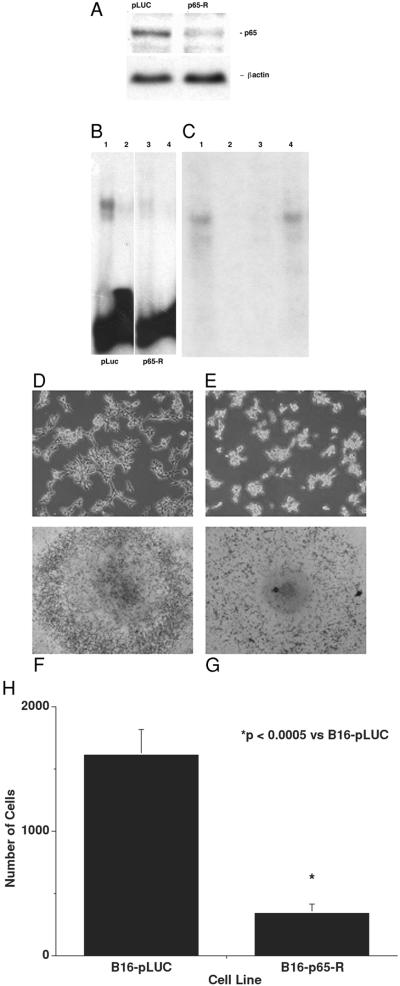
To assess the effects of tumor cell-specific blockade of NF-κB expression on the metastatic phenotype, B16-F10 clones were stably transfected with either the p65-R plasmid or with pLUC. The resultant stably transfected clones were screened for p65 protein levels by immunohistochemistry using anti-p65-specific monoclonal antibodies. Two clones were selected for further analysis: B16-p65-R, stably transfected with plasmid p65-R that showed 0% of cells strongly positive for p65, and B16-pLUC, stably transfected with pLUC that showed 50–80+% of cells strongly positive for p65 (similar to the level of p65 expression in wild-type B16-F10 melanoma cells). Further evidence of inhibition of p65 expression and functional activity in the B16-p65-R cell line was provided by Western blot analysis (Fig. 3A) and gel shift assay (Fig. 3 B and C). Specifically, Western analysis showed that p65 protein levels were significantly (3.3-fold, as quantitated by scanning densitometry) reduced in B16-p65-R cells when compared with B16-pLUC control cells. We also performed electrophoretic mobility shift assays using nuclear extracts from B16-pLuc and B16-p65-R cells. Only nuclear extracts from B16-pLuc, but not from B16-p65-R cells, contained a complex that associated with the NF-κB binding site (Fig. 3B). Gel shift assay also showed specific disruption of NF-κB DNA binding in the presence of either an excess of cold consensus oligonucleotide (Fig. 3B) or anti-p65 antibody (Fig. 3C). Suppression of p65 expression in the B16-p65-R clone significantly reduced (P < 0.0005) the ability of B16-F10 cells to metastasize in C57Bl6 mice (53 ± 10.2 lung tumor metastases) when compared with the B16-pLUC clone (117.6 ± 12.8 metastases). Thus, blocking NF-κB expression within tumor cells alone also was able to reduce tumor metastasis significantly.
Figure 3.
Analysis of p65 expression and functional activities of NFκB in B16-pLUC clone and B16-p65-R clone. (A) Western analysis of p65 expression in B16-pLUC and B16-p65-R clones. (B and C) Functional analysis of NFκB activity using gel shift (B) and supershift (C) assays. (B) Gel shows reduction in DNA binding activity in B16-p65-R cell line. Lane 1, nuclear extract from B16-pLUC cells (LUC-NE); lane 2, LUC-NE incubated with 200-fold molar excess of cold consensus oligo; lane 3, nuclear extract from B16-p65-R cells (p65-R-NE); and lane 4, p65-R-NE incubated with 200-fold molar excess of cold consensus oligo. (C) Gel shows specific disruption of NFκB DNA binding with anti-p65 antibody. Lane 1, p-65-R-NE with 32P-labeled consensus oligo; lane 2, p65-R-NE incubated with 200-fold molar excess of cold consensus oligo; lane 3, p65-R-NE with 32P-labeled consensus oligo and 2 μg of anti-p65 antibody; and lane 3, p65-R-NE with 32P-labeled consensus oligo and 2 μg of normal rabbit IgG. Degree of invasion of B16-pLUC clone (D) and B16-p65-R clone (E) into matrigel. The migrating and invasive B16-pLUC cells display an elongated morphology compared with the rounded B16-p65-R cells. Invasiveness of B16-pLUC cells (F) and B16-p65-R cells (G) as determined by Boyden chamber analysis. (H) Quantitation of tumor cell invasion into Boyden chambers.
Several different functional pathways essential to the metastatic phenotype, including tumor angiogenesis, apoptosis, and cell cycle, were evaluated in p65-R-treated and control mice to assess how ribozymes blocking NF-κB may suppress the metastatic spread of B16-F10 cells. When compared with CLDC-pLUC-treated control mice, mice treated with CLDC-p65-R did not show significantly different levels of tumor cell apoptosis [2.4 ± 0.5 (pLuc) vs. 2.5 ± 0.6 (p65-R) per 1,000 cells], tumor cell mitosis [3.5 ± 0.4 (pLuc) vs. 3.2 ± 0.7 (p65-R) per 1,000 cells] or tumor angiogenesis [16.8 ± 1.3 (pLuc) vs. 16.1 ± 3.5 (p65-R) total blood vessels per tumor]. The inability of p65-R to alter either tumor apoptosis or mitosis significantly was further demonstrated in cells stably transfected with either p65-R or pLUC. Specifically, B16-p65-R cells showed 2.8 ± 0.7 apoptotic cells per 1,000 cells counted vs. 1.5 ± 0.5 for B16-pLUC, and 6.4 ± 1.7 mitotic cells per 1,000 cells counted vs. 5.8 ± 1.5 for B16-pLUC. In contrast, tumor invasiveness was significantly inhibited by suppression of NF-κB expression. Tumor cell invasion, as assessed by both invasion into matrigel (Fig. 3 D and E), and by Boyden chamber analysis (Fig. 3 F–H), was significantly reduced (P < 0.0005) in B16-p65-R cells when compared with either B16-pLUC cells or wild-type B16-F10 cells. When plated on extracellular matrix, the migrating and invasive B16-pLUC cells (Fig. 3D) displayed an elongated morphology, compared with the rounded B16-p65R cells (Fig. 3E). The short 16-h incubation time of the Boyden chamber invasion assay ensured that the fraction of cells stained corresponded to invasive and migratory cells and not proliferative cells (compare Fig. 3F with Fig. 3G). Thus, blocking NF-κB expression altered the ability of tumor cells to invade their surrounding microenvironment but did not significantly alter the level of tumor cell apoptosis or mitosis, nor did it decrease tumor angiogenesis.
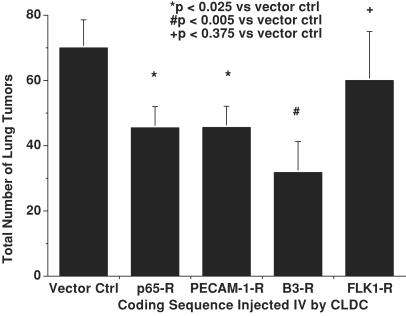
Based on these results, CLDC-based systemic delivery of ribozymes in tumor-bearing mice was used to identify genes whose NF-κB-regulated expression could play a role in promoting tumor invasiveness. Integrin αvβ3 and PECAM-1 were targeted because they form a ligand-receptor pair (25), and each are regulated by NF-κB (26, 27). In addition, αvβ3 has been shown to participate in the progression of human melanoma (28, 29), in part by virtue of its effects on melanoma cell invasion (30). Finally, given the role of vascular endothelial growth factor-mediated signaling in tumor invasion (31), CLDC-based delivery of an HCMV-driven plasmid expressing an anti-FLK-1 ribozyme also was investigated. Systemic delivery of the anti-β3 and anti-PECAM-1 ribozymes each reduced the metastasis of B16-F10 melanoma in C57Bl6 mice at least as effectively as did p65-R, the anti-p65 ribozyme (Fig. 4), suggesting that these adhesion molecules may participate in the invasive phenotype generated by activation of NF-κB. Conversely, the anti-FLK-1 ribozyme failed to reduce significantly metastatic spread, consistent with published results (24).
Figure 4.
Effects of i.v. treatment with CLDC containing ribozymes targeting p65, integrin β3, PECAM-1, FLK-1, and pVector on metastatic B16 lung tumors.
NF-κB can transactivate both genes known to promote metastasis and genes known to inhibit metastasis in tumor cells themselves as well as in critical normal cell types (1). Therefore, the ability to suppress the expression of NF-κB or important NF-κB-regulated genes in both tumor cells and in critical normal cells was used to dissect out specific genes and functional pathways that regulate the metastatic phenotype in tumor-bearing hosts. Suppression of target-gene expression in a variety of different cell types is important, because it is becoming clear that tumor metastasis is caused by alterations in gene expression both within tumor cells, as well as within surrounding normal cells (32). Systemic, plasmid-based ribozymes targeting either the p65 or p50 subunits of NF-κB, as well as i.v. injection of tumor cells stably transfected with an anti-p65 ribozyme, demonstrated that the expression of NF-κB plays an essential role in promoting the metastatic spread of melanoma. Ribozyme-mediated suppression of NF-κB in vivo resulted in antitumor effects against a cell line with wild-type p53 expression, contrary to what would have been predicted from in vitro studies (9). The loss of NF-κB expression in tumor cells decreased their capacities to invade the extracellular matrix. In addition, ribozymes targeting integrin β3 or PECAM-1, NF-κB-regulated genes involved in cell adhesion, also significantly reduced metastatic spread, suggesting that this ligand-receptor pair may, in part, mediate the role of NF-κB in promoting both invasion and metastasis. Furthermore, by using ribozymes to target PECAM-1 expression in tumor-bearing mice, our studies demonstrate that PECAM-1 plays an important role in regulating tumor metastasis. Although it is beyond the scope of these studies, the level of suppression of tumor metastasis achieved by a single injection of plasmid-based ribozymes suggests the possible therapeutic utility of this approach. In summary, these studies demonstrate the power of using in vivo, ribozyme-based gene targeting in adult animals for identifying the genes and functional pathways that regulate complex biologic phenotypes.
Acknowledgments
We thank Celia Hamilton for manuscript preparation. R.J.D. and P.-Y.D. are supported by National Cancer Institute Grants R-01-CA82575 and R-01-CA82548, respectively. R.J.D. also receives support from Cancer Research Program of California Grant CCRP-2II0022. M.K.S. was supported by the Herschel and Diana Zackheim Endowment Fund and the Leaders' Society Clinical Career Development Award of the Dermatology Foundation.
Abbreviation
- CLDC
cationic liposome:DNA complexes
References
- 1.Pahl H L. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 2.Stancovski I, Baltimore D. Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 3.Verma I M, Stevenson J. Proc Natl Acad Sci USA. 1997;94:11758–11760. doi: 10.1073/pnas.94.22.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C Y, Cusack J C J, Liu R, Baldwin A S., Jr Nat Med. 1999;5:412–417. doi: 10.1038/7410. [DOI] [PubMed] [Google Scholar]
- 5.Guttridge D C, Albanese C, Reuther J Y, Pestell R G, Baldwin A S., Jr Mol Cell Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins K A, Perez J R, Coleman T A, Dorshkind K, McComas W A, Sarmiento U M, Rosen C A, Narayanan R. Proc Natl Acad Sci USA. 1993;90:9901–9905. doi: 10.1073/pnas.90.21.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang S, Robinson J B, Deguzman A, Bucana C D, Fidler I J. Cancer Res. 2000;60:5334–5339. [PubMed] [Google Scholar]
- 8.Liu Y, Thor A, Shtivelman E, Cao Y, Heath T R, Debs R J. J Biol Chem. 1999;274:13338–13344. doi: 10.1074/jbc.274.19.13338. [DOI] [PubMed] [Google Scholar]
- 9.Ryan K M, Ernst M K, Rice N R, Vousden K H. Nature (London) 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 10.Stein C A. J Clin Invest. 2001;108:641–644. doi: 10.1172/JCI13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folkman J. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 12.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Nature (London) 1995;13:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Mounkes L C, Liggitt H D, Brown C S, Solodin I, Heath T D, Debs R J. Nat Biotechnol. 1997;15:167–173. doi: 10.1038/nbt0297-167. [DOI] [PubMed] [Google Scholar]
- 14.Tu G, Kirchmaier A L, Liggitt D, Liu Y, Liu S, Yu W H, Heath T D, Thor A, Debs R J. J Biol Chem. 2000;275:30408–30416. doi: 10.1074/jbc.M004782200. [DOI] [PubMed] [Google Scholar]
- 15.Scanlon K J, Jiao L, Funato T, Wang W, Tone T, Rossi J J, Kashani-Sabet M. Proc Natl Acad Sci USA. 1991;88:10591–10595. doi: 10.1073/pnas.88.23.10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scanlon K J, Kashani-Sabet M. J Natl Cancer Inst. 1998;90:558–559. doi: 10.1093/jnci/90.8.558. [DOI] [PubMed] [Google Scholar]
- 17.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerr J F, Wyllie A H, Currie A R. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Diest P J, Brugal G, Baak J P. J Clin Pathol. 1998;51:716–724. doi: 10.1136/jcp.51.10.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desprez P Y, Lin C Q, Thomasset N, Sympson C J, Bissell M J, Campisi J. Mol Cell Biol. 1998;18:4577–4588. doi: 10.1128/mcb.18.8.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nolan G P, Ghosh S, Liou H-C, Tempst P, Baltimore D. Cell. 1991;64:961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh S, Gifford A M, Riviere L R, Tempst P, Nolan G P, Baltimore D. Cell. 1990;62:1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- 23.Haseloff J, Gerlach W L. Nature (London) 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 24.Pavco P A, Bouhana K S, Gallegos A M, Agrawal A, Blanchard K S, Grimm S L, Jensen K L, Andrews L E, Wincott F E, Pitot P A, et al. Clin Cancer Res. 2000;6:2094–2103. [PubMed] [Google Scholar]
- 25.Piali L, Hammel P, Uherek C, Bachmann F, Gisler R H, Dunon D, Imhof B A. J Cell Biol. 1995;130:451–460. doi: 10.1083/jcb.130.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie C K, Giordano A, Khalili K. J Cell Physiol. 2000;184:214–221. doi: 10.1002/1097-4652(200008)184:2<214::AID-JCP9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 27.Botella L M, Puig-Kroger A, Almendro N, Sanchez-Elsner T, Munoz E, Corbi A, Bernabeu C. J Immunol. 2000;164:1372–1378. doi: 10.4049/jimmunol.164.3.1372. [DOI] [PubMed] [Google Scholar]
- 28.Abelda S M, Mette S A, Elder D E, Stewart R, Damjanovich L, Herlyn M, Buck C. Cancer Res. 1990;50:6757–6764. [PubMed] [Google Scholar]
- 29.Montgomery A M, Reisfeld R A, Cheresh D A. Proc Natl Acad Sci USA. 1994;91:8856–8860. doi: 10.1073/pnas.91.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seftor R E, Seftor E A, Hendrix M J. Cancer Metastasis Rev. 1999;18:359–375. doi: 10.1023/a:1006317125454. [DOI] [PubMed] [Google Scholar]
- 31.Millauer B, Shawver L K, Plate K H, Risau W, Ullrich A. Nature (London) 1994;367:576–579. doi: 10.1038/367576a0. [DOI] [PubMed] [Google Scholar]
- 32.Berns A. Nature (London) 2001;410:1043–1044. doi: 10.1038/35074238. [DOI] [PubMed] [Google Scholar]