Abstract
We report the identification of six patients with 3q29 microdeletion syndrome. The clinical phenotype is variable despite an almost identical deletion size. The phenotype includes mild-to-moderate mental retardation, with only slightly dysmorphic facial features that are similar in most patients: a long and narrow face, short philtrum, and high nasal bridge. Autism, gait ataxia, chest-wall deformity, and long and tapering fingers were noted in at least two of six patients. Additional features—including microcephaly, cleft lip and palate, horseshoe kidney and hypospadias, ligamentous laxity, recurrent middle ear infections, and abnormal pigmentation—were observed, but each feature was only found once, in a single patient. The microdeletion is ∼1.5 Mb in length, with molecular boundaries mapping within the same or adjacent bacterial artificial chromosome (BAC) clones at either end of the deletion in all patients. The deletion encompasses 22 genes, including PAK2 and DLG1, which are autosomal homologues of two known X-linked mental retardation genes, PAK3 and DLG3. The presence of two nearly identical low-copy repeat sequences in BAC clones on each side of the deletion breakpoint suggests that nonallelic homologous recombination is the likely mechanism of disease causation in this syndrome.
The history of detecting microdeletion syndromes started with the recognition of discrete syndromic phenotypes associated with mental retardation, such as Prader-Willi, Miller-Dieker, Angelman, and Williams syndromes. Patients with similar phenotypes were then found to have similar submicroscopic deletions, in discrete genomic regions, that then defined the condition (Ledbetter et al. 1981; Schwartz et al. 1988; Knoll et al. 1989; Ewart et al. 1993).
In 1995, Flint et al. developed a strategy to screen for the abnormal inheritance of some subtelomeric DNA polymorphisms in individuals who had mental retardation alone and no associated clinical syndrome (Flint et al. 1995). Initially, 3 of 99 patients each had a single deletion at one of the telomeres. Since then, the technology has been developed to provide screening of all telomeres as a clinical service for suitably selected patients with mental retardation and dysmorphic features. The range of diagnostic yield is ∼6%–11% (Knight et al. 1999; Slavotinek et al. 1999; Flint and Knight 2003; Koolen et al. 2004). The frequency of specific chromosome submicroscopic deletions or duplications reported in the literature varies from single case reports to >50 cases reported (De Vries et al. 2003). Conditions such as 1p36 or 2q37.3 microdeletions have been reported frequently, and these conditions now assume the status of recognizable syndromes, since common clinical features have emerged through the collection of patients with the same deletion (Shapira et al. 1997; Heilstedt et al. 2003; Aldred et al. 2004). However, several of the subtelomeric deletions have been reported only rarely to date, and the delineation of the associated clinical phenotype is therefore much more difficult.
We report here the identification of six patients with a 3q29 microdeletion. In five patients, the deletion arose de novo, and, for one patient, the parental origin could not be determined. The clinical phenotype is described in all six patients and is compared with that of a single case that was reported by Rossi et al. (2001). The molecular boundaries of the recurrent 1.5-Mb deletion are defined, and nonallelic homologous recombination is proposed as a mechanism of deletion formation in these patients.
Table 1 summarizes the clinical features of the six patients presented here and includes the few clinical details available on the patient analyzed in the study by Rossi et al. (2001). Figure 1 illustrates the patients' facial features and hands. To summarize, this condition is not associated with any antenatal abnormalities, and the birth history was uneventful in all patients. Birth measurements were within the normal range, although microcephaly was noted in patient 6. The level of developmental delay often was not fully recognized until after the 1st year of life, except in patient 6, in whom failure to thrive was noted at 5 mo of age. Growth parameters in all patients were at or below the 50th percentile but remained in the normal range. Speech delay was a particular feature, and all children needed special educational provision because of more general intellectual difficulties, although one child was in a mainstream school with substituted lessons in a specialist learning support unit. Autism was a feature of the behavior of two of the patients. Patient 1's behavior was obsessive and repetitive and met the ICD-10 (International Classification of Diseases, World Health Organization) diagnostic criteria for autism spectrum disorders; patient 3 presented at 3 years of age with moderate-to-severe cognitive learning difficulties with autistic traits but was not diagnosed as having a recognized form of autism. His psychological presentation was complex at that stage and remained so. The dysmorphology in the children is variable and not striking, but there are some facial similarities, and each child also has certain unique features. Generally, the face is long and narrow, especially in patients 2 and 4. The philtrum appears short in all patients, and a high nasal bridge is present in patients 1, 2, 3, and 4. The ears are generally large but well developed. The additional features found in at least two individuals were chest-wall deformity (pectus carinatum in one patient and pectus excavatum in another) and long and tapering fingers in two individuals, with fifth-finger clinodactyly in one of them. One additional child was noted to have fifth-finger brachydactyly, but X-ray assessment was not performed. An ungainly, ataxic gait was described in two patients, and, in patient 2, diagnoses of Angelman and Rett syndromes were considered and excluded. Each of the remaining features was identified only in a single patient: recurrent bilateral middle ear infections, requiring grommets, adenoidectomy, and removal of a cholesteatoma, in patient 2; ligamentous laxity in patient 3; downslanting palpebral fissures, posteriorly rotated ears, smooth philtrum, and prominent lower lip in patient 4; left-sided unilateral cleft lip and palate and nail hypoplasia in patient 5; progressive microcephaly and abnormal skin pigmentation in patient 6; and horseshoe kidney and hypospadias in the patient described by Rossi et al. (2001).
Table 1.
Summary of the Clinical Features Found in the Six New Patients with the 3q29 Deletion and in the Patient from the Study by Rossi et al. (2001)
| Clinical Feature | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient fromRossi et al.(2001) |
| Pregnancy | Normal | Normal | Normal | Normal | |||
| Birth weight (kg) | 3.1 | 3.120 | 2.930 | 2.778 | 2.3 | ||
| Length of gestation | 38 wk | Term | Term | 41 wk | 41 wk | ||
| Feeding | Poor suckling | Failure to thrive | |||||
| Age at sitting | 9 mo | 7 mo | |||||
| Age at walking | 18 mo | 16 mo | Normal | 21 mo | By 3 years | ||
| Age at first words | 28 mo | 19 mo | |||||
| Age when developmental delay was noted | 18 mo | 3 years | 18 mo | 3 years | 5 mo | ||
| Age at talking | 4–5 years | 6–7 years | 3 years | 2 words and Makaton at 8 years, 10 mo | |||
| Type of schooling | Special school | Special school | Special school | Special school (IQ = 70) | Learning support in mainstream school | Special school | Moderate mental retardation |
| Head circumference (percentile) | 9th–25th | 3rd | 50th | 10th | Progressive microcephalya | ||
| Height (percentile) | 25th | 25th–50th | 25th | 40th | 3rd–10th | .4th | |
| Weight (percentile) | 2nd–9th | 25th | 3rd–10th | .4th | |||
| Hands and feet | Long and tapering fingers | Fifth-finger brachydactyly | Long and tapering fingers; clinodactyly of fifth finger and third, fourth, and fifth toes | Long | |||
| Behavior | ICD-10 autistic | Autistic features | |||||
| Pectus | Excavatum | Carinatum | |||||
| Gait | Arm flapping when young | Ataxia and excited, stereotypical waving | Ungainly gait but neurologically normal | ||||
| Additional features | Chronic otitis media and cholesteatoma | Ligamentous laxity | Scaphoid skull (familial) and frontal bossing; downslanting palpebral fissures, smooth philtrum, and prominent lower lip; posteriorly rotated ears | Prominent metopic suture; left unilateral cleft lip and palate; nail hypoplasia | Increased pigmentation on dorsum of left foot; not toilet trained; frequent headaches | Horseshoe kidney and hypospadias |
Head circumference at birth: 32 cm (2 SD below the mean); at age 0.4 years: 37 cm (>4 SD below the mean); and at age 8.8 years: 46 cm (>5 SD below the mean).
Figure 1.
Photographs of patient 1, at age 9 years, 11 mo; patient 2, at age 14 years; patient 3, at age 6 years, 7 mo; patient 4, at age 6 years; patient 5, at age 7 years, 11 mo; and patient 6, at age 8 years, 10 mo. Written consent to publish these photographs was obtained from the parents of each child.
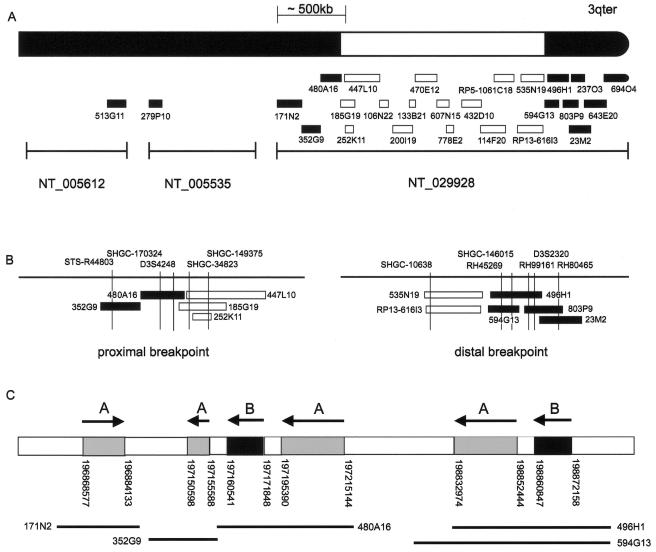
Molecular cytogenetic results are summarized in table 2 and figure 2. The samples were collected from laboratories in the United Kingdom and the Netherlands (Nijmegen), and FISH was performed on interphase or metaphase spreads, depending on the sample availability. G-banding at the 550 band or at a higher level, FISH probe preparation, and hybridization were performed using standard techniques. Parental samples were available for patients 1–5, and all deletions had arisen de novo. Patient 6 was adopted out of the birth family; thus, parental samples were unavailable for analysis. FISH analysis of patients 1 and 2 showed the presence of a terminal deletion of 3q29 by use of 3qter probe pVYS223B from the Totelvysion subtelomere screening kit (Vysis) on metaphase spreads. Deletions in patients 3 and 6 were initially suspected on high-resolution G-banding and were confirmed using the 3q subtelomeric probe CTC-196F4 from the study by the National Institutes of Health and Institute of Molecular Medicine Collaboration (1996). In patient 5, the deletion was detected using probe 3qtel106 from the Cytocell subtelomere screening kit. Probes pVYS223B, CTC-196F4, and 3qtel106 all map within BAC clone RP5-1061C18 (fig. 2A). In patient 4, the deletion was detected using multiplex ligation-dependent probe amplification (MLPA) (Koolen et al. 2004). The 3q29 deletion was not detectable with the older MLPA MRC (Medical Research Council) Holland telomere kit, PO19, which recognizes sequence in the nondeleted clone RP11-496H1, but is identifiable only with the new kit, PO36, which uses sequence within the BDH gene located within the deleted clones RP13-616I3 and RP11-535N19 as a MLPA probe. To refine the deletion, 26 overlapping BAC and PAC clones were identified using the then-available build 31 sequence (National Center for Biotechnology Information [NCBI] Map Viewer). The human genomic clones from the RPCI-5, -11, and -13 BAC and PAC libraries were obtained from the BACPAC Resources Center. Slides were analyzed using a fluorescence microscope (Leica DMRB), and images were recorded using SmartCapture 2 software (Digital Scientific). Published STS primers STS-R44803, SHGC-170324, D3S4248, SHGC-34823, SHGC-149375, SHGC-10638, RH45269, SHGC-146015, RH99161, D3S2320, and RH80465 were used to check the identity of the clones (fig. 2B). In patients 1–4 and patient 6, the deletion endpoints were refined to within a single BAC clone. The telomeric breakpoint was within BAC clones RP11-594G13 and RP11-496H1, and the centromeric breakpoint was within BAC clones RP11-185G19 and RP11-480A16. For patient 5, there was insufficient material available to define the centromeric breakpoint, but the telomeric breakpoint was within BAC clone RP11-496H1, as for patient 2. The centromeric breakpoint lay between BAC clones RP11-171N2 and RP11-185G19 (table 2). At the telomeric breakpoint, the two BAC clones that define the deletion endpoint in the six patients overlap, and both contain STSs RH45269 and SHGC-146015. Most of clone RP11-594G13 is also contained within RP11-496H1, but clone RP11-594G13 extends ∼60 kb further toward the centromere than RP11-496H1. On the basis of the resolution of FISH, the breakpoints in each of the patients are likely to be similar and to lie within 60–70 kb of each other.
Table 2.
Limits of Interstitial 3q29 Microdeletion in Six Unrelated Patients[Note]
|
Presence or Absenceof Probe in Patientb |
|||||||
| Contig andBAC/PAC Clonea | AccessionNumber | 1 | 2 | 3 | 4 | 5 | 6 |
| NT_005612: | |||||||
| 513G11 | AC117469 | P | P | P | P | P | P |
| NT_005535: | |||||||
| 279P10 | AC125362 | P | P | P | P | P | P |
| NT_029928: | |||||||
| 171N2 | AC069513 | P | P | P | P | NT | P |
| 352G9 | AC124944 | P | P | P | P | NT | P |
| 480A16 | AC024937 | P | D | D | D | NT | D |
| 185G19 | AC139666 | D | D | D | D | NT | D |
| 252K11 | AC026308 | D | D | D | D | D | D |
| 447L10 | AC069257 | D | D | D | D | NT | D |
| 106N22 | AC083822 | D | D | D | D | NT | D |
| 200I19 | AC092933 | D | D | D | D | NT | D |
| 133B21 | AC023797 | D | D | D | D | NT | D |
| 470E12 | AC055725 | D | D | D | D | NT | D |
| 607N15 | AC127904 | D | D | D | D | NT | D |
| 778E2 | AC016949 | D | D | D | D | D | D |
| 432D10 | AC068302 | D | D | D | D | NT | D |
| 114F20 | AC092937 | D | D | D | D | D | D |
| RP5-1061C18 | AL121981 | D | D | D | D | D | D |
| RP13-616I3 | AC128709 | D | D | D | D | D | D |
| 535N19 | AC126183 | D | D | D | D | D | D |
| 594G13 | AC132008 | P | D | P | P | D | P |
| 496H1 | AC024560 | P | P | P | P | P | P |
| 803P9 | AC055764 | P | P | P | P | P | P |
| 23M2 | AC022621 | P | P | P | P | P | P |
| 237O3 | AC144530 | P | P | P | P | NT | P |
| 643E20 | AC135893 | P | P | P | P | NT | P |
| 694O4 | AC073135 | P | P | P | P | NT | P |
Note.— All accession numbers listed in the table are from GenBank.
Unless otherwise stated, the BAC or PAC clones used were from the RP11 library.
P = probe was present on both chromosome 3 homologues by FISH; D = probe was deleted on one of the chromosome 3 homologues; NT = probe was not tested, because of insufficient chromosomal material available from the patient.
Figure 2.
A, Genomic map of chromosome 3q, showing which clones are present (blackened) and which clones are deleted (unblackened). All six patients have deletions of BAC clones RP11-252K11–RP11-535N19 (see text). The MLPA probe, which can be used to detect the deletions in all six patients, is located within the BDH gene within BAC clones RP13-616I3 and RP11-535N19. The commercially available FISH probes—Vysis 3qter probe pVYS223B (D3S3560), Cytocell 3qtel106, and CTC-196F4—are all located within BAC clone RP5-1061C18. Unless otherwise stated, the BAC or PAC clones used were from the RP11 library. Contig NT_029928 and the size of the BAC clones within it are approximately to scale. The three contigs, GenBank NT_029928 (2.6 Mb), GenBank NT_005535 (1.2 Mb), and GenBank NT_005612 (100 Mb), are separated by gaps of unknown length in the sequence. B, The detailed positioning of BAC clones at the proximal and distal breakpoints. The relative position of each BAC clone was determined by identifying the presence or absence of known STS markers within the BAC clones and the genomic sequence around the deletion boundaries. C, LCR sequences in the deletion breakpoint genomic region. The region-specific LCRs designated “A” and “B” are shown (see text). The arrows above the bar show the orientation of the repeats; the position of the repeats on chromosome 3 are given below the bar. From left to right on the diagram, the repeat sizes are 15 kb, 5 kb, 11 kb, 19 kb, 19 kb, and 11 kb. The BAC clones that contain the LCRs are marked below the genomic location. The diagram is not to scale.
The extent of the common microdeletion was determined using the finished sequence clones selected from the contigs on NCBI build 35.1 (NCBI Map Viewer). The microdeletion is estimated to be ∼1.5 Mb, and it contains at least 22 transcripts, 5 of which are known genes (PYT1A, PAK2 [MIM 605022], MFI2 [MIM 155750], DLG1 [MIM 601014], and BDH [MIM 603063]), 7 of which are incomplete cDNAs with two or more documented cDNA sequences, and 10 of which are hypothetical genes with no experimental evidence.
Since the deletion limits in the six patients were almost identical, the finished genomic sequence on each side of the deletion was investigated to determine whether any region-specific low-copy repeats (LCRs) were present. Nix analysis (HGMP-RC Nix Session Web site) of RP11-496H1 (GenBank accession number AC024560) found regions of sequence homology within BAC clones RP11-480A16, RP11-352G9, and RP11-171N2. Two separate LCR sequences, designated “repeat A” and “repeat B,” were identified on each side of the deletion region (fig. 2C). By use of BLAT analysis (Human BLAT Search Web site), repeat A was identified four times on chromosome 3, at positions 198832974–198852444, 197195390–197215144, and 197150598–197155588 in one orientation and at position 196868577–196884133 in the opposite orientation. Each repeat was >97.5% homologous and was ∼19 kb, ∼19 kb, ∼5 kb, and ∼15 kb long, respectively (fig. 2C). Repeat B occurred twice on chromosome 3; both repeats were in the same orientation and were located on chromosome 3 on each side of the breakpoint, at positions 198860847–198872158 and 197160541–197171848. These sequences were 98% homologous, and both were 11 kb in length (fig. 2C).
In summary, there is evidence of the presence of region-specific LCRs within the genomic sequence at each end of the microdeletion. It is likely that the formation of the similarly sized de novo microdeletions identified in the six patients was facilitated by these repeats and probably arose by nonallelic homologous recombination between LCRs on each side of the breakpoint, resulting in a single copy of the LCR and a genomic deletion between them. Alternatively, the presence of LCRs on each side of the deletion may not be directly involved in the deletion, per se, but they may act to predispose the genome to form a deletion. Both disease-causing mechanisms are now well recognized, although the presence of LCRs on each side of the deletion suggests that the former mechanism is more likely (Osborne et al. 2001; Shaw and Lupski 2004).
This analysis of six patients with an interstitial microdeletion of 3q29 is the first collation of cases to delineate 3q29 microdeletion syndrome. The clinical features of a patient from a previous report have been included for comparison, but samples from this patient were not available to establish the exact deletion breakpoints, and the family has been lost to clinical follow-up (E. Rossi, personal communication). The molecular deletion boundaries detected in the patients are strikingly similar, yet there is considerable clinical variability in the phenotype, with mild-to-moderate mental retardation being the only common and consistent feature. In addition, slightly dysmorphic facial features are similar in most patients: a long and narrow face, short philtrum, and high nasal bridge. As additional patients are screened for microdeletion syndromes on the basis of mental retardation and autism alone, it is likely that this syndrome will become increasingly well identified. Understanding the exact molecular mechanism of disease in these patients—that is, understanding how the deletion of some 22 genes in this region affects the development of an individual—is, of course, the next challenge. At this stage, it is impossible to attribute the phenotype to any one of the deleted genes, but two genes within the deleted area—PAK2 and DLG1—merit further interest, since both are autosomal homologues of known X-linked mental retardation genes PAK3 (MIM 300142) and DLG3 (MIM 300189). Loss-of-function mutations in either PAK3 or DLG3 result in moderate-to-severe mental retardation (Allen et al. 1998; Tarpey et al. 2004). The DLG1 protein SAP97, like SAP102 (DLG3), is a component of the postsynaptic density, and RNAi knockdown experiments of SAP97 result in reduced surface expression of GRIA1 (MIM 138248) and GRIA2 (MIM 138247) and a decrease in both AMPA (MIM 138248) and NMDA (MIM 138251) excitatory postsynaptic currents (Nakagawa et al. 2004). This suggests that loss of PAK2 or DLG1 may have a critical role in the development of mental retardation in these patients, but, clearly, this hypothesis needs further investigation.
Acknowledgments
We are grateful to the patients and families, for taking part in this study, and to the clinicians, for referring them. We thank Elizabeth Kerr for her expert technical assistance. J.C. is supported by the Medical Research Council, United Kingdom; H.M. is supported by the Department of Health, United Kingdom; B.B.A.d.V. is supported by a grant from the Netherlands Organization for Health Research and Development; and F.L.R. is supported by the Wellcome Trust.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BACPAC Resources Center, http://bacpac.chori.org/home.htm (for human genomic clones from the RPCI-5, -11, and -13 BAC and PAC libraries)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for all accession numbers of BAC and PAC clones and genomic contigs listed in )
- HGMP-RC Nix Session, http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix/ (for homology searching within BAC clone sequences)
- Human BLAT Search, http://www.genome.ucsc.edu/cgi-bin/hgBlat (for homology searching within the chromosome 3 sequence)
- NCBI Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/ (for BLAST sequence homology search and for transcripts and genes within the 3q29 deleted region)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PAK2, MFI2, DLG1, BDH, PAK3, and DLG3)
References
- Aldred MA, Sanford RO, Thomas NS, Barrow MA, Wilson LC, Brueton LA, Bonaglia MC, Hennekam RC, Eng C, Dennis NR, Trembath RC (2004) Molecular analysis of 20 patients with 2q37.3 monosomy: definition of minimum deletion intervals for key phenotypes. J Med Genet 41:433–439 10.1136/jmg.2003.017202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA (1998) PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet 20:25–30 10.1038/1675 [DOI] [PubMed] [Google Scholar]
- De Vries BB, Winter R, Schinzel A, van Ravenswaaij-Arts C (2003) Telomeres: a diagnosis at the end of the chromosomes. J Med Genet 40:385–398 10.1136/jmg.40.6.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, Keating MT (1993) Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet 5:11–16 10.1038/ng0993-11 [DOI] [PubMed] [Google Scholar]
- Flint J, Knight S (2003) The use of telomere probes to investigate submicroscopic rearrangements associated with mental retardation. Curr Opin Genet Dev 13:310–316 10.1016/S0959-437X(03)00049-2 [DOI] [PubMed] [Google Scholar]
- Flint J, Wilkie AO, Buckle VJ, Winter RM, Holland AJ, McDermid HE (1995) The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet 9:132–140 10.1038/ng0295-132 [DOI] [PubMed] [Google Scholar]
- Heilstedt HA, Ballif BC, Howard LA, Lewis RA, Stal S, Kashork CD, Bacino CA, Shapira SK, Shaffer LG (2003) Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am J Hum Genet 72:1200–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Regan R, Nicod A, Horsley SW, Kearney L, Homfray T, Winter RM, Bolton P, Flint J (1999) Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet 354:1676–1681 10.1016/S0140-6736(99)03070-6 [DOI] [PubMed] [Google Scholar]
- Knoll JH, Nicholls RD, Magenis RE, Graham JM Jr, Lalande M, Latt SA (1989) Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet 32:285–290 [DOI] [PubMed] [Google Scholar]
- Koolen DA, Nillesen WM, Versteeg MH, Merkx GF, Knoers NV, Kets M, Vermeer S, van Ravenswaaij CM, de Kovel CG, Brunner HG, Smeets D, de Vries BB, Sistermans EA (2004) Screening for subtelomeric rearrangements in 210 patients with unexplained mental retardation using multiplex ligation dependent probe amplification (MLPA). J Med Genet 41:892–899 10.1136/jmg.2004.023671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter DH, Riccardi VM, Airhart SD, Strobel RJ, Keenan BS, Crawford JD (1981) Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N Engl J Med 304:325–329 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Futai K, Lashuel HA, Lo I, Okamoto K, Walz T, Hayashi Y, Sheng M (2004) Quaternary structure, protein dynamics, and synaptic function of SAP97 controlled by L27 domain interactions. Neuron 44:453–467 10.1016/j.neuron.2004.10.012 [DOI] [PubMed] [Google Scholar]
- National Institutes of Health and Institute of Molecular Medicine Collaboration (1996) A complete set of human telomeric probes and their clinical application. Nat Genet 14:86–89 10.1038/ng0996-86 [DOI] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW (2001) A 1.5 million–base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 10.1038/ng753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi E, Piccini F, Zollino M, Neri G, Caselli D, Tenconi R, Castellan C, Carrozzo R, Danesino C, Zuffardi O, Ragusa A, Castiglia L, Galesi O, Greco D, Romano C, Pierluigi M, Perfumo C, Di Rocco M, Faravelli F, Dagna Bricarelli F, Bonaglia M, Bedeschi M, Borgatti R (2001) Cryptic telomeric rearrangements in subjects with mental retardation associated with dysmorphism and congenital malformations. J Med Genet 38:417–420 10.1136/jmg.38.6.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CE, Johnson JP, Holycross B, Mandeville TM, Sears TS, Graul EA, Carey JC, Schroer RJ, Phelan MC, Szollar J, Flannery DB, Stevenson RE (1988) Detection of submicroscopic deletions in band 17p13 in patients with the Miller-Dieker syndrome. Am J Hum Genet 43:597–604 [PMC free article] [PubMed] [Google Scholar]
- Shapira SK, McCaskill C, Northrup H, Spikes AS, Elder FF, Sutton VR, Korenberg JR, Greenberg F, Shaffer LG (1997) Chromosome 1p36 deletions: the clinical phenotype and molecular characterization of a common newly delineated syndrome. Am J Hum Genet 61:642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13 Spec No 1:R57–R64 10.1093/hmg/ddh073 [DOI] [PubMed] [Google Scholar]
- Slavotinek A, Rosenberg M, Knight S, Gaunt L, Fergusson W, Killoran C, Clayton-Smith J, Kingston H, Campbell RH, Flint J, Donnai D, Biesecker L (1999) Screening for submicroscopic chromosome rearrangements in children with idiopathic mental retardation using microsatellite markers for the chromosome telomeres. J Med Genet 36:405–411 [PMC free article] [PubMed] [Google Scholar]
- Tarpey P, Parnau J, Blow M, Woffendin H, Bignell G, Cox C, Cox J, et al (2004) Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet 75:318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]