Abstract
Biotin-responsive basal ganglia disease (BBGD) is a recessive disorder with childhood onset that presents as a subacute encephalopathy, with confusion, dysarthria, and dysphagia, and that progresses to severe cogwheel rigidity, dystonia, quadriparesis, and eventual death, if left untreated. BBGD symptoms disappear within a few days with the administration of high doses of biotin (5–10 mg/kg/d). On brain magnetic resonance imaging examination, patients display central bilateral necrosis in the head of the caudate, with complete or partial involvement of the putamen. All patients diagnosed to date are of Saudi, Syrian, or Yemeni ancestry, and all have consanguineous parents. Using linkage analysis in four families, we mapped the genetic defect near marker D2S2158 in 2q36.3 (LOD=5.9; θ=0.0) to a minimum candidate region (∼2 Mb) between D2S2354 and D2S1256, on the basis of complete homozygosity. In this segment, each family displayed one of two different missense mutations that altered the coding sequence of SLC19A3, the gene for a transporter related to the reduced-folate (encoded by SLC19A1) and thiamin (encoded by SLC19A2) transporters.
Introduction
Biotin-responsive basal ganglia disease (BBGD [MIM 607483]) was first described in 1998 in a number of families in Saudi Arabia (Ozand et al. 1998). Patients presented in childhood with a subacute encephalopathic picture of undefined origin, including confusion, vomiting, and a vague history of febrile illness. They progressed to acute encephalopathy, with sudden loss of developmental milestones; inability to swallow; loss of speech (or slurred speech); loss of motor function, with development of quadriparesis or quadriplegia; and seizures. Left untreated, the disorder results in a chronic or slowly progressive encephalopathy, with an akinetic mute state, permanent loss of speech and comprehension, and eventual death. Magnetic resonance imaging (MRI) examination of patients with BBGD revealed specific bilateral necrosis in the head of the caudate nucleus and in the putamen. However, results of tests to uncover evidence of infection, storage or mitochondrial diseases, or toxicological damage were all negative. Since all affected individuals had consanguineous parents, it was concluded that BBGD is likely due to a recessive genetic defect. Fortunately, administration of high doses of biotin (5–10 mg/kg/d) early in the disorder was found to eliminate the symptoms of BBGD within a few days, with no recurrence unless biotin treatment was discontinued. In the latter circumstance, symptoms returned within a month. Treatment later in the disorder left residual paraparesis, mild mental retardation, or dystonia. Even when biotin administration resolved the clinical symptoms of BBGD, the abnormal MRI findings remained but showed no further progression.
Biotin is essential for the activity of a number of critical carboxylases, including propionyl coenzyme A (CoA), acetyl CoA, 3-methylcrotonyl CoA, and pyruvate carboxylases (Moss and Lane 1971). These are converted from apoenzymes to active holoenzymes by transfer of biotin from holocarboxylase synthetase. Specific carboxylase deficiencies have been described that are due to mutations in the corresponding structural genes. Mutation in the holocarboxylase synthetase gene leads to multiple carboxylase deficiency (Suzuki et al. 1994; Aoki et al. 1995; Dupuis et al. 1996; Narisawa et al. 1996), which may, in some cases, be biotin responsive (Wolf and Feldman 1982). Since biotin is not synthesized de novo in humans, multiple carboxylase deficiency may also result from mutation in biotinidase (Pomponio et al. 1995), which is essential for recycling endogenous biotin. The patients with BBGD did not display deficiency of any of these enzymes, indicating that their genetic defect lay in some additional component of biotin metabolism. We have used linkage mapping and positional cloning to identify mutations in the transporter gene SLC19A3 as the cause of BBGD in four families.
Material and Methods
Families with BBGD
Four families with seven affected members are shown in figure 1. After institutional review board approval, informed consent to participate and blood samples were obtained from patients and family members; standard methods were used for extraction of DNA. Families 11, 30, and 44 are of Saudi origin; these families each involve a first-cousin mating, and they each comprise affected individuals described in the study by Ozand et al. (1998) (patients 1 [patient 1103 in the present study], 2 [1102], 3 [4407], 4 [4408], and 7 [3003]). Related kindreds 12 and 14 are of Yemeni origin. Affected individual 1403 was described as “patient 6” in the study by Ozand et al. (1998). Affected individual 1203 is a previously undescribed affected family member with features of BBGD; this individual has an affected sibling not available for the present study.
Figure 1.
Homozygosity mapping and 2q haplotypes in four consanguineous families with BBGD. Blackened symbols indicate patients; unblackened symbols indicate unaffected individuals. Marker alleles, numbered as described in the “Material and Methods” section, are given for seven affected individuals and eight parents within four families with BBGD. Boxes highlight regions of complete homozygosity in affected individuals, indicating disease-chromosome haplotypes. Borders of the minimum candidate interval are denoted by the connected arrows. Genotypes that were not determined are shown by square brackets. Not shown, for confidentiality, are seven unaffected siblings in three of the families who were genotyped and included in the LOD score calculations. None of these samples showed homozygosity for the candidate region. On the basis of the results of mutation analysis, the first three families (11, 20, and 44), all of which involve first-cousin matings, likely share a common ancestor, with the difference in the allele for D2S1344 in family 44 likely being due to a change in the microsatellite marker repeat.
Genotyping
PCR was performed in a total volume of 7.5 μl, with 30 ng/μl of genomic DNA, 0.1–1.0 μl (of a 5-μM concentration) of each dye-labeled primer pair (PerkinElmer/Applied Biosystems and Research Genetics), 0.06 μl (of a 5-U/μl concentration) AmpliTaq Gold DNA polymerase (PerkinElmer/Applied Biosystems), 0.75 μl 10× GeneAmp PCR buffer II (PerkinElmer/Applied Biosystems), 0.75 μl GeneAmp dNTP mix (2.5 mM [PerkinElmer]), 0.75 μl MgCl2 (25 mM), and sufficient dH2O to bring the final volume to 7.5 μl. PCR was performed in the MJ Research DNA Engine tetrad with the following conditions: initial denaturation at 95°C for 12 min; 10 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 60 s; 25 cycles at 89°C for 30 s, 55°C for 30 s, and 72°C for 60 s; and 1 cycle at 72°C for 40 min. Analysis of marker D2S1256 used reagents as stated above, except for the final MgCl2 concentration, which was 1.0 mM, not 2.5 mM. The PCR conditions were also different: initial denaturation at 95°C for 15 min; 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s; and 1 cycle at 72°C for 10 min. PCR products were run on a 4.5% acrylamide gel (4.5% AutoPAGE Plus [Sigma]) on the ABI 377XL DNA Sequencer (PerkinElmer/Applied Biosystems) for 2 h, with a size standard (GS350 TAMRA or GS500 TAMRA [PerkinElmer/Applied Biosystems]) run in each lane to ensure lane-to-lane and gel-to-gel size-calling consistency. The data from the gels were analyzed with ABI GeneScan, version 3.1, and Genotyper, version 2.1, software. The genome scan used 302 highly polymorphic di-, tri-, and tetranucleotide repeat microsatellite markers from the ABI Linkage Mapping Set, versions 1 and 2 (PerkinElmer), and from the Weber Human Screening Set, version 9 (Research Genetics). After 2q36-37 was implicated in the genome scan, we chose additional markers in this vicinity using the Human Genome Database and the UCSC Genome Browser. Sequences for primers used for markers in figure 1 are listed in table 1. Allele sizes were related to previous typing on the CEPH panel by regenotyping CEPH individual 133101 (Fondation Jean Dausset CEPH Web Server). Alleles were numbered in figure 1, consistent with the allele designations given in the Human Genome Database, except for the following alleles: alleles 1–9 of D2S1344 were 280–304 bp, differing by 3 bp; alleles 1–11 of D2S1256 were 285–325 bp, differing by 4 bp; and alleles 1 and 2 of D2S260 were 269 bp and 267 bp, respectively.
Table 1.
Primers Sequences Used for Markers on Chromosomes 2 and 19
| Marker | Primer Sequences |
| D2S325 | 5′-AGCTTCAGAAATGCCGTTG-3′, 5′-CCAGTCTTCAGAGATAACAGGG-3′ |
| D2S164 | 5′-GTCCTAACAGGCCACAGACC-3′, 5′-GCTGGCAGTATCACATGACA-3′ |
| D2S126 | 5′-TGCCTAGCACAGAGCCTACT-3′, 5′-TCCATGGCATAGAACAAAAGT-3′ |
| D2S266 | 5′-AATCCAAATTAACGCAGGGTA-3′, 5′-GTACCTCCCTCCTTTATCC-3′ |
| D2S2204 | 5′-CCACAGTGCCTGGCAT-3′, 5′-TGTGACTTTTCTAAAACTGGCTTC-3′ |
| D2S2354 | 5′-TGGGAATGAAGCCCAA-3′, 5′-GAGAAACCAGAAACCAGACTTAACT-3′ |
| D2S2389 | 5′-GCCTCTGTAGGGACTTGTGA-3′, 5′-TTTGTAGAGTACCCTGGAAGTTACC-3′ |
| D2S1349 | 5′-TTTGAAGGTTTTGTGTTT AATCC-3′, 5′-GGACATAAGGAGGGGAACAT-3′ |
| D2S159 | 5′-AGGGATTGGGGACTGG-3′, 5′-GCAGTTTGAGTTTAGGATGA-3′ |
| D2S2158 | 5′-AGCTGTCATCTCCTATGATAAA-3′, 5′-CGTGCAGTTATGTACGG-3′ |
| D2S1344 | 5′-GGCTTTAAGGGTGAGAAAGC-3′, 5′-TCCATCAGTGCTTAAGGAGC-3′ |
| D2S401 | 5′-GGGAGATTTGGAGAAACA-3′, 5′-CAGAAGTTGCAGAACCAT-3′ |
| D2S439 | 5′-AGGAACAAAGTCTCATTCTCTTG-3′, 5′-ACAGATATCATTTACAACACATGTG-3′ |
| D2S1256 | 5′-ATTCACACTACCAGCCTGGC-3′, 5′-TTTCCTCAGTCCTGCCCTG-3′ |
| D2S260 | 5′-ATCAACATTTTTCCCCTGTAC-3′, 5′-TGCCGCTCTTTTCAGATGG-3′ |
| D2S427 | 5′-ATGTAGACTGCTAACAGTGTCTGC-3′, 5′-TTGGTCAACATGTCAATTTAGG-3′ |
| D2S206 | 5′-TTAAAAATTAAGTAGGCTTTTGGTT-3′, 5′-GTCCTCATGTGTTTATGCTGT-3′ |
| D19S596 | 5′-GAATCCGAGAGGTGGG-3′, 5′-GCCAGAGCCACTGTGT-3′ |
| D19S867 | 5′-CAATGAAAATGCTTTGTAAAAC-3′, 5′-CCTTCAGAGGTGACCAG-3′ |
Two microsatellite markers, D19S596 and D19S867 (table 1), flanking the 1.2-Mb critical region in 19q13 that was defined for infantile bilateral striatal necrosis, were specifically genotyped to exclude this genomic segment. PCR cycling conditions were the same as for genome scan markers, except that the second set of cycles was performed 21 times and 23 times, respectively, for D19S596 and D19S867. These products were analyzed on Applied Biosystems 3730xl DNA Analyzer, with size standard GS500 LIZ. The data were analyzed with GeneMapper, version 3.5 (Applied Biosystems).
Statistical Analysis
Two-point LOD scores between the disease locus and the individual markers in our screening set were calculated using the MLINK program of the FASTLINK 3.0P software package (Lathrop and Lalouel 1984; Lathrop et al. 1984, 1986; Cottingham et al. 1993; Schaffer et al. 1994; Schaffer 1996). Haplotypes were constructed manually through visual inspection of the markers under consideration. Fine mapping of the markers in our linked region was accomplished by consulting known linkage and physical maps.
Mutation Scanning
From patient genomic DNA, we amplified coding exons of SLC19A3; three exons of a neighboring hypothetical gene, DKFZp547H025 (GenBank accession number NM_020161); and two other related sequences, named “alt3” and “alt4” (see the “Results” section and fig. 2), and, with primers located in flanking intron and UTR sequences, we performed standard 30-μl PCR (in 10 mM Tris [pH 8.9], 50 mM KCl, 1.5–2.5 mM MgCl2, 10 pmol of each primer, and 200 μM of each dNTP, including 50–100 ng DNA and 1 U Taq polymerase [Invitrogen]). The oligonucleotide primers were designed using Primer3 and are available on request. The PCR products were assessed by SSCP analysis; when an abnormal band shift was observed, the sample was analyzed by direct sequencing by using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) on an ABI 377XL DNA Sequencer (PerkinElmer). Screening of controls was performed by PCR-SSCP analysis of the SLC19A3 68G→T/G23V mutation: the 219-bp exon 2 product was amplified with primers 5′-TCAGCCATGGATTGTTACAGA-3′ and 5′-TCCTTGTTCAAGTCCCATAAAA-3′ for 30 cycles at 94°C for 45 s, 58°C for 45 s, and 72°C for 1 min and a final 10-min extension at 72°C. The PCR product was labeled by incorporation of [32P]-dGTP, was displayed on 8% nondenaturing polyacrylamide gels containing 10% glycerol, and was then detected by autoradiography with the use of XAR film (Kodak). Screening of controls was performed by PCR-RFLP analysis of the SLC19A3 exon 5 1264A→G/T422A mutation: a 291-bp PCR product was amplified with primers 5′-CCCTGTGGCCAATATGTTCT-3′ and 5′-TTGCTTGTTGTAAGGTTGAGAAA-3′ for 30 cycles at 94°C for 45 s, 57°C for 45 s, and 72°C for 1 min and a final 10-min extension at 72°C. The PCR product was digested with BspHI to produce fragments of 170 bp and 121 bp (wild type) and 291 bp (mutant) and was resolved by electrophoresis on 2% agarose gels (Invitrogen). White control chromosomes were from 162 unrelated grandparents or parents from the extended CEPH pedigrees (Fondation Jean Dausset CEPH Web Server). Arab control chromosomes were from the Coriell Cell Repositories (two panels of 10 individuals each [catalog IDs HD05 and HD27]) and from 42 unrelated members of other Arab disease pedigrees collected for mapping/polymorphism studies.
Figure 2.
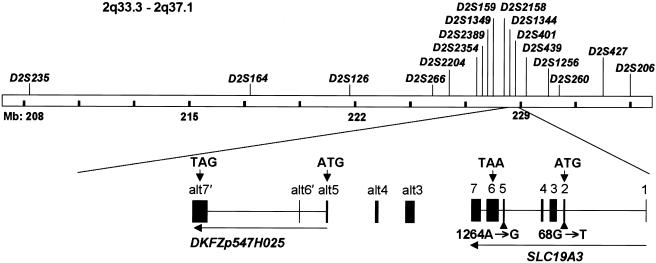
SLC19A3 genomic region, gene structure, and BBGD point mutations. Locations of the microsatellite markers used for the linkage study of the 2q33.3-2q37.1 region and the corresponding gene region for SLC19A3 and DKFZp547H025 are shown. The human SLC19A3 gene is located in 2q36.3, spans 32.91 kb of genomic DNA, and consists of seven exons with a start codon in exon 2 and a TAA stop codon in exon 6. The SLC19A3 cDNA includes a predicted 1,488-bp ORF encoding a 496-aa protein of 55.6 kDa molecular weight. One mutation, 68 G→T, was identified in exon 2, and another mutation, 1264 A→G, was found in exon 5. Sequences alt3, alt4, and alt5 were detected by TBLASTN search (see the BLAST Web site) and were found to be similar to SLC19A3 exons 3, 4, and 5, respectively. The nearby hypothetical protein DKFZp547H025, which is based on a sequenced cDNA, contains three exons (alt5, along with sequences designated “alt6′” and “alt7′,” which are unrelated to SLC19A3), with start and stop codons indicated by arrows. Nine novel SNPs were identified in the 5′ UTR, alt3, alt6′, and alt7′ in SLC19A3 and DKFZp547H025; the SNPs were submitted to dbSNP and were assigned accession numbers 32476166–32476174.
Hydrophobicity Predictions
Hydrophobicity plots in figure 3 were determined using HMMTOP 2.0 (Tusnady and Simon 1998, 2001). SLC19A3 (National Center for Biotechnology Information [NCBI] Entrez Protein accession number NP_079519) was submitted for analysis.
Figure 3.
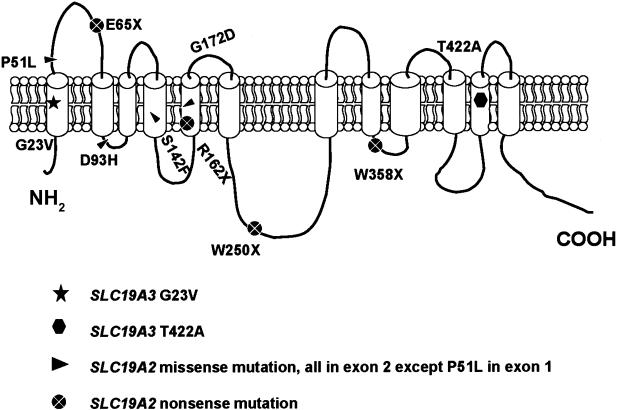
Schematic model of the reduced-folate transporter family. SLC19A3 encodes a predicted protein containing 12 putative transmembrane domains. The relative positions of the mutations identified in SLC19A3 in patients with BBGD and of the missense and nonsense mutations in SLC19A2, reported in patients with Rogers syndrome, are shown (Diaz et al. 1999; Labay et al. 1999; Raz et al. 2000; Scharfe et al. 2000; Lagarde et al. 2004). The positions of mutations in SLC19A2 were assigned in accordance with hydrophobicity plots that were determined by HMMTOP 2.0 (Tusnady and Simon 1998, 2001). SLC19A2 and SLC19A3 (NCBI accession numbers NP_008927 and NP_079519) were used for analysis.
Results
Genome Scan
For our initial genome scan, we genotyped 12 individuals, including 6 affected subjects, from four families with 302 markers (fig. 1 [excluding family 12]). On the basis of initial positive LOD scores on the long arm of chromosome 2, we added additional markers to saturate this region, genotyping all 22 available samples: the 12 original individuals, 3 members of family 12, and 7 unaffected siblings in these families (not shown, for confidentiality). Marker D2S2158 demonstrated significant linkage, with a maximum LOD score of 5.9 at θ=0.0 (table 2). In each family, the affected individuals were homozygous for the region surrounding D2S2158, consistent with the known consanguinity and recessive inheritance pattern (fig. 1). The minimum region of complete homozygosity was between D2S2354 and D2S1256, a segment of ∼2 Mb. The families did not share identical haplotypes across the entire minimum candidate region, indicating either that more than one founder mutation was represented or that a single-founder defect could be further localized by examining regions of identity in all seven affected individuals.
Table 2.
LOD Scores for Linkage of BBGD to 2q36-37[Note]
|
LOD at θ= |
||||||||
| Marker | Locationa(Mb) | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| D2S325 | 208.096 | −∞ | −.06 | .85 | .93 | .62 | .28 | .08 |
| D2S164 | 217.786 | 4.43 | 4.30 | 3.78 | 3.15 | 1.98 | 1.03 | .38 |
| D2S126 | 221.843 | 1.47 | 1.88 | 2.08 | 1.89 | 1.28 | .67 | .22 |
| D2S266 | 225.111 | .89 | 2.20 | 2.38 | 2.08 | 1.30 | .64 | .20 |
| D2S2204 | 226.302 | −1.01 | .99 | 1.85 | 1.91 | 1.48 | .88 | .36 |
| D2S2354 | 227.175 | 3.29 | 3.66 | 3.68 | 3.28 | 2.24 | 1.25 | .49 |
| D2S2389 | 227.372 | 3.52 | 3.42 | 3.03 | 2.55 | 1.65 | .91 | .36 |
| D2S1349 | 227.636 | 4.04 | 3.91 | 3.43 | 2.85 | 1.77 | .90 | .31 |
| D2S159 | 227.649 | 4.20 | 4.08 | 3.62 | 3.04 | 1.97 | 1.07 | .41 |
| D2S2158 | 227.886 | 5.90 | 5.74 | 5.10 | 4.30 | 2.76 | 1.43 | .48 |
| D2S1344 | 227.912 | 4.94 | 4.80 | 4.27 | 3.60 | 2.34 | 1.26 | .47 |
| D2S401 | 228.084 | 5.66 | 5.50 | 4.86 | 4.06 | 2.55 | 1.29 | .42 |
| D2S439 | 229.006 | 4.39 | 4.26 | 3.75 | 3.13 | 2.00 | 1.06 | .40 |
| D2S1256 | 229.241 | 1.36 | 2.84 | 2.98 | 2.62 | 1.69 | .88 | .33 |
| D2S260 | 230.090 | −1.78 | −.22 | .24 | .30 | .20 | .11 | .04 |
| D2S427 | 232.032 | 1.06 | 1.59 | 1.83 | 1.67 | 1.13 | .58 | .17 |
| D2S206 | 233.534 | −∞ | .23 | 1.17 | 1.29 | 1.00 | .58 | .20 |
Note.— The peak LOD score is shown in bold italics.
Marker location is based on the May 2004 human genome assembly (UCSC Genome Browser).
Analysis of the Candidate Region
We examined the region for known gene candidates using the UCSC Genome Browser, as well as using 20-kb chunks of genomic BAC sequence in BLASTX searches (see the BLAST Web site) of GenBank, to identify additional conserved sequences. The candidate region contained genes for the insulin-receptor substrate (IRS1), collagens 4A3 and 4A4 (COL4A3 and COL4A4), a nucleoporin family member (HRB), chemokine ligand 20 (CCL20), several anonymous cDNAs, and SLC19A3. We first focused on the latter, since it is related to THTR-1 (encoded by SLC19A2), the high-affinity transporter for thiamin, another water-soluble vitamin. Mutations in SLC19A2 underlie thiamin-responsive megaloblastic anemia (or “Rogers syndrome”) (Diaz et al. 1999; Fleming et al. 1999; Labay et al. 1999).
SLC19A3 is predicted to comprise seven exons spanning 32.9 kb in 2q36.3, with the coding sequence confined to exons 2–6 (fig. 2). To ensure that we identified all SLC19A3-related potential coding sequences in the vicinity, we used the THTR-1 protein sequence and the predicted SLC19A3 coding sequence to search the candidate region with TBLASTN (see the BLAST Web site). This search identified another set of related sequences, which begin ∼40 kb 3′ to the gene and correspond to sequences related to exons 3, 4, and 5; we designated these sequences as “alt3,” “alt4,” and “alt5,” respectively. The latter two represent ORFs encoding amino acids that are 68% identical to those encoded by the equivalent segment of SLC19A3. However, alt3 predicts 71% identity but has two separate stop codons that interrupt the ORF. No reported human ESTs connect alt3, alt4, and alt5 in a cDNA. However, the cDNA DKFZp547H025 has been reported to encode a hypothetical protein of 140 aa that begins from the ORF of alt5 and continues into two additional exons with ORFs unrelated to SLC19A3. We designated these putative coding exons “alt6′” and “alt7′.” Since SLC19A3 and the other potential exons are all in the same orientation, it is conceivable that alternative splicing could produce a hybrid transcript. Consequently, to search for the BBGD defect, we performed mutation scanning of exons 1–6 of SLC19A3, as well as alt3, alt4, alt5, alt6′, and alt7′.
Mutations in SLC19A3
Two putative mutations were identified in the four families, and both of these mutations predicted missense changes in the protein structure, as shown in figure 4. The mutation in exon 2 was found in two patients (1203 and 1403) from a Yemeni family. Sequence analysis disclosed a homozygous G→T change at nucleotide 68, which is predicted to alter a highly conserved glycine at codon 23 to valine (c.68G→T; p.G23V) (fig. 2). An equivalent glycine residue is present at this position in mouse Slc19a3 and in both SLC19A2 and SLC19A1, the two other members of this transporter family in humans. This putative mutation is predicted to alter the first transmembrane domain (fig. 3) of the SLC19A3 protein.
Figure 4.
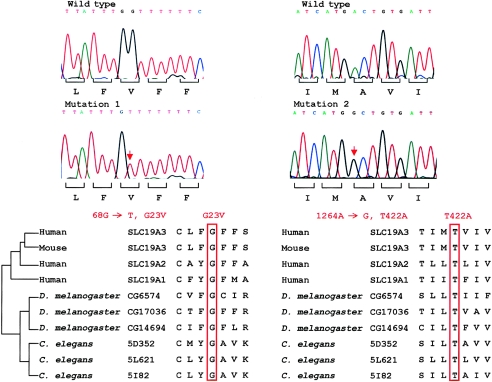
SLC19A3 point mutations at sites conserved in the reduced-folate transporter family. Sequence chromatograms of the two missense mutations and the corresponding wild-type alleles are shown in the upper panel. These mutations altered glycine (“Mutation 1”) and threonine (“Mutation 2”) residues at codon 23 and codon 422, respectively, that are highly conserved in related transporters in humans, mouse, and lower organisms (lower panel).
In the three other families, all of Saudi origin, we identified an A→G missense mutation at nucleotide position 1264 in exon 5, that alters a BspHI restriction site (fig. 4). This sequence change predicts a substitution of the threonine at codon 422 of the SLC19A3 protein with alanine (c.1264A→G, p.T422A) (fig. 2). The equivalent threonine is also present in the penultimate transmembrane domain of the SLC19A1- and SLC19A2-encoded proteins (fig. 3).
BLASTP analysis (see the BLAST Web site) of Drosophila melanogaster and Caenorhabditis elegans proteins with any of the human SLC19A1, SLC19A2, or SLC19A3 proteins detects three related genes in each species: CG6574, CG14694, and CG17036 in D. melanogaster and 5D352, 5L621, and 5I82 in C. elegans. These appear to be the only three members of the reduced-folate carrier transporter family in each organism, and they are comparable to the three genes found in mammals. However, since the transporters show greater sequence identity within a species than across species, it is not possible to conclusively assign an orthologue for each of the human tranporters by sequence comparison. For example, the SLC19A3 protein shows 33%, 33%, and 35% identity with the three Drosophila proteins, respectively, whereas the SLC19A2 protein shows 35%, 34%, and 34% identity with these same proteins. Interestingly, however, all three members of this gene family, in both D. melanogaster and C. elegans, display conserved glycine and threonine residues at positions comparable to the sites in SLC19A3 that are altered in the families with BBGD (fig. 4).
To establish that these two alterations do not represent benign polymorphisms, we examined 364 chromosomes from unrelated whites in the extended CEPH collection (Fondation Jean Dausset CEPH Web Server) and 124 chromosomes from unrelated individuals of Arab ancestry. None of these 488 control chromosomes possessed either alteration, arguing strongly that these missense alterations cause BBGD in these families. Unfortunately, since we do not have access to cell lines from these subjects, we have not been able to determine whether these mutations have any effect on SLC19A3 mRNA splicing in addition to their direct effect on the protein sequence.
Discussion
SLC19A1, SLC19A2, and SLC19A3 are genes on chromosomes 21q22.3, 1q24.2, and 2q36.3, respectively, encoding closely related proteins, termed the “reduced-folate carrier family of transporters” on the basis of the function of the SLC19A1 protein. The SLC19A2 protein is a thiamin transporter that is defective in thiamin-responsive megaloblastic anemia (Rogers syndrome) (Diaz et al. 1999; Fleming et al. 1999; Labay et al. 1999). The SLC19A3 protein shares 39% identity and 56% similarity with SLC19A1 and 48% identity and 64% similarity with SLC19A2, including a single reduced-folate carrier–related domain (aa 11–439) (Eudy et al. 2000). Hydrophobicity analysis of all three proteins reveals a predicted topology with 12 putative transmembrane domains (fig. 3), which is consistent with a transporter function. Northern blot and RT-PCR analyses have showed the presence of 3.5-, 2.6-, and 2.0-kb SLC19A3 transcripts in many tissues, including brain, but with greatest abundance in placenta, kidney, and liver. Interestingly, in the mouse, a single Slc19a3 mRNA species is seen and is relatively more abundant in the mouse brain than has been reported for humans (Eudy et al. 2000). Our RT-PCR analysis of the SLC19A3 transcript indicates that both exon 2 and exon 5 are expressed in the transcript in both brain and peripheral tissues (data not shown), suggesting that the brain-specific pathology of BBGD is not due to alternative splicing in peripheral tissues and that the differences in mRNA size detected by northern blot are not due to differences in the coding region. Refined expression studies in human intestinal epithelial cells have shown SLC19A3 protein to be expressed at the brush-border membrane but not at the basolateral membrane (Said et al. 2004), consistent with a transporter function.
Our detection—in four families with BBGD, representing two different disease haplotypes—of two different missense changes that segregate with the disorder but are not found on 488 control chromosomes implicates SLC19A3 as the disease gene. Furthermore, these mutations alter amino acid residues that are identical in all of the similar transporters in humans, mouse, and lower organisms, suggesting a strong functional pressure for evolutionary conservation. The c.68G→T mutation (p.G23V), found in a family of Yemeni origin, substitutes a hydrophobic valine residue for a more flexible glycine residue in the first transmembrane domain (fig. 3). A missense mutation that changes glycine to aspartic acid at codon 172 of SLC19A2 has also been reported in patients with Rogers syndrome (Labay et al. 1999; Raz et al. 2000), and two recent studies showed that the mutant protein failed in normal-cell targeting to the plasma membrane, with consequent loss of the thiamin transporter activity (Balamurugan and Said 2002; Baron et al. 2002). Indeed, the SLC19A3 G23V mutations and the clustering of Rogers syndrome mutations in exon 2 of SLC19A2 support the view that this exon encodes a region that is critical for successful transport function (Diaz et al. 1999; Fleming et al. 1999; Labay et al. 1999; Raz et al. 2000; Scharfe et al. 2000; Gritli et al. 2001; Lagarde et al. 2004). The other BBGD mutation, c.1264A→G, found in three families of Saudi origin, also alters a transmembrane domain, substituting an alanine residue for the hydrophilic threonine at position 422 (p.T422A). Again, the extreme evolutionary conservation of this threonine residue suggests that it is critical for SLC19A3 function.
Since the symptoms of BBGD may be reversed by high doses of biotin therapy, it is reasonable to suspect that SLC19A3 may act normally as a biotin transporter. Humans lack the ability to synthesize biotin and therefore depend entirely on dietary sources (Combs 1992). Biotinidase can recycle endogenous biotin so that most of the biotin can be preserved in the body, but biotinidase activity is low in human brain and central spinal fluid, restricting the recycling of biotin in the CNS (Suchy et al. 1985; Fois et al. 1986). Normal carboxylase and biotinidase values in patients with BBGD are consistent with the disorder being caused by a defect in the biotin transporter system, possibly across cerebral capillaries at the blood-brain barrier or, alternatively, in the neuronal cells themselves (Ozand et al. 1998). The caudate neurodegeneration evident in patients with BBGD suggests that striatal neurons may be particularly susceptible to a lack of adequate biotin. The high dosage of biotin used to treat BBGD may then act simply by increasing passive diffusion, making sufficient biotin available to the caudate nucleus (Spector and Mock 1987). Recently, investigation of humans with induced marginal biotin deficiency has shown a much greater reduction in levels of SLC19A3 mRNA in blood leukocytes than of mRNAs for 10 known biotin-related genes (Vlasova et al. 2005). These findings implicate SLC19A3 in biotin metabolism and indicate that it is a more sensitive indicator of biotin deficiency than the known biotin carboxylases, holocarboxylase synthetase, biotinidase, and the sodium-dependent multivitamin transporter.
In seeming contradiction with this hypothesis, SLC19A3 has recently been reported to be a second thiamin transporter (Rajgopal et al. 2001), like SLC19A2. These investigators used HeLa cells that were transiently transfected with SLC19A3 cDNA to assess the transport activity for radiolabeled thiamin, folic acid, and methotrexate, but they did not test biotin. To date, the genes encoding the putative pyridoxine (vitamin B6) and niacin transporters also have not been identified. An uptake assay to reveal a novel transporter function should include those micronutrients to also test for the possibility of multivitamin transport via the same protein. Interestingly, Na+-dependent multivitamin transporter, the intestinal biotin transporter, is also used by two other micronutrients, pantothenic acid and lipoate (Prasad et al. 1998, 1999; Wang et al. 1999). Additional work is required to establish whether SLC19A3 is capable of transporting biotin in addition to thiamin or, alternatively, whether the positive effect of biotin therapy in BBGD acts indirectly to rescue the effects of altered SLC19A3 transport of thiamin or another micronutrient.
In either of the above circumstances, explaining the brain-specific effects in patients with BBGD may require a fuller delineation and understanding of the options for biotin and thiamin transport, of the relative cellular distributions of the various transporters, of the specific requirements of different cells and tissues for these vitamins, and of the specific roles of biotin in different cells. Notably, global biotin deficiency produces many symptoms not seen in patients with BBGD, such as dry skin, seborrheic dermatitis, fungal infections, erythematous periorifacial macular rashes, nausea, and anorexia. The absence of these symptoms in patients with BBGD suggests that sufficient biotin is available in all regions except the brain. Moreover, the brain MRI findings in patients with biotinidase deficiency and those with multiple carboxylase deficiency are also different from the findings observed in patients with BBGD, implying that biotin may have other potential roles in the striatum beyond that of cofactor for carboxylases (McMahon 2002). Interestingly, several studies suggest that biotin may be involved directly or indirectly in the regulation of gene expression (Rodriguez-Melendez and Zempleni 2003), raising another option for the mechanism by which SLC19A3 mutation may lead to the basal ganglia necrosis of BBGD.
As a result of the wider use of advanced brain imaging technologies such as MRI and CT, bilateral striatal necrosis has been recognized in a number of disorders (Fujita et al. 1994; Okiyama et al. 1994; Sola Martinez et al. 1994). Extrapyramidal symptoms are the main clinical correlates of striatal necrosis and can be observed in mitochondrial encephalopathies (Martin et al. 1988; Medina et al. 1990; Leuzzi et al. 1992; Thyagarajan et al. 1995), glutaric acidemia type I, 3-methylglutaconic academia (Gascon et al. 1994), Huntington disease (Finch 1980; Buck et al. 1981), Wilson disease (Sener 1993), and CNS infection (Larsen and Crisp 1996; Zambrino et al. 2000). Detailed clinical evidence excluded those diagnoses in patients with BBGD who were described elsewhere (Ozand et al. 1998). However, since the description of BBGD, this pathology has been described in patients with a novel disorder, infantile bilateral striatal necrosis (IBSN), in which symmetric degeneration of the caudate and putamen was observed. Interestingly, three patients were reported to have moderate response to biotin (Straussberg et al. 2002). In one Israeli Bedouin kindred showing an autosomal recessive inheritance pattern, linkage analysis refined the critical candidate region on 19q13 (Basel-Vanagaite et al. 2004). In view of the similarity of IBSN to BBGD, we specifically tested the IBSN locus–flanking markers D19S596 and D19S867, to examine the possible involvement of this locus in BBGD. Linkage of BBGD to 19q13 was specifically excluded, indicating that these similar disorders are not allelic. However, careful comparison of these disorders and their degree of response to biotin may reveal new information concerning the role that biotin plays in the striatum. Similarly, the mechanism of striatal necrosis in IBSN and BBGD may indicate that an assessment of the potential participation of biotin metabolism in other neurodegenerative disorders of the striatum, such as Huntington disease, should be considered.
Although we identified the BBGD gene using Arab kindreds, it is likely that this disorder exists in other populations but that it may not have been recognized because it has only recently been formally described. Identification of mutations in the SLC19A3 gene will help to differentiate and confirm the clinical diagnosis of BBGD and will open an avenue for prenatal and presymptomatic diagnosis. Early molecular diagnosis of BBGD is particularly important to prevent basal ganglia damage, since the progression of the pathology and of the clinical course of symptoms can be prevented simply by the administration of biotin. It is important to assess whether patients with phenotypic similarilites to those with BBGD are known within the neurogenetics community in the United States and elsewhere, since identification of such patients will permit their effective treatment and will prevent unnecessary debilitation and premature death.
Acknowledgments
We are grateful to the families with BBGD who participated in this study, as well as to the clinicians at the King Faisal Specialist Hospital and Research Centre. We would also like to thank Jianmin Chen and the Massachusetts General Hospital Genomics Core Facility, for their assistance in genotyping services; Dr. Tony Castelli, for technical advice; and Dr. Mei Sun, for help with northern blot. This work was supported by the King Faisal Specialist Hospital and Research Centre, National Institutes of Health grant NS16367, and the Huntington’s Disease Society of America’s Coalition for the Cure. W.Q.Z. was the recipient of a fellowship from the Harvard Center for Neurodegeneration and Repair.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/blast/ (for BLASTX, TBLASTN, and BLASTP)
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for nine novel SNPs in the 5′ UTR, alt3, alt6′, and alt7′ of SLC19A3 and DKFZp547H025 [accession numbers 32476166–32476174])
- Fondation Jean Dausset CEPH Web Server, http://www.cephb.fr/ (for CEPH family genotypes used in map construction)
- HMMTOP 2.0, http://www.enzim.hu/hmmtop/index.html
- Human Genome Database, http://www.gdb.org
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for DKFZp547H025 mRNA [accession number NM_020161])
- NCBI Entrez Protein Database, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=protein&cmd=search&term= (for SLC19A3 [accession number NP_079519])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BBGD) [PubMed]
- Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- Aoki Y, Suzuki Y, Sakamoto O, Li X, Takahashi K, Ohtake A, Sakuta R, Ohura T, Miyabayashi S, Narisawa K (1995) Molecular analysis of holocarboxylase synthetase deficiency: a missense mutation and a single base deletion are predominant in Japanese patients. Biochim Biophys Acta 1272:168–174 [DOI] [PubMed] [Google Scholar]
- Balamurugan K, Said HM (2002) Functional role of specific amino acid residues in human thiamine transporter SLC19A2: mutational analysis. Am J Physiol Gastrointest Liver Physiol 283:G37–G43 [DOI] [PubMed] [Google Scholar]
- Baron D, Assaraf YG, Cohen N, Aronheim A (2002) Lack of plasma membrane targeting of a G172D mutant thiamine transporter derived from Rogers syndrome family. Mol Med 8:462–474 [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Straussberg R, Ovadia H, Kaplan A, Magal N, Shorer Z, Shalev H, Walsh C, Shohat M (2004) Infantile bilateral striatal necrosis maps to chromosome 19q. Neurology 62:87–90 [DOI] [PubMed] [Google Scholar]
- Buck SH, Burks TF, Brown MR, Yamamura HI (1981) Reduction in basal ganglia and substantia nigra substance P levels in Huntington’s disease. Brain Res 209:464–469 [DOI] [PubMed] [Google Scholar]
- Combs GF (1992) Biotin. In: The vitamins: rundamental aspects in nutrition and health. Academic, San Diego, pp 329–343 [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Diaz GA, Banikazemi M, Oishi K, Desnick RJ, Gelb BD (1999) Mutations in a new gene encoding a thiamine transporter cause thiamine-responsive megaloblastic anaemia syndrome. Nat Genet 22:309–312 [DOI] [PubMed] [Google Scholar]
- Dupuis L, Leon-Del-Rio A, Leclerc D, Campeau E, Sweetman L, Saudubray JM, Herman G, Gibson KM, Gravel RA (1996) Clustering of mutations in the biotin-binding region of holocarboxylase synthetase in biotin-responsive multiple carboxylase deficiency. Hum Mol Genet 5:1011–1016 [DOI] [PubMed] [Google Scholar]
- Eudy JD, Spiegelstein O, Barber RC, Wlodarczyk BJ, Talbot J, Finnell RH (2000) Identification and characterization of the human and mouse SLC19A3 gene: a novel member of the reduced folate family of micronutrient transporter genes. Mol Genet Metab 71:581–590 [DOI] [PubMed] [Google Scholar]
- Finch CE (1980) The relationships of aging changes in the basal ganglia to manifestations of Huntington’s chorea. Ann Neurol 7:406–411 [DOI] [PubMed] [Google Scholar]
- Fleming JC, Tartaglini E, Steinkamp MP, Schorderet DF, Cohen N, Neufeld EJ (1999) The gene mutated in thiamine-responsive anaemia with diabetes and deafness (TRMA) encodes a functional thiamine transporter. Nat Genet 22:305–308 [DOI] [PubMed] [Google Scholar]
- Fois A, Cioni M, Balestri P, Bartalini G, Baumgartner R, Bachmann C (1986) Biotinidase deficiency: metabolites in CSF. J Inherit Metab Dis 9:284–285 [DOI] [PubMed] [Google Scholar]
- Fujita K, Takeuchi Y, Nishimura A, Takada H, Sawada T (1994) Serial MRI in infantile bilateral striatal necrosis. Pediatr Neurol 10:157–160 [DOI] [PubMed] [Google Scholar]
- Gascon GG, Ozand PT, Brismar J (1994) Movement disorders in childhood organic acidurias: clinical, neuroimaging, and biochemical correlations. Brain Dev Suppl 16:94–103 [DOI] [PubMed] [Google Scholar]
- Gritli S, Omar S, Tartaglini E, Guannouni S, Fleming JC, Steinkamp MP, Berul CI, Hafsia R, Jilani SB, Belhani A, Hamdi M, Neufeld EJ (2001) A novel mutation in the SLC19A2 gene in a Tunisian family with thiamine-responsive megaloblastic anaemia, diabetes and deafness syndrome. Br J Haematol 113:508–513 [DOI] [PubMed] [Google Scholar]
- Labay V, Raz T, Baron D, Mandel H, Williams H, Barrett T, Szargel R, McDonald L, Shalata A, Nosaka K, Gregory S, Cohen N (1999) Mutations in SLC19A2 cause thiamine-responsive megaloblastic anaemia associated with diabetes mellitus and deafness. Nat Genet 22:300–304 [DOI] [PubMed] [Google Scholar]
- Lagarde WH, Underwood LE, Moats-Staats BM, Calikoglu AS (2004) Novel mutation in the SLC19A2 gene in an African-American female with thiamine-responsive megaloblastic anemia syndrome. Am J Med Genet A 125:299–305 [DOI] [PubMed] [Google Scholar]
- Larsen PD, Crisp D (1996) Acute bilateral striatal necrosis associated with Mycoplasma pneumoniae infection. Pediatr Infect Dis J 15:1124–1126 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, White RL (1986) Construction of human linkage maps: likelihood calculations for multilocus linkage analysis. Genet Epidemiol 3:39–52 [DOI] [PubMed] [Google Scholar]
- Leuzzi V, Bertini E, De Negri AM, Gallucci M, Garavaglia B (1992) Bilateral striatal necrosis, dystonia and optic atrophy in two siblings. J Neurol Neurosurg Psychiatry 55:16–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JJ, Van de Vyver FL, Scholte HR, Roodhooft AM, Ceuterick C, Martin L, Luyt-Houwen IE (1988) Defect in succinate oxidation by isolated muscle mitochondria in a patient with symmetrical lesions in the basal ganglia. J Neurol Sci 84:189–200 [DOI] [PubMed] [Google Scholar]
- McMahon RJ (2002) Biotin in metabolism and molecular biology. Annu Rev Nutr 22:221–239 [DOI] [PubMed] [Google Scholar]
- Medina L, Chi TL, DeVivo DC, Hilal SK (1990) MR findings in patients with subacute necrotizing encephalomyelopathy (Leigh syndrome): correlation with biochemical defect. Am J Neuroradiol 11:379–384 [PMC free article] [PubMed] [Google Scholar]
- Moss J, Lane MD (1971) The biotin-dependent enzymes. Adv Enzymol Relat Areas Mol Biol 35:321–442 [DOI] [PubMed] [Google Scholar]
- Narisawa K, Suzuki Y, Aoki Y (1996) [Cloning of the holocarboxylase synthetase cDNA and identification of mutations prevalent in Japanese HCS-deficient patients.] Nippon Rinsho 54:259–267 [PubMed] [Google Scholar]
- Okiyama R, Mitani M, Hayashi H, Tanabe H (1994) [Acute encephalopathy with symmetrical lesions of the thalamus, the putamen and the cerebellum on magnetic resonance imaging.] No To Shinkei 46:579–583 [PubMed] [Google Scholar]
- Ozand PT, Gascon GG, Al Essa M, Joshi S, Al Jishi E, Bakheet S, Al Watban J, Al-Kawi MZ, Dabbagh O (1998) Biotin-responsive basal ganglia disease: a novel entity. Brain 121:1267–1279 [DOI] [PubMed] [Google Scholar]
- Pomponio RJ, Reynolds TR, Cole H, Buck GA, Wolf B (1995) Mutational hotspot in the human biotinidase gene causes profound biotinidase deficiency. Nat Genet 11:96–98 [DOI] [PubMed] [Google Scholar]
- Prasad PD, Wang H, Huang W, Fei YJ, Leibach FH, Devoe LD, Ganapathy V (1999) Molecular and functional characterization of the intestinal Na+-dependent multivitamin transporter. Arch Biochem Biophys 366:95–106 [DOI] [PubMed] [Google Scholar]
- Prasad PD, Wang H, Kekuda R, Fujita T, Fei YJ, Devoe LD, Leibach FH, Ganapathy V (1998) Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J Biol Chem 273:7501–7506 [DOI] [PubMed] [Google Scholar]
- Rajgopal A, Edmondnson A, Goldman ID, Zhao R (2001) SLC19A3 encodes a second thiamine transporter ThTr2. Biochim Biophys Acta 1537:175–178 [DOI] [PubMed] [Google Scholar]
- Raz T, Labay V, Baron D, Szargel R, Anbinder Y, Barrett T, Rabl W, Viana MB, Mandel H, Baruchel A, Cayuela JM, Cohen N (2000) The spectrum of mutations, including four novel ones, in the thiamine-responsive megaloblastic anemia gene SLC19A2 of eight families. Hum Mutat 16:37–42 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Melendez R, Zempleni J (2003) Regulation of gene expression by biotin (review). J Nutr Biochem 14:680–690 [DOI] [PubMed] [Google Scholar]
- Said HM, Balamurugan K, Subramanian VS, Marchant JS (2004) Expression and functional contribution of hTHTR-2 in thiamin absorption in human intestine. Am J Physiol Gastrointest Liver Physiol 286:G491–G498 [DOI] [PubMed] [Google Scholar]
- Schaffer AA (1996) Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered 46:226–235 [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Scharfe C, Hauschild M, Klopstock T, Janssen AJ, Heidemann PH, Meitinger T, Jaksch M (2000) A novel mutation in the thiamine responsive megaloblastic anaemia gene SLC19A2 in a patient with deficiency of respiratory chain complex I. J Med Genet 37:669–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sener RN (1993) Wilson’s disease: MRI demonstration of cavitations in basal ganglia and thalami. Pediatr Radiol 23:157 [DOI] [PubMed] [Google Scholar]
- Sola Martinez MT, Pierot L, Noseda G, Martin-Duverneuil N, Harpey JP, Roy C, Chiras J (1994) Acute bilateral striatal necrosis in an infant: CT and MRI. Neuroradiology 36:245–246 [DOI] [PubMed] [Google Scholar]
- Spector R, Mock D (1987) Biotin transport through the blood-brain barrier. J Neurochem 48:400–404 [DOI] [PubMed] [Google Scholar]
- Straussberg R, Shorer Z, Weitz R, Basel L, Kornreich L, Corie CI, Harel L, Djaldetti R, Amir J (2002) Familial infantile bilateral striatal necrosis: clinical features and response to biotin treatment. Neurology 59:983–989 [DOI] [PubMed] [Google Scholar]
- Suchy SF, McVoy JS, Wolf B (1985) Neurologic symptoms of biotinidase deficiency: possible explanation. Neurology 35:1510–1511 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Aoki Y, Ishida Y, Chiba Y, Iwamatsu A, Kishino T, Niikawa N, Matsubara Y, Narisawa K (1994) Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat Genet 8:122–128 [DOI] [PubMed] [Google Scholar]
- Thyagarajan D, Shanske S, Vazquez-Memije M, De Vivo D, DiMauro S (1995) A novel mitochondrial ATPase 6 point mutation in familial bilateral striatal necrosis. Ann Neurol 38:468–472 [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (1998) Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 283:489–506 [DOI] [PubMed] [Google Scholar]
- ——— (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850 [DOI] [PubMed] [Google Scholar]
- Vlasova TI, Stratton SL, Wells AM, Mock NI, Mock DM (2005) Biotin deficiency reduces expression of SLC19A3, a potential biotin transporter, in leukocytes from human blood. J Nutr 135:42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang W, Fei YJ, Xia H, Yang-Feng TL, Leibach FH, Devoe LD, Ganapathy V, Prasad PD (1999) Human placental Na+-dependent multivitamin transporter: cloning, functional expression, gene structure, and chromosomal localization. J Biol Chem 274:14875–14883 [DOI] [PubMed] [Google Scholar]
- Wolf B, Feldman GL (1982) The biotin-dependent carboxylase deficiencies. Am J Hum Genet 34:699–716 [PMC free article] [PubMed] [Google Scholar]
- Zambrino CA, Zorzi G, Lanzi G, Uggetti C, Egitto MG (2000) Bilateral striatal necrosis associated with Mycoplasma pneumoniae infection in an adolescent: clinical and neuroradiologic follow up. Mov Disord 15:1023–1026 [DOI] [PubMed] [Google Scholar]