Abstract
The E2F family of transcriptional regulators consists of six different members. Analysis of E2F-regulated promoters by using cultured cells suggests that E2Fs may have redundant functions. However, animal studies have shown that loss of individual E2Fs can have distinct biological consequences. Such seemingly conflicting results could be due to a difference in E2F-mediated regulation in cell culture vs. animals. Alternatively, there may be genes that are specifically regulated by an individual E2F which have not yet been identified. To investigate this possibility further, we have analyzed gene expression in E2F1 nullizygous mice. We found that loss of E2F1 did not cause changes in expression of known E2F target genes, suggesting that perhaps E2F1-specific promoters are distinct from known E2F target promoters. Therefore, we used oligonucleotide microarrays to identify mRNAs whose expression is altered on loss of E2F1. We demonstrate by chromatin immunoprecipitation that several of the promoters that drive expression of the deregulated mRNAs selectively recruit E2F1, but not other E2Fs, and this recruitment is via an element distinct from a consensus E2F binding site. To our knowledge, these are as yet undocumented examples of promoters being occupied in asynchronously growing cells by a single E2F family member. Interestingly, the E2F1-specific target genes that we identified encode proteins having functions quite different from the function of known E2F target genes. Thus, whereas E2F1 may share redundant functions in cell growth control with other E2F family members, it may also play an important biological role distinct from the other E2Fs.
The E2F family of transcription factors is thought to be a key regulator of cell growth control. To date, six different E2Fs have been identified, E2F1 to -6, each of which can heterodimerize with DP1 or DP2 to form 12 different DNA binding transcriptional regulators (1–3). Members of the E2F family bind to and regulate the promoters for genes whose products are important for cell cycle progression and DNA synthesis. In almost all cases, multiple different E2Fs have been shown to activate these promoters (4). In addition to the several dozen cell cycle-dependent promoters whose regulation by E2F has been well-characterized, recent studies have suggested that a much larger set of promoters may be E2F targets. In one study in which mRNA from cells harboring overexpressed E2F1, E2F2, or E2F3 was analyzed by using cDNA microarrays, the authors report that 1,240 of the 19,000 mRNAs (7%) were altered in expression (5). However, in almost every case, if expression of an mRNA was altered by overexpression of one E2F family member, it was also altered by overexpression of another family member. The authors state that the few genes that appeared to respond selectively to one family member in the microarray analysis did not show the same selectivity when analyzed by Northern blots. Similarly, a second study identified ≈60 mRNAs that were altered on overexpression of E2F1 or E2F2. All 60 mRNAs responded similarly to E2F1 and E2F2 (6). Finally, we and others have shown that all known or suspected E2F target promoters are bound by multiple E2Fs in vivo (7–10).
Taken together, the experiments described above would seem to suggest that there is very little target gene specificity among the E2F family members. However, loss of E2F1 does have biological consequences. For example, loss of E2F1 can lead to tumor development in the reproductive tract, lung, and lymphatic system (11, 12). Also, overexpression of E2F1 can enhance neoplasia in the skin of transgenic mice (13) and can cause dysplasia and tumor formation in the liver of transgenic mice (14). These animal model studies suggest that there may be genes that are more dependent and/or sensitive to the effects of E2F1 than to other E2F family members. Therefore, we have undertaken an investigation focused on the identification of E2F1-specific target genes.
Materials and Methods
Mouse Husbandry.
C3H/HeJ and C57BL/6J [wild-type (wt) and E2F1 nullizygous animals] were housed in the McArdle Laboratory Animal Facilities in plastic cages on corncob bedding from Bed-O'Cobs (Anderson Cob Division, Maumee, OH) and fed Breder Blox (Harlan, Madison, WI). Food and acidified water were available ad libitum. Newborn mice, 14-day embryos, and adult mice were used, as indicated for each experiment, to obtain the tissues needed for RNA analysis and chromatin immunoprecipitation experiments. After sacrifice, the tissues were immediately frozen in liquid nitrogen and stored at −70°C.
Analysis of RNA.
Total RNA was extracted from the liver by the guanidine thiocyanate/CsCl method, as described previously (15). Reverse transcription–PCR analysis was performed as described previously (16); the information needed to design the primers used to analyze the dihydrofolate reductase (dhfr), thymidine kinase (TK), cyclin E, and retinoblastoma (Rb) promoters can be found at our web site, http://mcardle.oncology.wisc.edu/farnham/. All primers were synthesized at the University of Wisconsin Biotechnology Center DNA Synthesis Facility.
Oligonucleotide Microarrays.
Total RNA was extracted from liver by using guanidine thiocyanate/CsCl as described previously (15). For the oligonucleotide microarrays, mRNA was purified twice by using Oligotex mRNA Kit (Qiagen, Chatsworth, CA) and electrophoresed on a 1% agarose/1× MAE buffer [50% formamide/2.2 M formaldehyde/1 mM 4-morpholinopropanesulfonic acid (Mops), pH 7.0/0.4 M NaOAc/0.05 mM EDTA] gel to examine for degradation. For oligonucleotide microarray experiments, poly(A) RNA samples were pooled from eight newborn livers from either wt or E2F1 nullizygous mice. A complete protocol for converting RNA into “target” for hybridization to microarrays is available at our web site. In brief, twice-purified poly(A) RNA from quiescent, regenerating, and newborn livers, or liver tumors was used to create cDNA with a T7-poly(T) primer and reverse transcriptase Superscript II (Invitrogen). Approximately 1 μg of cDNA was subjected to in vitro transcription (Ambion, Austin, TX) in the presence of biotinylated UTP and CTP (Enzo Diagnostics). The cRNA was fragmented and combined with BSA (0.5 mg/ml) in a buffer containing 2× Mes, 1.7 M NaCl, 40 mM EDTA, and 0.02% Tween 20. Target cRNA (10 μg) was hybridized for 16 h at 40°C to each oligonucleotide array (Mu6500 tetraset; Affymetrix, Santa Clara, CA) containing probes for more than 6,500 murine genes and expressed sequence tags. Arrays were washed in the Affymetrix Fluidics Station at 50°C with 6× SSPE-T (0.9M NaCl/60 mM Na2HPO4/6 mM EDTA/0.005% Triton X-100, pH 7.6) then at 40°C with 0.5× SSPE-T. Arrays were then stained with streptavidin phycoerythrin (Molecular Probes) and washed with 6× SSPE-T. Fluorescent intensities were measured with a laser confocal scanner (Hewlett-Packard) and analyzed with genechip software (Affymetrix).
Chromatin Immunoprecipitation.
The chromatin immunoprecipitation assay was performed as previously described (7–9) with modifications to allow use of the protocol with mouse embryos and tissues. A detailed protocol, containing these modifications and primer sequences, can be found at our web site.
Results
Loss of E2F1 Has Minor Effects on Expression of Known Target Genes.
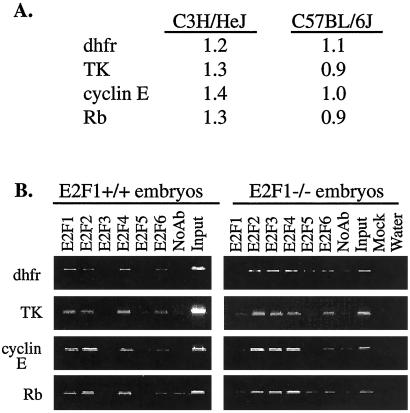
We have previously examined binding of E2F1 to -6 to a large set of known E2F target genes by using a chromatin immunoprecipitation assay (7–9). These previous studies have all shown that, in asynchronously growing cells, multiple E2Fs bind to each of the tested promoters. However, all of our previous studies, as well as those of other labs (10), have examined target gene specificity of the E2F family by using established cell lines. It remained possible that individual E2Fs would have distinct roles in the expression of target genes in the context of a living animal. For example, the expression of an E2F1-specific target gene might be affected in mice nullizygous for E2F1. Therefore, as a first approach in the identification of E2F1-specific target genes, we compared the levels of mRNA of several known E2F target genes in embryos from wt mice and mice nullizygous for E2F1. As shown in Fig. 1A, the expression of four well-characterized E2F target promoters was not affected by the loss of E2F1 in C3H/HeJ mice. To confirm these results, we also used mice in which the E2F1 null allele was transferred to the C57BL/6J background. Again, no difference in expression of dhfr, TK, cyclin E, or Rb mRNA was observed between the wt and E2F1 nullizygous mice.
Figure 1.
Loss of E2F1 has minor effects on expression of known target genes. (A) Reverse transcription–PCR was used to analyze the levels of dhfr, TK, cyclin E, and Rb mRNA in RNA prepared from the livers of C3H/HeJ and C57BL/6J wt vs. E2F1 nullizygous newborn mice. (B) Chromatin immunoprecipitations were performed from C57BL/6J wt and nullizygous E2F1 embryos using antibodies to E2F1 to -6, and a no antibody control. The chromatin was then analyzed by PCR using primers specific for the dhfr, TK, cyclin E, and Rb promoters. For this experiment, as well as those shown in Figs. 3 and 4, the amount of chromatin used in the PCR reaction of the input lane represents 0.4% of the starting material, and the amount of sample used in the PCR reaction of the immunoprecipitated lane represents 6.7% of the precipitated chromatin. Therefore, approximately equal signals in the sample and input lanes suggest that about 6% of the starting material was precipitated by the antibody.
It seemed likely that the expression of the E2F target genes was not influenced by loss of E2F1 because of the functional redundancy of the E2F family members on these promoters. To test this hypothesis, we adapted the chromatin immunoprecipitation protocol for use with 14-day mouse embryos. Briefly, this procedure required initial mincing of the embryos in PBS, crosslinking in formaldehyde for a longer time than used for tissue culture cells, and then processing with a Medi-machine to achieve single cells (details concerning the adaptation of the formaldehyde crosslinking and chromatin immunoprecipitation protocol for use with embryos and organs can be found at our web site). As shown in Fig. 1B, each of the tested E2F target promoters was bound by multiple E2Fs in the embryos from both wt and nullizygous mice. Therefore, functional redundancy of the E2F family members prevented the loss of E2F1 from causing a change in expression of these particular genes.
Loss of E2F1 Has Minor Effects on a Global Gene Expression Profile.
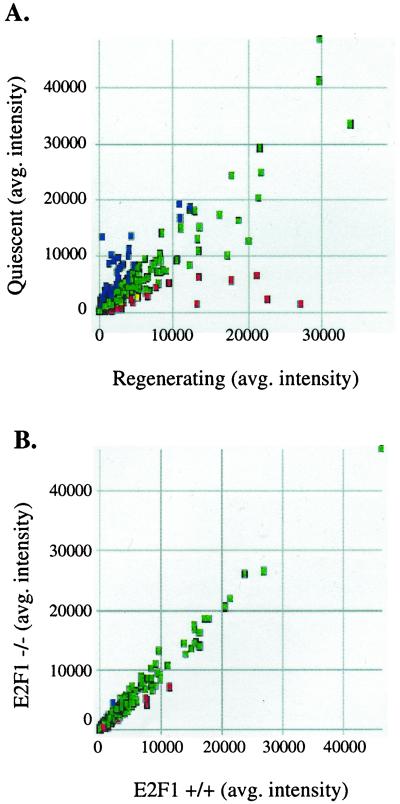
In addition to the genes shown in Fig. 1B, we have also tested the expression and promoter occupancy of additional known E2F target genes in wt vs. E2F1 nullizygous mice. In all cases, we found that the promoters were occupied by multiple E2Fs and that loss of E2F1 caused very little change in mRNA levels of the target genes (E. R. Lukas and J.W., unpublished data). However, because altered expression of E2F1 can result in neoplasia (11–14), it remained possible that there did exist E2F1-specific target promoters. Testing many different genes individually was not practical. Therefore, as a second approach, we used oligonucleotide microarrays. We prepared mRNA from the livers of newborn C3H/HeJ mice wt or nullizygous for E2F1 and used this mRNA to probe a series of Affymetrix GeneChips. As a control to ensure that we could detect changes in gene expression by using oligonucleotide microarrays, we compared gene expression in quiescent livers to regenerating livers. As expected, we observed many differences in the gene expression profile between the quiescent vs. regenerating liver (Fig. 2A). For example, mRNA levels of cell cycle-regulated genes such as cdc2, cyclin A, and cyclin B were 65-, 6-, and 5-fold higher, respectively, in the regenerating liver (the complete data set for this experiment, and for the comparison of wt to E2F1 nullizygous mice, can be found on our web site). In contrast, as shown in Fig. 2B, there are almost no differences in the overall gene expression profile of livers containing or lacking E2F1. This degree of identity between the mRNAs expressed in the two samples was surprising. However, it must be considered that only 6,500 mRNAs were represented on the microarray and that these did not necessarily correspond to the mRNAs that would be most responsive to loss of E2F1. Although few differences between wt and E2F1 nullizygous mice were detected, we did find that the expression of several genes was affected greater than 3-fold by the loss of E2F1 (Table 1). Interestingly, none of these genes are known E2F target genes nor do they encode proteins involved in cell cycle control or DNA synthesis. However, we do note that E2F1 has previously been shown to repress the plasminogen activator inhibitor-1 (PAI-1) gene in tissue culture overexpression assays (17). Thus, our studies that show an increase in PAI-1 in the absence of E2F1 and the previous overexpression studies both suggest that E2F1 may, directly or indirectly, repress the PAI-1 promoter.
Figure 2.
Loss of E2F1 has minor effects on expression of all genes. (A) mRNA was prepared from quiescent or regenerating livers from 6-week-old C3H/HeJ mice and used to probe murine Affymetrix DNA microarrays. The colored boxes represent the difference call for each gene in the Mu6500 tetraset made by the Affymetrix genechip software algorithm (green, no change; red, mRNAs that are higher in regenerating liver; blue, mRNAs that are higher in quiescent liver). (B) mRNA was prepared from the livers of newborn wt or E2F1 nullizygous C3H/HeJ mice and used to probe murine Affymetrix DNA microarrays. The colored boxes represent the difference call for each gene in the Mu6500 tetraset made by the Affymetrix genechip software algorithm (green, no change; red, mRNAs that are higher in wt liver; blue, mRNAs that are higher in null liver). A complete data set for this experiment can be found at our web site.
Table 1.
Table of genes deregulated by loss of E2F1 in C3H/HeJ mice
| Accession no. | Entrez definition | Fold change |
|---|---|---|
| mRNAs that decrease in the absence of E2F1 | ||
| L27121 | Mus musculus (10-1) hydroxysteroid sulfotransferase (mSTa2) mRNA, complete cds | 21.1 |
| M88694 | M. musculus thioether S-methyltransferase mRNA, complete cds | 9.7 |
| M77497 | M. musculus cytochrome P-450 naphthalene hydroxylase mRNA, complete cds | 4.4 |
| M77015 | Mouse 3-β-hydroxysteroid dehydrogenase/delta-5-δ-4-isomerase mRNA sequence | 3.9 |
| D00926 | Mouse mRNA for transcription factor S-II-related protein | 3.7 |
| D45850 | Mouse mRNA for estradiol 17-β-dehydrogenase (A-specific), complete cds | 3.4 |
| X83202 | M. musculus mRNA for 11-β-hydroxysteroid dehydrogenase/carbonyl reductase | 3.3 |
| M57960 | Mouse carboxylesterase mRNA, complete cds | 3.2 |
| X70398 | Mouse pentylenetetrazol-related mRNA PTZ-17 | 3.2 |
| L10106 | M. musculus protein tyrosine phosphate mRNA, complete cds | 3.1 |
| NM_019646 | M. musculus eukaryotic translation initiation factor 3, subunit 8 | 3.1 |
| mRNAs that increase in the absence of E2F1 | ||
| M12347 | Mouse mRNA for skeletal muscle α-actin | 3.4 |
| M73490 | M. musculus apolipoprotein E mRNA, 3′ end | 3.5 |
| M63660 | Mouse G-α-13 protein mRNA, complete cds | 3.5 |
| M34398 | M. musculus loricrin gene, complete cds | 3.8 |
| M33960 | Mouse PAI-1 mRNA, complete cds | 5.7 |
| NM_016908 | M. musculus synaptotagmin 5 (Syt5) mRNA | 6.4 |
Listed are the genes that showed the greatest response to the loss of E2F1 in the experiment shown in Fig. 2B. mRNAs that decrease in the absence of E2F1 are those which require E2F1 for high level expression; mRNAs that increase in the absence of E2F1 are those which are normally repressed by E2F1.
The Identification of E2F1-Specific Target Promoters.
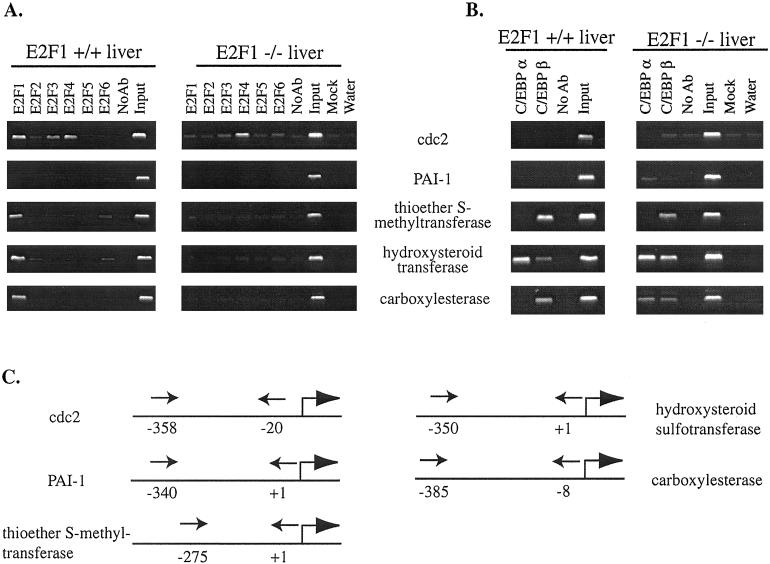
The analysis of mouse promoter regions remains difficult because of the incomplete nature of the mouse genome database. However, we were able to identify genomic DNA corresponding to the regions just upstream of the transcription start site of several of the genes listed in Table 1. Therefore, we performed chromatin immunoprecipitation experiments by using liver from wt C3H/HeJ mice and analyzed the promoters of the hydroxysteroid sulfotransferase, thioether-S-methyltransferase, carboxylesterase, and PAI-1 genes; promoter sequences were not available in the public database for the other identified mRNAs. As a control, we monitored binding of the E2Fs to the cdc2 promoter, a well-characterized E2F target gene. We found that the promoter regions of the hydroxysteroid sulfotransferase, thioether-S-methyltransferase, and carboxylesterase genes, all of which showed decreased mRNA levels on loss of E2F1, were in fact bound by E2F1 in vivo (Fig. 3A). Interestingly, these promoters were not bound by other E2F family members. A previous study has shown that E2F1-mediated repression of the PAI-1 promoter requires multiple elements located within several hundred bp of the start site of transcription (18). Our primer set would allow detection of E2F binding to these elements. However, in contrast to the other tested promoters, the PAI-1 promoter was not directly bound by any E2F family member, suggesting that the effects of loss of E2F1 on PAI-1 are indirect.
Figure 3.
The identification of E2F1-specific target promoters. For both A and B, the controls and amounts of chromatin used are as described in Fig. 1; the chromatin was analyzed by using primers shown in C. (A) A chromatin immunoprecipitation experiment was performed from liver isolated from adult C3H/HeJ wt or E2F1 nullizygous mice using antibodies to the E2F family members, as well as a no antibody control. (B) A chromatin immunoprecipitation experiment was performed from liver isolated from adult C3H/HeJ wt and E2F1 nullizygous mice using antibodies to C/EBPα and C/EBPβ, as well as a no antibody control. (C) Shown is a schematic of the E2F1-specific promoters and the approximate location of the primers (arrows) used in the chromatin immunoprecipitation experiments. The numbers are relative to +1 being the 5′ end of the mRNA (indicated by a bent arrow).
The E2F1-specific target promoters were identified because of the fact that the mRNAs driven by these promoters were decreased on loss of E2F1. We would thus predict that the other E2Fs cannot compensate for loss of E2F1 by increased binding to the promoter regions. To test this prediction, we performed chromatin immunoprecipitation experiments by using antibodies to E2F1 to -6 and liver from C3H/HeJ mice nullizygous for E2F1. As a control, we monitored the binding of E2Fs to the cdc2 promoter. As expected, we saw no E2F1 binding to promoters in the E2F1 nullizygous mice, indicating that the signal detected by the E2F1 antibody in the wt mice is indeed due to E2F1 binding to the identified promoters. Although we found that other E2F family members were bound to the cdc2 promoter in the livers of the E2F1 nullizygous mice, there were no E2F family members bound to the hydroxysteroid sulfotransferase, thioether-S-methyltransferase, or carboxylesterase promoters. We conclude that the loss of E2F1 is not compensated for by increased binding of other E2F family members to the sulfotransferase, thioether-S-methyltransferase, or carboxylesterase promoters, thus leading to a decreased expression of these mRNAs.
The promoters that are bound only by E2F1 and not by other E2Fs do not contain consensus E2F sites, suggesting that E2F1 is recruited to the promoter regions either by binding to a novel sequence element or by interaction with another DNA binding protein. All of the E2Fs have a very similar DNA binding domain, making it likely that any sequence bound directly by E2F1 would also be bound by the other E2Fs. Therefore, we have investigated the possibility that E2F1 is being recruited to certain promoters via protein–protein interactions. E2F1 has been reported to interact with two other DNA binding proteins, Sp1 (19, 20) and CCAAT/enhancer binding protein (C/EBP) α (21–23). A large number of promoters contain Sp1 sites, and we have previously shown that at least some of these promoters are not bound by E2F in cells (e.g., the cad promoter (24) and a dhfr promoter having a mutated E2F site (7). Also, the identified E2F1-specific target promoters are not GC rich and do not resemble typical Sp1-regulated promoters. However, we note that C/EBPα is a very important transcription factor in the liver (25, 26) and that the hydroxysteroid sulfotransferase, thioether-S-methyltransferase, and carboxylesterase promoters all have C/EBP consensus sites. Therefore, we have investigated whether E2F1 might be recruited to promoters via C/EBPα. We performed chromatin immunoprecipitation experiments by using livers of wt and E2F1 nullizygous mice and antibodies to C/EBPα and C/EBPβ. The cdc2 promoter does not contain a C/EBP binding site, and, as expected, the C/EBP family members tested did not bind to the promoter (Fig. 3B). Although we did observe C/EBPα binding to certain of the E2F1 target promoters (e.g., hydroxysteriod sulfotransferase), other E2F1 target promoters were not bound by C/EBPα in either the wt or E2F1 nullizygous mice. For example, in the thioether S-methyltransferase promoter, the C/EBP binding sites are occupied by C/EBPβ, not C/EBPα (C/EBPβ has never been shown to interact with E2F1). Thus, we conclude that, at least in certain cases, E2F1 must be recruited to the promoter via a mechanism distinct from interaction with C/EBPα.
Examination of the Tissue Specificity for Recruitment of E2F1 to Target Promoters.
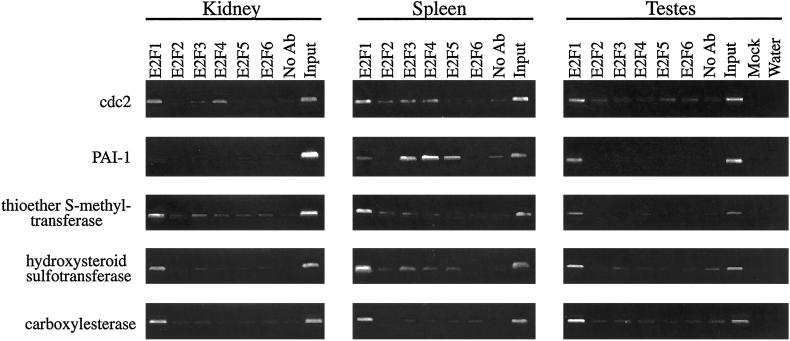
We identified the E2F1-specific target promoters by using the mouse liver as a model system. It was of interest to determine whether these promoters are E2F1 targets in other tissues. Therefore, we performed a chromatin immunoprecipitation experiment by using antibodies to the different E2F family members and chromatin derived from mouse kidney, spleen, and testis. Several interesting conclusions can be drawn from these experiments. First, the target promoters that are activated by E2F1 (hydroxysteroid sulfotransferase, thioether-S-methyltransferase, and carboxylesterase) are all bound by E2F1, but not by appreciable levels of the other E2Fs, in the kidney, spleen, and testes (Fig. 4) as well as in the liver (Fig. 3). Therefore, these genes are likely to be regulated by E2F1 in multiple tissues. Second, although E2F family members are not bound to the PAI-1 promoter in liver (Fig. 3A) or kidney (Fig. 4), E2Fs are bound to this promoter in the spleen and testes (Fig. 4). This result does raise the possibility that E2Fs may directly regulate the PAI-I promoter in certain tissues. Third, it is interesting to note that, in the testis, E2F1 is the predominant E2F on all promoters that we have examined using chromatin immunoprecipitation, including promoters having consensus E2F sites (e.g., cdc2). Previous studies of E2F1 nullizygous mice showed tumor development in reproductive organs and testicular atrophy (11, 12). Our results suggest that the severe effect of loss of E2F1 on the adult testes may be due to the fact that E2F1 plays a major role in transcriptional regulation of E2F target genes in that tissue.
Figure 4.
Analysis of the binding of E2F1 to target promoters in different tissues. A chromatin immunoprecipitation experiment was performed using the indicated tissues and antibodies to the different E2Fs. The chromatin was analyzed by using primers specific for the indicated promoters. The controls and amounts of chromatin used are as described in Fig. 1.
Discussion
We have performed a three-pronged approach toward the identification of E2F1-specific target genes. First, we examined several known E2F target promoters but found that, even in a living animal, multiple different E2Fs bound to each promoter analyzed. Second, we performed a microarray analysis by using mRNA from wt and E2F1 nullizygous mice and found that the mRNAs that were altered by the loss of E2F1 were not previously known to be regulated by the E2F family. Third, we showed that several of the promoters of the genes that we found to require E2F1 for high level expression were in fact bound by E2F1, but not other E2F family members, in vivo. Thus, the combined use of E2F1 nullizygous mice, oligonucleotide-based microarrays, and chromatin immunoprecipitation has allowed the identification of promoters bound specifically by E2F1. We realize that neither the microarray analysis or the chromatin immunoprecipitation assay provide direct evidence that E2F1 regulates the identified genes by changes in transcription rate. Direct evidence of an alteration in the transcription rate of a particular gene is quite difficult to obtain when using animal model systems. However, the fact that loss of E2F1 results in a decrease in the levels of the mRNAs, in combination with the observation that E2F1, but not other E2Fs, is bound to the proximal promoter regions, strongly suggests that we have identified genes that are directly regulated by E2F1.
To our knowledge, our studies provide as yet undocumented examples of promoters being occupied in vivo by E2F1, but not by the other E2Fs. Previous studies have shown that the E2F family typically regulates genes that are involved in cell cycle progression or DNA replication. Interestingly, the E2F1-specific target genes that we have now identified encode proteins having functions quite distinct from the function of most E2F target genes. Thus, whereas E2F1 may share redundant functions in cell growth control with other E2F family members, it may also play an important biological role distinct from other E2Fs. For example, thioether S-methyltransferase is an important enzyme in the metabolism of sulfur and selenium-containing compounds in animals. It is also involved in the biochemical detoxification of sulfur mustards (27). Hydroxysteroid sulfotransferase catalyzes the sulfation of diverse drugs, endogenous compounds, and xenobiotics. Although involved in detoxification reactions, this compound can also lead to the bioactivation of certain carcinogens (28). Carboxylesterase is also involved in the detoxification of foreign compounds and in the esterification of free cholesterol (29). Clearly, altered levels of these E2F1-specific target genes could affect an animal's response to toxic agents, and further investigations of the effects of loss of E2F1 on such responses are warranted.
Having shown that promoters do exist that are bound by E2F1, but not by other E2Fs, it is now important to expand our analysis to identify a larger set of such promoters and to determine whether E2F1 regulates different sets of promoters in different tissues. The promoters that we have identified do not have a consensus E2F binding site. Therefore, we propose that E2F1 is recruited to these promoters by other DNA binding proteins. Our studies suggest that simple in vitro DNA-protein analyses and/or inspection of a promoter for E2F consensus sites will not be adequate for the identification of genes regulated specifically by E2F1. It should be useful to develop high throughput screens for the identification of such promoters that are based on a combination of chromatin immunoprecipitation, genomic microarray analysis, and a battery of different tissues and cell types. On identification of a larger set of E2F1-specific promoters, DNA sequence analysis may reveal common elements that could be used to identify a novel E2F1-interacting protein.
Acknowledgments
We thank Tim Jatkoe for assistance with the microarray analysis and Zobeida Diaz and Sarah Harkins-Perry for assistance with mouse husbandry. We also acknowledge Erika Lukas for performing preliminary experiments that were critical for the success of this study, and we thank Lili Yamasaki for the original E2F1 nullizygous B6/129Sv mouse strain. This work was supported in part by research grants from the National Cancer Center/National Institutes of Health (CA22484 and CA07175), and the National Institute of General Medical Sciences/National Institutes of Health (GM08349).
Abbreviations
- dhfr
dihydrofolate reductase
- Rb
retinoblastoma
- TK
thymidine kinase
- C/EBP
CCAAT/enhancer binding protein
- PAI-1
plasminogen activator inhibitor-1
- wt
wild type
References
- 1.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 2.Farnham P J, Slansky J E, Kollmar R. Biochim Biophys Acta. 1993;1155:125–131. doi: 10.1016/0304-419x(93)90001-s. [DOI] [PubMed] [Google Scholar]
- 3.Slansky J E, Farnham P J. In: Transcriptional Control of Cell Growth: the E2F Gene Family. Farnham P J, editor. Vol. 208. New York: Springer; 1996. pp. 1–30. [Google Scholar]
- 4.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller H, Bracken A P, Vernell R, Moroni M C, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner J D, Helin K. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins J R. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells J, Boyd K E, Bartley S M, Farnham P J. Mol Cell Biol. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kel A E, Kel-Margoulis O V, Farnham P J, Bartley S M, Wingender E, Zhang M Q. J Mol Biol. 2001;309:99–120. doi: 10.1006/jmbi.2001.4650. [DOI] [PubMed] [Google Scholar]
- 9.Weinmann A S, Bartley S M, Zhang M Q, Zhang T, Farnham P J. Mol Cell Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi Y, Rayman J B, Dynlacht B D. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 11.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 12.Field S J, Tsai F-Y, Kuo F, Zubiaga A M, Kaelin W G, Jr, Livingston D M, Orkin S H, Greenberg M E. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 13.Pierce A M, Fisher S M, Conti D J, Johnson D G. Oncogene. 1998;16:1267–1276. doi: 10.1038/sj.onc.1201666. [DOI] [PubMed] [Google Scholar]
- 14.Conner E A, Lemmer E R, Omori M, Wirth P J, Factor V M, Thorgeirsson S S. Oncogene. 2000;19:5054–5062. doi: 10.1038/sj.onc.1203885. [DOI] [PubMed] [Google Scholar]
- 15.Lukas E R, Bartley S M, Graveel C R, Diaz Z M, Dyson N, Harlow E, Yamasaki L, Farnham P J. Mol Carcinog. 1999;25:295–303. doi: 10.1002/(sici)1098-2744(199908)25:4<295::aid-mc8>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 16.Graveel C R, Jatkoe T, Madore S J, Holt A L, Farnham P J. Oncogene. 2001;20:2704–2712. doi: 10.1038/sj.onc.1204391. [DOI] [PubMed] [Google Scholar]
- 17.Koziczak M, Krek W, Nagamine Y. Mol Cell Biol. 2000;20:2014–2022. doi: 10.1128/mcb.20.6.2014-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koziczak M, Muller H, Helin K, Nagamine Y. Eur J Biochem. 2001;268:4969–4978. doi: 10.1046/j.0014-2956.2001.02428.x. [DOI] [PubMed] [Google Scholar]
- 19.Karlseder J, Rotheneder H, Wintersberger E. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin S Y, Black A R, Kostic D, Pajovic S, Hoover C N, Azizkhan J C. Mol Cell Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timchenko N A, Wilde M, Darlington G J. Mol Cell Biol. 1999;19:2936–2945. doi: 10.1128/mcb.19.4.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slomiany B A, D'Arigo K L, Kelly M M, Kurtz D T. Mol Cell Biol. 2000;20:5986–5987. doi: 10.1128/mcb.20.16.5986-5997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen L M, Iwama A, Lodie T A, Sasaki K, Felsher D W, Golub T R, Tenen D G. Mol Cell Biol. 2001;21:3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi Y, Wang W, Ninomiya T, Nagano H, Ohta K, Itoh H. Mol Pathol. 1998;52:19–24. doi: 10.1136/mp.52.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaret K. In: The Liver: Biology and Pathobiology. 3rd Ed. Arias I M, Boyer J L, Fausto N, Jakoby W B, Schachter D A, Shafritz D A, editors. New York: Raven; 1994. pp. 53–64. [Google Scholar]
- 27.Mozier N M, Hoffman J L. FASEB J. 1990;4:3329–3333. doi: 10.1096/fasebj.4.15.2253846. [DOI] [PubMed] [Google Scholar]
- 28.Banoglu E. Curr Drug Metab. 2000;1:1–30. doi: 10.2174/1389200003339234. [DOI] [PubMed] [Google Scholar]
- 29.Becker A, Bottcher A, Lackner K J, Fehringer P, Notka F, Aslanidis C, Schimitz G. Arterioscler Thromb. 1994;14:1346–1355. doi: 10.1161/01.atv.14.8.1346. [DOI] [PubMed] [Google Scholar]