Abstract
Elevated plasma levels of C-reactive protein (CRP), an inflammation-sensitive marker, have emerged as an important predictor of future cardiovascular disease and metabolic abnormalities in apparently healthy men and women. Here, we performed a systematic survey of common nucleotide variation across the genomic region encompassing the CRP gene locus. Of the common single-nucleotide polymorphisms (SNPs) identified, several in the CRP promoter region are strongly associated with CRP levels in a large cohort study of cardiovascular risk in European American and African American young adults. We also demonstrate the functional importance of these SNPs in vitro.
Introduction
C-reactive protein (CRP) was initially defined as a protein in serum that binds to the C polysaccharide of Streptococcus pneumoniae (Tillet and Francis 1930) and is the prototypical protein up-regulated as part of the acute phase response in humans (Abernathy and Avery 1941). More recently, elevated CRP has been identified as a biomarker for cardiovascular disease (CVD) risk, supplementary to traditional risk factors such as BMI, smoking, diabetes, and cholesterol levels (Ridker et al. 1997, 2000, 2005; Danesh et al. 2000, 2004; Hackam and Anand 2003). Family and twin studies suggest that additive genetic factors account for as much as 40% of the variance in plasma CRP levels (Pankow et al. 2001; Retterstol et al. 2003; MacGregor et al. 2004), but the specific genetic determinants remain poorly characterized.
Human genetic association studies of CRP include several that have assessed the relationship between genotype and plasma CRP levels (Szalai et al. 2002; Zee and Ridker 2002; Brull et al. 2003; Russell et al. 2004) or disease risk (Zee and Ridker 2002; Wolford et al. 2003; Russell et al. 2004). These generally have suffered from lack of functional data on DNA-sequence variation. Polymorphisms reported elsewhere as being associated with CRP levels include a polymorphism in the promoter region (Kovacs et al. 2005), a synonymous polymorphism in exon 2 (Russell et al. 2004; Suk et al. 2005), a polymorphism in the 3′ UTR (Brull et al. 2003), and a second polymorphism in the 3′ UTR (Russell et al. 2004). These studies have been restricted to a small number of polymorphisms within the CRP gene locus, without consideration of the patterns of variation across the locus as a whole. Thus, although the reported associations suggest that variation at the CRP locus is significantly correlated with basal CRP levels, it remains possible that the observed associations were attributable to other polymorphisms in strong linkage disequilibrium (LD) with the reported polymorphisms, because none of the prior work demonstrated functional differences between alleles. Prior analyses have also been largely restricted to European populations, with no prior study including >100 African Americans (AAs).
To explore whether common genetic variants at the human CRP gene locus influence plasma CRP, we used an approach that starts with genomic resequencing to identify all common patterns of nucleotide diversity and to define common haplotypes in the European American (EA) and AA populations. We then genotyped a set of polymorphisms representative of the patterns of common variation identified in a much larger population-based study sample, and we identified associations with specific haplotypes. Lastly, we experimentally validated the observed associations with polymorphisms in the promoter region, using in vitro CRP promoter analysis in a human hepatocyte cell line.
Material and Methods
SNP Discovery, tagSNP Selection, and Genotyping
Polymorphic sites were identified by resequencing of PCR products tiled across the CRP gene region by use of standard BigDye Terminator v3.1 chemistry from Applied Biosystems (ABI). The gene and flanking region were resequenced in 47 samples: 24 unrelated AAs from the Coriell AA100 panel and 23 unrelated EAs from the CEPH pedigrees. Coriell sample identifiers are available at the Web site of the SeattleSNPs Program for Genomic Applications. We resequenced 6,800 bp in each sample, covering the two exons, the intervening intron, and 1.7 kbp 5′ and 2.8 kbp 3′ of the gene. SNP numbering is based on the GenBank accession number AF449713. Bins of common SNPs (minor-allele frequency [MAF] >10%) in strong LD, as defined by r2>0.64, were identified within each population by use of LDselect v1.0 (Carlson et al. 2004). Seven tagSNPs were selected for coverage of all bins in the two sequenced populations. Relative to AF449713, the seven tagSNPs positions were 790 (dbSNP rs3093058), 1440 (rs3091244), 1919 (rs1417938), 2667 (rs1800947), 3006 (rs3093066), 3872 (rs1205), and 5237 (rs2808630).
The previous literature has generally used a nomenclature for polymorphism positions relative to the ATG codon. Thus, the previously reported promoter polymorphism has been referred to as −286 (Kovacs et al. 2005), the synonymous polymorphism in exon 2 has been referred to as +1059 (Russell et al. 2004; Suk et al. 2005), and the polymorphisms in the 3′ UTR have been referred to as +1444 (Brull et al. 2003) and +2147 (Russell et al. 2004). Because our resequencing efforts identified several insertion/deletion polymorphisms in the CRP gene region, numbering relative to an origin such as the ATG codon would be ambiguous, so we have chosen to refer to each polymorphism relative to its position in GenBank accession number AF449713, which details the results of our polymorphism-discovery survey. For convenience, here is a brief list of pseudonyms for polymorphisms reported elsewhere: −717 is 1009 (rs2794521), −286 is 1440 (rs3091244), +1059 is 2667 (rs1800847), +1444 is 3014 (rs1130864), and +2147 is 3872 (rs1205).
tagSNPs were genotyped using TaqMan Assays By Design (ABI) under standard conditions, with the exception of the triallelic tagSNP (detailed below). Probe and primer sequences for each assay are listed in table 1. End-point fluorescence values were genotyped manually within the SDS software (ABI) for tagSNPs 790, 3872, and 5237. Threshold cycle (Ct) values from real-time reads were exported to Excel (Microsoft) and were used to cluster and manually score genotypes for 1919, 2667, and 3006. The triallelic SNP (1440) was genotyped in a single tube assay, by use of reagents, at the following final concentrations in a 5-μl reaction: three TaqMan probes at 0.2 μM each, two PCR primers at 1 μM each, 1× Universal Mastermix (ABI), 1 M betaine (Sigma-Aldrich), and 1 ng/μl genomic DNA. Thermal cycling reactions were performed as follows: 50° for 2 min, 95° for 10 min, and then 45 cycles at 95° for 15 s, 53° for 30 s, and 60° for 1 min. Real-time data were acquired on the ABI 7900HT instrument. Ct values from real-time data for all three probes were exported from the SDS 2.1 software (ABI) and were used to visualize and manually call genotypes in Excel. Haplotypes for cladistic analysis were inferred using PHASE v2.0 (Stephens and Donnelly 2003).
Table 1.
TaqMan Primer and Probe Sequences
|
Primer(5′→3′) |
||||
| SNP | Forward | Reverse | Allelea | Allelic Probe |
| 790 | GACAGTGTGGAGGGATTACTTGAAT | GTAAAGCTTCAGGCAGGAGTCA | Ab | VIC ATCTGATTCTACTCTTTCC MGBNFQ |
| Tb | 6FAM CTGATTCTACACTTTCC MGBNFQ | |||
| 1440 | TGTTGGAGAGGCAGCTACCA | TCCTGCGAAAATAATGGGAAA | A | NED ATGGCCACTAGTTTAA MGBNFQ |
| T | VIC ATGGCCACTTGTTTAA MGBNFQ | |||
| C | 6FAM TGGCCACTCGTTTAA MGBNFQ | |||
| 1919 | TGCTTTTGGCCAGACAGGTAA | TTAGACCCCACCCCCATACC | A | VIC ATGGGAGAGATTTGAT MGBNFQ |
| T | 6FAM CTATGGGAGAGTTTTG MGBNFQ | |||
| 2667 | CCGCCAAGATAGATGGTGTTAATC | CCTGGTGGGAGACATTGGAA | Cb | 6FAM CTGGTGAGAGCACAAA MGBNFQ |
| Gb | VIC CTGGTGACAGCACAA MGBNFQ | |||
| 3006 | CCTGAGAATGGAGGTAAAGTGTCT | AAACACCTCAAATTCTGATTCTTTTGGAC | A | VIC CTCGTTAACTATGATGGGAA MGBNFQ |
| C | 6FAM CTCGTTAACTATGCTGGGAA MGBNFQ | |||
| 3872 | GCCATCTTGTTTGCCACATG | GCTCCTCCACTTCCAGTTTGG | Ab | VIC TGTCCTCATAGTCTCT MGBNFQ |
| Gb | 6FAM CCTCACAGTCTCTC MGBNFQ | |||
| 5237 | TCTTCAGAATTCAGTTGCTTGCAT | AAATTATTAAGGCCAGAGGCTGTCT | Ab | VIC ACCAGACTATGTATAGTAAG MGBNFQ |
| Gb | 6FAM CCAGACTACGTATAGTAA MGBNFQ | |||
Probe allele is recorded relative to GenBank accession number AF449713.
Probes are designed to the reverse strand.
CARDIA Population Study Sample
The Coronary Artery Risk Development in Young Adults (CARDIA) study is a population-based study, initiated in 1985, to investigate the evolution of cardiovascular risk factors in a large, biracial cohort of young adults (Hughes et al. 1987; Friedman et al. 1988). Participants aged 18–30 years were recruited from four geographic locations by community-based sampling (Birmingham, AL; Chicago; and Minneapolis) and through the membership of a large prepaid health plan (Oakland). In 1985–1986, baseline examinations of 5,115 participants, or 51% of the eligible persons contacted, were performed. Participants were contacted every 6 mo and were re-examined at five follow-up examinations (year 2 in 1987–1988, year 5 in 1990–1991, year 7 in 1992–1993, year 10 in 1995–1996, and year 15 in 2000–2001).
The initial study population was approximately balanced with respect to age (45% were aged 18–24 years; 55% were aged 25–30 years), sex (46% men; 54% women), race (52% AA; 48% EA), and education (40% had completed ⩽12 years of education; 60% had completed >12 years). Overall retention rates for follow-up examinations among surviving participants were 91% at year 2, 86% at year 5, 81% at year 7, 79% at year 10, and 74% at year 15. The CARDIA database includes repeated measures of lifestyle, physiologic, and metabolic risk factors, including smoking, obesity, and physical activity. Participants eligible for the current study included those who consented to isolation of genomic DNA from peripheral-blood leukocytes obtained at the year 10 examination and for which an adequate DNA sample was available (n=3,790).
Phenotypic Measurements and Statistical Analysis
Plasma CRP levels were assayed using BNII immunonephelometry (BNII Nephelometer 100 Analyzer [Dade Behring]) on plasma, stored at year 7, from 3,382 returning CARDIA participants who also consented to DNA testing and on plasma from 3,146 consenting participants who returned at year 15. The latter number represents 86% of the 3,672 original CARDIA subjects who attended the year 15 examination. Two CRP measurements were available at year 15, BNII and ELISA. BNII is a more sensitive assay than ELISA (Wener et al. 2000), so only the results of analysis of the BNII data are presented, although results from an analysis of the ELISA data at year 15 were consistent (data not shown). BNII plasma CRP measurements had a range of 0–333 mg/liter. Samples with ELISA-based CRP >10 mg/liter have been suggested to represent acute illness, stress, or infection (Shine et al. 1981; Pearson et al. 2003); 48 such samples were observed at year 15, comparable to the 50 samples with BNII CRP >18 mg/liter at year 15. Exclusion of samples with BNII >18 mg/liter had negligible impact on results at either year 7 or year 15 (data not shown). Plasma-CRP measurements at years 7 and 15 were significantly correlated for samples above this threshold, so samples with high CRP measurements were not removed from the analysis.
Stepwise linear regression was run in the R statistics program v2.0.1, with ln(plasma CRP) (the natural log of plasma CRP) at year 7, ln(plasma CRP) at year 15, or change in ln(plasma CRP) between years 7 and 15 as the dependent variable. The following phenotypic measures were allowed to enter the model as independent variables: ln(BMI), age, sex, ethnicity, systolic blood pressure, diastolic blood pressure, ln(glucose), ln(insulin), ln(total cholesterol), ln(HDL), ln(LDL), and ln(insulin/glucose). The Bayesian information criterion (Hastie et al. 2001), a measure of how well each model fits the data that is penalized for the number of parameters in the model, was used for all model comparisons.
For each of the dependent variables (CRP at year 7 and CRP at year 15), independent variables retained in the best-fit model at both time points were included as covariables in an analysis of the effects of haplotype on ln(plasma CRP). Analyses were run using the haplo.stats package for R (Lake et al. 2003), a generalized linear-regression framework that incorporates haplotype phase uncertainty by inferring a probability matrix of haplotype likelihoods for each individual by use of the expectation-maximization (EM) haplotype-inference algorithm rather than by assignment of a most likely haplotype phase resolution to each sample. Haplo.stats analysis was performed on the complete CARDIA cohort, with race and sex included as covariables, as well as on subsets of the CARDIA cohort stratified by sex and race. Haplo.stats analysis was restricted to haplotypes with an inferred frequency >1%, representing ∼33 chromosomes in this cohort.
In Vitro Promoter-Variant Vector Construction
PCR products from the CRP promoter region representing several haplotypes were cloned into a modified pGL3-basic luciferase expression vector (Promega). Short and extended clones of the native promoter represent bps 540–1726 and bps 1023–1726, respectively, relative to GenBank AF449713 (corresponding to positions −1186 to +1 and −703 to +1, relative to the transcription start site (TSS) discussed by Li et al. [1990]). To reduce interleukin 6 (IL6) dependence, the proximal CRP promoter (−107 to +1, relative to TSS) was replaced with the SV40 minimal promoter (bps 2349–2513 of pDsRed2-C1 [Promega]). Construct sequences were verified by standard fluorescent resequencing, and all construct sequences have been deposited into GenBank. Maxipreps of each construct were made (Plasmid Maxi Kit, part #12162 [QIAGEN]) and were diluted to a concentration of 200 ng/μl by UV densitometry, verified by ethidium bromide staining of serial dilutions in agarose gels.
Luciferase Assays
Transient transfection assays were conducted in a human liver hepatoma cell line (PLC/PRF/5 cells from ATCC [Manassas]) in 24-well plates, with 24 replicate wells per construct per experiment. Cells were seeded in serum-containing media. After 24 h, cells were transfected in serum-free media (so that transfection time also served as serum-starvation time). In each well, 200 ng of plasmid was transfected, with Fugene 6 transfection reagent (Roche Diagnostics), per protocols provided with the reagent. After 18 h, transfection media was removed, and cells either were induced with 200 U/ml human IL6 (Stem Cell Technologies) in serum-free media or were mock induced with non-IL6–containing serum-free media. Cell lysates were prepared 24 h later, and luciferase activity was assayed in a Wallac plate luminometer, per protocols of the Luciferase Assay System (Promega).
For each 24-well data set, the two highest and the two lowest values were dropped to remove outliers possibly related to variation in transfection efficiency. For the remaining data, luciferase values were normalized to the average activity of the promoter-less empty vector in native promoter experiments or to the SV40 minimal promoter alone in replacement promoter experiments, to yield data reflecting fold-activity increase over baseline levels for each CRP promoter construct. Standard error measurements and two-tailed student's t tests were used to assess the significance of differences between haplotype constructs.
Results
SNP Discovery and Selection of CRP Gene tagSNPs
Polymorphisms in the CRP gene region were identified by direct resequencing of 47 individuals: 24 AAs and 23 EAs. We identified 31 SNPs overall, 30 polymorphic in AAs and 13 polymorphic in EAs (fig. 1). No amino acid–changing SNPs were identified in either population. There were 18 common SNPs (i.e., SNPs with a relative MAF >10% in at least one of the two populations). Ten unique CRP haplotypes were inferred from the 18 common SNPs by use of a Bayesian statistical method for haplotype reconstruction (PHASE v2.0) (Stephens et al. 2001).
Figure 1.
A GeneSNPs image of CRP, showing the region scanned for polymorphism. Coding regions of exons are shown in blue, and the UTR is shown in green. Polymorphic positions are indicated with ticks below the axis, and the length of the tick represents MAF in the combined population. A highly repetitive region of the intron (shown in pink) could not be screened for polymorphism by resequencing. Nonsyn = nonsynonymous; Synon = synonymous; CDS - coding sequence.
We next used an LD-based tagSNP-selection algorithm (LDselect) (Carlson et al. 2004) at a threshold of r2>0.64, and we identified five tagSNPs that summarize common patterns of variation across the CRP gene in the sequenced samples (fig. 2). Two additional SNPs—a common triallelic SNP from the promoter region and a rare, synonymous SNP that previously was associated with CRP level (Zee and Ridker 2002)—were selected for analysis independent of the LDselect results. The seven tagSNPs resolved the 10 unique haplotypes observed in the sequenced samples as eight clades of related haplotypes (fig. 3), with each tagSNP allele tagging either a specific haplotype (SNPs 790, 1919, 2667, 3006, and 5237) or a clade of evolutionarily related haplotypes (SNP 1440 and 3872).
Figure 2.
Visual genotype diagram of patterns of common variation at the CRP locus. Each row corresponds to an individual DNA sample in the polymorphism-discovery panel; E001–E023 are EAs, and D001–D040 are AAs. Each column corresponds to a polymorphic site. Sites are ordered by LD, not position along the chromosome, with sites showing similar patterns of genotype shown adjacent to one another. Site numbers are relative to GenBank accession number AF449713. Selected tagSNPs are indicated with arrows.
Figure 3.
Phylogeny of common CRP haplotypes. Haplotypes were inferred for all 18 common SNPs (MAF >10% in either AA or EA sequenced samples) by use of PHASE v 2.0. A neighbor-joining phylogenetic tree of the major CRP haplotypes resolved using the seven selected tagSNPs is shown, with branch lengths representing evolutionary distance as the proportion of alleles shared at all 18 common SNPs. The tagSNP alleles associated with each resolved haplotype are shown.
CRP Genotype-Phenotype Association Study
We subsequently genotyped the seven selected tagSNPs in 3,790 U.S. adults recruited as part of the CARDIA study (Hughes et al. 1987; Friedman et al. 1988). The subjects, who were aged 18–30 years at baseline, have been followed prospectively for 15 years and have undergone serial assessment for various cardiovascular risk factors (table 2). Allele frequencies at all tagSNPs were consistent between the sequenced samples and the CARDIA cohort (table 3), although tagSNP allele frequencies differed significantly between AAs and EAs. Inferred haplotype frequencies were also similar between the sequenced samples and the CARDIA cohort (table 4).
Table 2.
CARDIA Participant Characteristics at Year 15, by Race and Sex[Note]
| AA |
EA |
|||
| Characteristic | Male(n=603) | Female(n=851) | Male(n=815) | Female(n=895) |
| Age, in years | 39.5 (3.70) | 39.6 (3.85) | 40.6 (3.34) | 40.6 (3.37) |
| BMI, in kg/m2 | 29.0 (6.15) | 31.6 (7.89) | 27.7 (4.81) | 26.7 (6.60) |
| Total cholesterol, in mg/dl | 184.9 (40.71) | 179.2 (33.21) | 191.2 (38.47) | 182.2 (32.05) |
| Cholesterol, in mg/dl: | ||||
| LDL | 115.4 (35.70) | 108.5 (31.09) | 121.4 (33.01) | 107.1 (29.10) |
| HDL | 47.6 (13.72) | 54.1 (13.65) | 43.1 (11.56) | 56.1 (14.76) |
| Triglycerides, mg/dl | 106.1 (66.99) | 83.1 (56.53) | 140.7 (149.43) | 94.1 (59.36) |
| Diabetes (%) | 4.8 | 4.7 | 3.1 | 1.6 |
| Fasting glucose, in mg/dl | 88.2 (17.84) | 86.9 (26.85) | 89.4 (18.77) | 82.6 (13.88) |
| Fasting insulin level, mU/ml | 15.3 (12.25) | 16.6 (12.83) | 14.0 (9.98) | 11.8 (7.44) |
| Blood pressure, in mmHg: | ||||
| Systolic | 118.8 (15.78) | 116.1 (16.17) | 113.7 (12.82) | 106.4 (12.22) |
| Diastolic | 79.0 (12.50) | 75.6 (12.41) | 75.5 (10.12) | 69.5 (9.57) |
| Current smokers (%) | 32.7 | 25.5 | 17.1 | 15.1 |
| Daily alcohol consumption, in ml | 16.5 (35.48) | 6.1 (24.60) | 15.1 (25.84) | 8.0 (13.42) |
| C-reactive protein, in μg/ml | 3.1 (5.11) | 4.8 (6.38) | 2.0 (4.70) | 2.8 (4.28) |
Note.— Values represent mean (±SD) unless otherwise noted.
Table 3.
tagSNP Allele Frequencies in SNP Discovery and CARDIA Populations
|
Allele Frequencies (%) in |
||||||
| AA |
EA |
|||||
| SNP | dbSNP Number | Alleles | Sequenced Samples(n=24) | CARDIA Cohort(n=1,557) | Sequenced Samples(n=23) | CARDIA Cohort(n=1,820) |
| 790 | rs309358 | A/T | 77/23 | 83/17 | 100/0 | 100/0 |
| 1440 | rs3091244 | A/C/T | 33/26/41 | 26/45/29 | 5/62/33 | 7/64/30 |
| 1919 | rs1417938 | A/T | 83/17 | 88/12 | 70/30 | 70/30 |
| 2667 | rs1800947 | C/G | 2/98 | 1/99 | 4/96 | 6/94 |
| 3006 | rs3093066 | A/C | 29/71 | 23/77 | 0/100 | 0/100 |
| 3872 | rs1205 | A/G | 15/85 | 19/81 | 25/75 | 34/66 |
| 5237 | rs2808630 | A/G | 94/6 | 82/18 | 65/35 | 72/28 |
Table 4.
tagSNP Haplotype Frequencies in SNP Discovery and CARDIA Populations
| tagSNP Haplotype Frequencies (%) in |
||||
| AA |
EA |
|||
| Haplotype | Sequenced Samples(n=24) | CARDIA Cohort(n=1,557) | Sequenced Samples(n=23) | CARDIA Cohort(n=1,820) |
| H1 | 2 | 1 | 4 | 6 |
| H2 | 15 | 18 | 24 | 28 |
| H3 | 4 | 9 | 0 | 2 |
| H4 | 6 | 17 | 35 | 28 |
| H5 | 17 | 12 | 30 | 29 |
| H6 | 23 | 17 | 0 | 0 |
| H7 | 4 | 3 | 7 | 6 |
| H8 | 29 | 22 | 0 | 0 |
| Other | 0 | 1 | 0 | 1 |
Stepwise linear regression of log-transformed CRP values on selected clinical, demographic, and lifestyle measurements identified six statistically significant predictors at both year 7 and year 15—BMI, race, sex, triglycerides, insulin level, and current smoking—as well as two significant interactions on CRP level, between sex and triglycerides (P<.0001) and between sex and smoking (P<.0001). Best-fit models at each time point are shown in table 5 (year 7) and table 6 (year 15). At both time points, the interaction between sex and triglycerides reflects a strong association with triglyceride levels in women and a weak association in men, and the interaction between sex and smoking reflects a strong association with smoking in men but not in women (data not shown).
Table 5.
| Variable | Coefficient | SE | t | P |
| Intercept | −10.94494 | 2.88816 | … | … |
| ln(BMI) | 5.18221 | .75648 | 6.85 | <.00001 |
| Male | −3.56098 | .87599 | −4.065 | .00005 |
| ln(triglycerides) | 1.64048 | .59597 | 2.753 | .00594 |
| EA | −.19726 | .04202 | −4.694 | <.00001 |
| Current smoker | .83452 | .14108 | 5.915 | <.00001 |
| ln(insulin) | .18478 | .05211 | 3.546 | .00040 |
| ln(HDL) | −1.0518 | .2822 | −3.727 | .00020 |
| Sex × ln(triglycerides) | .50974 | .07917 | 6.439 | <.00001 |
| Sex × smoking | −.39441 | .0877 | −4.497 | <.00001 |
| ln(BMI) × ln(triglycerides) | −.6605 | .17643 | −3.744 | .00019 |
| Sex × ln(HDL) | .51058 | .17352 | 2.942 | .00328 |
Note.— t statistics and P values were calculated from the coefficients and SEs within the best-fit multivariate model by the haplo.glm function of the haplo.stats R package.
Subjects with missing data at any predictor variable were excluded from this analysis.
Table 6.
| Variable | Coefficient | SE | t | P |
| Intercept | −6.97033 | .71399 | … | … |
| ln(BMI) | 2.64426 | .12112 | 21.832 | <.00001 |
| Sex = Male | −1.83951 | .33939 | −5.42 | <.00001 |
| ln(triglycerides) | −.32598 | .12674 | −2.572 | .01016 |
| Race = EA | −.1184 | .04667 | −2.537 | .01123 |
| Current smokers | 5.43416 | .88041 | 6.172 | <.00001 |
| ln(insulin) | −.07988 | .12149 | −.658 | .51090 |
| Sex × ln(triglycerides) | .40566 | .08014 | 5.062 | <.00001 |
| ln(BMI) × smoking | −.93057 | .21364 | −4.356 | .00001 |
| Race × smoking | −.37527 | .09451 | −3.971 | .00007 |
| Sex × smoking | −.31565 | .09251 | −3.412 | .00065 |
| Sex × ln(insulin) | .19499 | .07463 | 2.613 | .00902 |
Note.— t statistics and P values were calculated from the coefficients and SEs within the best-fit multivariate model by the haplo.glm function of the haplo.stats R package.
Subjects with missing data at any predictor variable were excluded from this analysis.
There was significant variation in CRP level among CRP haplotypes (P<10-6), and the patterns of association were similar at both time points (fig. 4 and tables 7 and 8) as well as in analyses stratified by sex and race (data not shown). The results of a similar analysis without covariables were also consistent (data not shown). Haplotype 1 (H1) (tagged by SNP 2667) was associated with significantly reduced CRP levels relative to H2, the referent, at both time points. In addition, the clade of H1–H4, which carry the C allele of the triallelic SNP 1440, was associated with lower CRP levels than was the clade of H5–H8. Of the latter group, H6 (tagged by SNP 790) was associated with the highest CRP levels relative to all other haplotypes. The results of a similar analysis without covariables were also consistent (data not shown). Models fitted under the assumption of allelic independence showed a better fit than did recessive or dominant models at any of the loci (data not shown). Models fitted without interactions between haplotype and phenotypic variables other than race showed a better fit than did models including such interactions (data not shown). Exclusion of the 48 subjects with BNII CRP values >18 mg/liter did not alter these results. Similarly, exclusion of the upper and lower 5% of CRP values did not alter the results, so it is unlikely that the observed associations were attributable to a small number of influential data points. Comparison of the residual variance of the full model with the residual variance of models excluding single predictors demonstrated that only BMI explained a greater proportion of variance than did the tagSNPs at both time points (table 9). Results were similar for analyses stratified by race (data not shown).
Figure 4.
Relative effects of tagSNP haplotypes on ln(CRP). Regression coefficients from haplo.glm analysis were estimated for each haplotype, with phenotypic variables included as covariables. Coefficients reflect difference in mean ln(CRP) per copy relative to H2, the most frequent haplotype. H1 was significantly lower than H2, and H5–H8 were significantly higher (see P values in tables 7 and 8).
Table 7.
Association of CRP Haplotypes with Plasma CRP Level at Year 7[Note]
| Variable | tagSNP Alleles | Haplotype Frequencya | Coefficient | SE | t | P |
| Intercept | … | … | −8.0161 | .0703 | … | … |
| EA | … | … | −.0332 | .0502 | −.661 | .509 |
| Male | … | … | −1.0083 | .1817 | −5.549 | <.00001 |
| ln(BMI) | … | … | 2.4367 | .1008 | 24.18 | <.00001 |
| ln(insulin) | … | … | .1963 | .0511 | 3.845 | .00012 |
| ln(triglycerides) | … | … | −.2996 | .0722 | −4.151 | .00003 |
| Current smoker | … | … | .7819 | .1406 | 5.561 | <.00001 |
| H1 | ACACCAA | .0381 | −.1262 | .0728 | −1.734 | .0831 |
| H2 | ACAGCAA | .2306 | Referent | … | … | … |
| H3 | ACAGCGA | .0398 | .1387 | .0725 | 1.912 | .056 |
| H4 | ACAGCGG | .2215 | .0563 | .0393 | 1.432 | .152 |
| H5 | ATTGCGA | .2053 | .2818 | .0398 | 7.085 | <.00001 |
| H6 | TTAGCGA | .0717 | .4498 | .0592 | 7.602 | <.00001 |
| H7 | AAAGCGA | .0481 | .3128 | .0669 | 4.677 | <.00001 |
| H8 | AAAGAGA | .0948 | .1749 | .054 | 3.236 | .00122 |
| Otherb | … | .0502 | .1908 | .0705 | 2.706 | .00685 |
| Sex × ln(triglycerides) | … | … | .367 | .0441 | 8.324 | <.00001 |
| Sex × smoking | … | … | −.3587 | .0877 | −4.091 | .00004 |
Note.— t statistics and P values were calculated from the coefficients and SEs within the best-fit multivariate model by the haplo.glm function of the haplo.stats R package.
Haplotype frequencies were inferred using the EM algorithm within the haplo.stats package.
Haplotypes with frequency <1% were pooled as “Other.”
Table 8.
Association of CRP Haplotypes with Plasma CRP Level at Year 15[Note]
| Variable | tagSNP alleles | Haplotype Frequencya | Coefficient | SE | t | P |
| Intercept | … | … | −7.1203 | .0745 | … | … |
| EA | … | … | −.0405 | .0495 | −.819 | .413 |
| Male | … | … | −1.69 | .1824 | −9.265 | <.00001 |
| ln(BMI) | … | … | 2.4717 | .0963 | 25.657 | <.00001 |
| ln(insulin) | … | … | .2016 | .0438 | 4.605 | <.00001 |
| ln(triglycerides) | … | … | −.4544 | .0671 | −6.771 | <.00001 |
| Current smoker | … | … | .7411 | .1456 | 5.09 | <.00001 |
| H1 | ACACCAA | .0378 | −.2275 | .0717 | −3.172 | .00153 |
| H2 | ACAGCAA | .2262 | Referent | |||
| H3 | ACAGCGA | .0414 | −.0335 | .0706 | −.475 | .635 |
| H4 | ACAGCGG | .2231 | .0534 | .0386 | 1.381 | .167 |
| H5 | ATTGCGA | .205 | .2125 | .0391 | 5.437 | <.00001 |
| H6 | TTAGCGA | .0733 | .5219 | .0573 | 9.107 | <.00001 |
| H7 | AAAGCGA | .0475 | .3152 | .066 | 4.778 | <.00001 |
| H8 | AAAGAGA | .096 | .2044 | .0531 | 3.851 | .00012 |
| Otherb | … | .0496 | .1552 | .0725 | 2.142 | .0323 |
| Sex × ln(triglycerides) | … | … | .4801 | .0418 | 11.487 | <.00001 |
| Sex × smoking | … | … | −.3583 | .0914 | −3.918 | .00009 |
Note.— t statistics and P values were calculated from the coefficients and SEs within the best-fit multivariate model by the haplo.glm function of the haplo.stats R package.
Haplotype frequencies were inferred using the EM algorithm within the haplo.stats package.
Haplotypes with frequency <1% were pooled as “Other.”
Table 9.
Proportion of Variance Explained
|
Data at Year 15 |
Data at Year 7 |
|||
| Modela | Proportion of Variance Explained | Change from Full Model | Proportion of Variance Explained | Change from Full Model |
| Full model | .3757 | … | .3208 | … |
| Full −haplotype | .3427 | .0330 | .3047 | .0161 |
| Full −ln(BMI) | .2637 | .1120 | .2187 | .1021 |
| Full −race | .3760 | −.0003 | .3204 | .0003 |
| Full −sex | .3406 | .0351 | .2785 | .0423 |
| Full −ln(insulin) | .3721 | .0036 | .3174 | .0033 |
| Full −ln(triglycerides) | .3554 | .0203 | .3057 | .0151 |
Full model contains all six predictors: haplotype, ln(BMI), race, sex, ln(insulin), and ln(triglycerides). Each of the nested models contains all but one of the predictors from the full model. For example, “full −haplotype” contains five predictors: ln(BMI), race, sex, ln(insulin), and ln(triglycerides) (i.e., all but haplotype).
The availability of high-sensitivity BNII CRP measurements at time points 8 years apart allowed us to assess the potential influence of CRP genotype on the temporal change in CRP level. Overall, the intrasubject correlation in ln(CRP) between years 7 and 15 was r=0.60 (P≪.001). On average, there was a 0.33 mg/liter, or 11.6%, increase during the 8-year interval. Although change in BMI and change in triglycerides were significantly correlated with change in CRP level, there were no significant associations between CRP haplotype and change in level (data not shown).
Contribution of CRP Genotype to Population Differences in CRP Levels
Median CRP levels were 1.0 mg/liter among EAs and 2.0 mg/liter among AAs (P<10-6 by t test on log-transformed data). Since the low-level CRP variant (H1) was more common among EAs and the high-level variant (H6) was more common among AAs, we assessed the extent to which CRP genotype or other known predictors accounted for variation in CRP levels between AAs and EAs in our population sample. To this end, we analyzed a series of nested, multivariable regression models that variably included or excluded terms for race/ethnicity, sex, BMI, triglycerides, insulin level, smoking status, and CRP haplotypes or tagSNP genotypes. The nongenotypic covariables sex, triglycerides, insulin level, and smoking status all contributed relatively little to the mean difference in CRP level between racial groups, estimated from the regression model—mean difference in ln(CRP) level=0.55±0.04 unadjusted; 0.43±0.04 adjusted for sex, triglycerides, insulin level, and smoking status. The further addition of BMI to the regression model had a more substantial effect—mean difference in ln(CRP) level=0.20±0.04—suggesting that more than half of the difference in CRP levels between ethnic groups was attributable to differences in BMI between AAs and EAs. The addition of CRP genotypes to the model resulted in an even greater reduction in the coefficient for ethnicity (e.g., mean difference in ln(CRP) level=-0.04±0.05 at year 15 [table 8]), to the point where there was no longer a statistically significant difference between racial groups at either time point (P=.51 [table 7]; P=.41 [table 8]).
CRP Promoter Analysis In Silico
Comparison of the human and mouse genomic sequence of the CRP locus (Mayor et al. 2000) showed a strongly conserved promoter region 5′ of the exons (fig. 5, upper panel) that contains response elements for several hepatic transcription factor families (HNF, C/EBP, and NFKB) involved in induction of CRP by IL6 (Li et al. 1990). Two SNPs map to the 5′ edge of this region, tagSNP 1440 and a SNP perfectly associated with tagSNP 790, 1421. A transcription factor–motif analysis of the region (TFSEARCH; Heinemeyer et al. 1998) identified two upstream stimulating factor 1 (USF1)–binding motifs altered by SNPs 1421 and 1440 (fig. 5, lower panel). Only two other USF1-binding motifs were predicted within the CRP transcript or upstream region, at 1630 and 2650.
Figure 5.
CRP promoter and predicted USF1 sites. Upper panel, A vista plot (Mayor et al. 2000) of mouse homology across the human CRP locus. The CRP transcript and repeat elements are annotated across the top of the plot (see key at left), and strong evolutionary conservation of the coding region in the second exon is apparent as the shaded blue peak. A putative promoter region (75% conserved) is visible as a pink peak stretching from 1444 to 1650; the two putatively functionally polymorphic USF1 sites (1421 and 1440) are on the edge of this region. Lower panel, Haplotypes of the polymorphisms at 1421 and 1440, with the putative USF1-binding sites indicated in uppercase letters. Alleles predicted to disrupt USF1 binding are shown in red, and alleles predicted to retain USF1 binding are shown in green. H1–H3 represents the ancestral haplotype, as inferred from the chimpanzee alleles at these SNPs. LINE = long interspersed nuclear element; LTR = long terminal repeat; SINE = short interspersed nuclear element.
In Vitro Gene-Reporter Assays
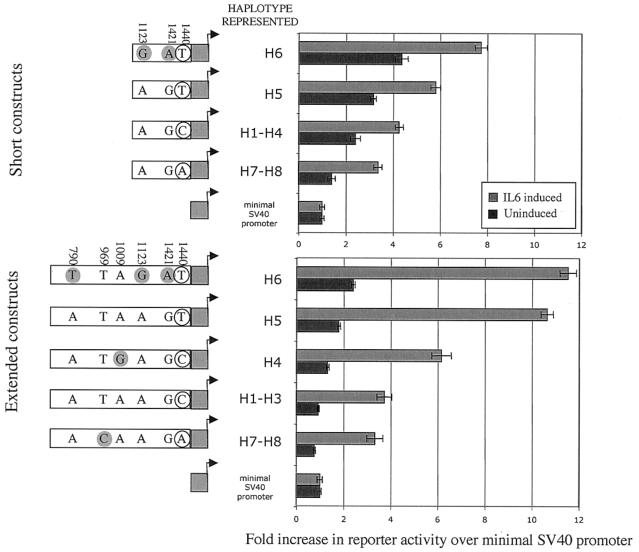
In light of the observed associations between promoter-region polymorphism and CRP levels, as well as the in silico promoter analysis, in vitro assays were generated to explore the variation identified in promoter polymorphisms in CRP. Each of the five unique promoter-region haplotypes was cloned into a luciferase-reporter plasmid for in vitro analysis. Promoter activity in the native promoter constructs was weak without IL6 induction (fig. 6). Therefore, we also analyzed each promoter haplotype with the native TSS replaced by the SV40 TSS to generate detectable signal without IL6 induction (fig. 7). The IL6-induced native H7/H8 constructs showed elevated promoter activity relative to H1–H3 (fig. 6) but not in the SV40-replacement constructs (fig. 7), suggesting that the elevated CRP levels associated with H7/H8 require the proximal IL6 responsive element. By comparison, in the native promoter, there was no evidence of increased IL6-induced promoter activity from H5 or H6.
Figure 6.
Native promoter construct activity. Short (−703 to +1, relative to TSS) and extended (−1186 to +1) versions of the CRP promoter region were cloned into a luciferase-reporter–gene plasmid. Transient transfection data show the increase in transcription levels for each construct over that of the plasmid-lacking promoter in both uninduced and IL6-induced environments. Shaded circles identify minor alleles at a given SNP. Unshaded circles identify one of the triallelic SNP variants at bp 1440. Error bars reflect SEM in 20 replicate experiments. The short construct group of H1–H4 is divided into two categories of long clones on the basis of the unique 1009 SNP in H4 that is not included in the short-clone constructs. Within the IL6-induced extended constructs, t tests show that H4 activity is significantly reduced, whereas H7–H8 activity is significantly increased (P<.05), consistent with the induced short constructs in which t tests show that H1–H4 activity is significantly reduced, whereas H7–H8 activity is significantly increased (P<.05). Uninduced activities showed similar trends, but the signal was relatively weak.
Figure 7.
SV40-replacement promoter construct activity. To increase the level of uninduced expression, an IL6-responsive negative regulatory element in the proximal segment of the CRP promoter (−107 to +1) was replaced with the SV40 minimal promoter. Transient transfection data shows the increase in transcription levels for each construct over that of the SV40 minimal promoter alone. Shaded circles identify minor alleles at a given SNP. Unshaded circles identify one of the triallelic SNP variants at bp 1440. Error bars reflect SE measurements. Within each combination of construct length and induction condition, t tests show that all differences between haplotypes’ behavior are significant (P<.05), except for H1–H3 versus H7–H8 in IL6-induced extended constructs.
Dramatically stronger uninduced expression was observed in the SV40-replacement constructs, and significant variation was observed between haplotypes (fig. 7). Consistent with the CRP genotype-phenotype association observed in the population study, the H6 promoter showed the highest activity, and the promoter activity associated with the H5 construct was also significantly increased. Interestingly, the magnitude of the difference between H5/H6 and H1–H4 SV40-replacement promoter activities was similar both with and without IL6 induction, suggesting that, although the H5 and H6 promoter haplotypes associate with elevated CRP levels, this association is not IL6-dependent but, rather, reflects a change in basal promoter activity.
Discussion
To determine how common genetic variants at the human CRP gene locus influence plasma CRP, we used a three-stage experimental design. In the first phase, all common genetic variation across the gene region was defined by resequencing the region in a multiethnic variation discovery panel, and tagSNPs representative of all common patterns in each ethnicity were selected for genotyping in a larger panel of clinically phenotyped samples. Seven tagSNPs were selected for genotyping, and these defined eight common haplotypes (fig. 3). In the second phase, associations between haplotypes of tagSNPs and CRP levels were investigated using standard regression models, and significant associations were identified for several SNPs in the putative promoter region. In the third phase, the observed associations were experimentally validated by in vitro CRP promoter analysis in human hepatocytes.
The results from our large population sample suggest that CRP haplotypes can be grouped into four categories on the basis of associated CRP level: lowest (H1), low (H2–H4), high (H5, H7, and H8), and highest (H6) (fig. 4). These results were consistent between year 7 and year 15. Relative to the covariable-adjusted population mean of 3.1 mg/liter at year 15, the haplotype associated with the lowest levels (H1) decreased plasma CRP an average of 1.5 mg/liter per copy, whereas the haplotype associated with the highest levels (H6) increased plasma CRP 1.5 mg/liter per copy.
An advantage of using tag SNPs defined on the basis of detailed sequence variation and LD data is that mapping the observed haplotype associations to potentially functional polymorphisms (or combinations thereof) can be straightforward. For example, the synonymous SNP at 2667 was the only observed difference between H1 and H2 in the SNP discovery samples. Therefore, it is likely that this polymorphism directly influences CRP levels, consistent with previous reports (Szalai et al. 2002; Zee and Ridker 2002; Brull et al. 2003; Russell et al. 2004), although the mechanism for a direct effect is unclear, so it remains possible that 2667 is in strong LD with a polymorphism in a distal regulatory element not within the region scanned for polymorphisms. Similarly, the sole consistent difference between the clade of haplotypes associated with low CRP levels (H1–H4) and the clade of haplotypes associated with higher CRP levels (H5–H8) appears to be the genotype at the triallelic promoter SNP 1440 (fig. 1); the low clade is associated with the C allele and the high clade with A or T alleles. Finally, the rare alleles at five SNPs (tagSNP 790 and unassayed SNPs 1123, 1421, 4741, and 6333) are restricted to the haplotype associated with highest CRP levels (H6), so the observed effect of this haplotype could be attributable to a haplotype of several of these SNPs.
Interestingly, stepwise regression analysis with phenotype and tagSNP genotype as independent variables identified exactly the same set of tagSNPs as being functionally important, because the functionally important haplotypes were uniquely associated with the rare allele of a single tagSNP. A haplotype-based analysis would have proven advantageous only if one of the haplotypes without such a tagSNP (H2, H3, or H7) showed a significant effect on CRP. In this case, haplotype analysis could even be considered a disadvantage because of the effort required to map the observed haplotype effects to specific tagSNPs, whereas direct tagSNP analysis immediately identified the key tagSNPs. Haplotype analysis may prove more powerful in genes with more complex recombination histories or when haplotype-tagging SNPs have not been comprehensively described, but our example demonstrates that haplotype-based analysis is not necessarily superior to direct tagSNP analyses.
To address the possibility that SNPs in the upstream region are functionally important in regulation of the CRP gene, haplotypes of the minimal (−703 to +1, relative to the TSS) and (−1186 to +1 ) extended promoter region were cloned into a luciferase-reporter plasmid for promoter-activity analysis. Some of the tagSNP haplotypes across the CRP region did not differ in the promoter region (e.g., H1–H3), so a single promoter construct represented these haplotypes. The relative basal and IL6-induced expression from each of the five unique promoter haplotypes was assessed in cultured human hepatoma cells. Basal expression levels for the native CRP promoter revealed no clear trend, because basal expression was quite weak (fig. 6). However, IL6-induced promoter activity from the H7/H8 constructs (tagged by the 1440 A allele) was marginally higher than all other haplotype constructs (P<.05 [uncorrected for multiple tests]). These results are consistent with the higher CRP levels associated with H7/H8 observed in the human population data. IL6-induced activity is a better model for CRP levels during an acute phase response than are basal CRP levels. Nonetheless, over time, we did not observe any apparent influence of H7/H8 on change in CRP levels.
The weak constitutive activity of the CRP promoter appears to be due to a negative regulatory element located within the proximal promoter region (Li et al. 1990). Therefore, we performed additional experiments in which we replaced the 107 bp adjacent to the TSS (including the proximal IL6-responsive elements present within the CRP promoter region) with the SV40 minimal promoter. As expected, the SV40-replacement promoter construct showed dramatically higher activity (fig. 7). For some haplotype comparisons, it was possible to assign the functional difference to a single SNP, because the constructs differed at only one SNP. For example, the significant difference observed between the H5 and H1–H3 constructs (fig. 7) is entirely attributable to the allele at SNP 1440 (C vs. T), which was the only sequence difference between these constructs. Similarly, the significant difference between the H4 and H1–H3 constructs is entirely attributable to SNP 1009. Thus, the marginally significant trend toward higher CRP levels that we observed for H4 in our population sample (fig. 3) may truly represent a functional effect of the H4-associated SNP 1009, as suggested elsewhere (Wolford et al. 2003). The significant difference observed between the H6 and H5 constructs might be attributable to any one of several polymorphic sites, but data from the shorter SV40-replacement constructs suggest that this difference is attributable to either 1123 or 1421 (fig. 7).
Of particular interest are the short replacement promoter constructs that reveal significantly different transcriptional activity that is based solely on the allele at triallelic SNP position 1440. Previous reports of increased CRP levels associated with SNP 3014, which is restricted to H5 in Europeans (Brull et al. 2003; Russell et al. 2004), might reflect LD with the functional 1440T allele, although the 3014 polymorphism could conceivably be functional in addition to the demonstrated functional changes at 1440, because it is in the 3′ UTR and might affect RNA structure or turnover.
In contrast to the H5 and H6 results, in which the in vitro assays lend strong support to the hypothesis that the observed association between these haplotypes is largely attributable to functional polymorphisms within the promoter region, the in vitro results for H7 and H8 provide, at best, weak support for the observed associations. This might reflect one of several possibilities: the 1440A allele may indeed be functionally different from the 1440C allele but not under the conditions assayed. That is, in addition to IL6, IL1B and tumor necrosis factor α are known to regulate CRP, and H7 and H8 may behave differentially in response to these or other as-yet-unidentified signals but not in the basal activity assay or under IL6 induction. Alternatively, the association for these haplotypes may be the result of a polymorphism that is in strong LD with the 1440 A allele but falls outside of the region assayed for promoter activity. Curiously, there were only four SNP alleles restricted to H7 and H8: 969C, 1440A, 6192A, and 6469G. The first two were within the analyzed promoter region for the long clones, whereas the latter two SNPs are >2,000 bp past the polyadenylation site of the transcript and do not appear to lie in regions of strong evolutionary conservation that would suggest a distal regulatory element. Thus, at this time, we have not identified a good candidate SNP outside of the promoter region to account for the elevated activities of H7 and H8.
These experimental results support the hypothesis that the CRP haplotype-phenotype associations observed in the CARDIA cohort are at least partially attributable to functional changes at promoter sites 1421 and 1440, each of which is also predicted to alter binding motifs for the transcription factor USF1. The clustering of USF1 sites in the promoter region (fig. 5) suggests that USF1 may play an important role in regulation of the CRP gene. The increased promoter activities associated with H5–H8 are particularly interesting in light of recent publications highlighting the potential importance of USF1 as a regulator of glucose and lipid-metabolism genes (Pajukanta et al. 2004).
African-descent populations have higher average CRP levels than do European-descent populations (Wener et al. 2000), although there is broad overlap in the range of levels across ethnic groups. Self-reported ethnicity in the United States is also associated with differences in morbidity and mortality from CVD and metabolic diseases, for a variety of medical, social, behavioral, and economic reasons (Watkins 2004). Thus, the genetic portion of the interethnic variance appeared to be attributable to frequency differences at three CRP SNPs (790, 1440, and 2667), in which the allele associated with lower CRP levels (790A, 1440C, and 2667C) was more frequent in EAs than in AAs. These results confirm recent observations that metabolic factors such as obesity account for a large portion of the variation in CRP levels associated with race/ethnicity (Albert et al. 2004; Anand et al. 2004) and suggest that the majority of the remaining variation in CRP levels associated with race/ethnicity in young adults is accounted for by genetic variants within the CRP locus.
It remains unclear whether CRP is a risk marker for inflammatory processes or is causally involved in the pathogenesis of CVD. The strong associations between CRP genotype and plasma-CRP level described here do not resolve this question directly but may prove useful in testing the hypothesis that CRP levels play a causal role in CVD pathogenesis. The functional CRP promoter variants appear to alter CRP levels independent of other CRP correlates, particularly BMI, and do so consistently within individuals over time. Thus, we expect that if CRP is causally involved in disease pathogenesis, then alleles associated with high CRP levels should be associated with greater risk of CVD. Currently, the average age in the CARDIA cohort is 42 years, so the number of incident CVD events is as yet insufficient to test this hypothesis. However, large, multiethnic human studies of older adults that assess atherosclerotic disease progression or occurrence of clinical cardiovascular events (such as myocardial infarction) can be used to address this important question. Such studies are currently in progress. Commonly prescribed cholesterol-lowering drugs, such as statins and PPARα activators, may additionally reduce cardiovascular risk through anti-inflammatory effects, including attenuation of constitutive and induced CRP gene expression (Kleemann et al. 2004; Ridker et al. 2005). Therefore, further characterization of the influence and interaction of common cis-acting nucleotide substitutions on transcriptional regulation of the CRP gene could have broad implications for the treatment and prevention of cardiovascular, metabolic, and related inflammatory disorders.
Note added in proof.—Functional associations between the 1440 and 1421 promoter SNPs and basal CRP levels have been confirmed in an independent study by Szalai et al. (in press).
Acknowledgments
This work was supported by Inflammatory Genomics and Atherosclerosis Prevention ancillary CARDIA grant HL71017 (to D.S.), by CARDIA contracts N01-HC-95095, N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-45134, and N01-HC-05187 from NHLBI, and by NHLBI Program for Genomic Applications grants HL66682 and HL66642 (to D.N. and M.R.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/index.html (for tagSNPs 790 [rs3093058], 1440 [rs3091244], 1919 [rs1417938], 2667 [rs1800947], 3006 [rs3093066], 3872 [rs1205], and 5237 [rs2808630])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the CRP gene [accession number AF449713])
- SeattleSNPs Program for Genomic Applications, http://pga.gs.washington.edu/protocols/dnapanel_protocol.html
- TFSEARCH, http://www.cbrc.jp/research/db/TFSEARCH.html
References
- Abernathy TJ, Avery OT (1941) The occurrence during acute infections of a protein not normally present in the blood. I. Distribution of the reactive protein in patient’s sera and the effect of calcium on the flocculation reaction with C-polysaccharide of pneumococcus. J Exp Med 73:173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert MA, Glynn RJ, Buring J, Ridker PM (2004) C-reactive protein levels among women of various ethnic groups living in the United States (from the Women’s Health Study). Am J Cardiol 93:1238–1242 [DOI] [PubMed] [Google Scholar]
- Anand SS, Razak F, Yi Q, Davis B, Jacobs R, Vuksan V, Lonn E, Teo K, McQueen M, Yusuf S (2004) C-reactive protein as a screening test for cardiovascular risk in a multiethnic population. Arterioscler Thromb Vasc Biol 24:1509–1515 [DOI] [PubMed] [Google Scholar]
- Brull DJ, Serrano N, Zito F, Jones L, Montgomery HE, Rumley A, Sharma P, Lowe GD, World MJ, Humphries SE, Hingorani AD (2003) Human CRP gene polymorphism influences CRP levels: implications for the prediction and pathogenesis of coronary heart disease. Arterioscler Thromb Vasc Biol 23:2063–2069 [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh J, Wheeler JG, Hirschfield GM, Eda S, Eiriksdottir G, Rumley A, Lowe GD, Pepys MB, Gudnason V (2004) C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med 350:1387–1397 [DOI] [PubMed] [Google Scholar]
- Danesh J, Whincup P, Walker M, Lennon L, Thomson A, Appleby P, Gallimore JR, Pepys MB (2000) Low grade inflammation and coronary heart disease: prospective study and updated meta-analyses. BMJ 321:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr, Liu K, Savage PJ (1988) CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 41:1105–1116 [DOI] [PubMed] [Google Scholar]
- Hackam DG, Anand SS (2003) Emerging risk factors for atherosclerotic vascular disease: a critical review of the evidence. JAMA 290:932–940 [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J (2001) The elements of statistical learning: data mining, inference and prediction. Springer-Verlag, New York [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GH, Cutter G, Donahue R, Friedman GD, Hulley S, Hunkeler E, Jacobs DR Jr, Liu K, Orden S, Pirie P, Tucker B, Wagenknecht L (1987) Recruitment in the Coronary Artery Disease Risk Development in Young Adults (CARDIA) Study. Control Clin Trials 8:68S–73S [DOI] [PubMed] [Google Scholar]
- Kleemann R, Verschuren L, de Rooij BJ, Lindeman J, de Maat MM, Szalai AJ, Princen HM, Kooistra T (2004) Evidence for anti-inflammatory activity of statins and PPARα activators in human C-reactive protein transgenic mice in vivo and in cultured human hepatocytes in vitro. Blood 103:4188–4194 [DOI] [PubMed] [Google Scholar]
- Kovacs A, Green F, Hansson LO, Lundman P, Samnegard A, Boquist S, Ericsson CG, Watkins H, Hamsten A, Tornvall P (2005) A novel common single nucleotide polymorphism in the promoter region of the C-reactive protein gene associated with the plasma concentration of C-reactive protein. Atherosclerosis 178:193–198 [DOI] [PubMed] [Google Scholar]
- Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ (2003) Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered 55:56–65 [DOI] [PubMed] [Google Scholar]
- Li SP, Liu TY, Goldman ND (1990) cis-acting elements responsible for interleukin-6 inducible C-reactive protein gene expression. J Biol Chem 265:4136–4142 [PubMed] [Google Scholar]
- MacGregor AJ, Gallimore JR, Spector TD, Pepys MB (2004) Genetic effects on baseline values of C-reactive protein and serum amyloid A protein: a comparison of monozygotic and dizygotic twins. Clin Chem 50:130–134 [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047 [DOI] [PubMed] [Google Scholar]
- Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XJ, Soro-Paavonen A, Naukkarinen J, Saarela J, Laakso M, Ehnholm C, Taskinen MR, Peltonen L (2004) Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1). Nat Genet 36:371–376 [DOI] [PubMed] [Google Scholar]
- Pankow JS, Folsom AR, Cushman M, Borecki IB, Hopkins PN, Eckfeldt JH, Tracy RP (2001) Familial and genetic determinants of systemic markers of inflammation: the NHLBI family heart study. Atherosclerosis 154:681–689 [DOI] [PubMed] [Google Scholar]
- Pearson TA, Mensah GA, Alexander RW, Anderson JL, Cannon RO 3rd, Criqui M, Fadl YY, Fortmann SP, Hong Y, Myers GL, Rifai N, Smith SC Jr, Taubert K, Tracy RP, Vinicor F (2003) Markers of inflammation and cardiovascular disease: application to clinical and public health practice: a statement for healthcare professionals from the Centers for Disease Control and Prevention and the American Heart Association. Circulation 107:499–511 [DOI] [PubMed] [Google Scholar]
- Retterstol L, Eikvar L, Berg K (2003) A twin study of C-reactive protein compared to other risk factors for coronary heart disease. Atherosclerosis 169:279–282 [DOI] [PubMed] [Google Scholar]
- Ridker PM, Cannon CP, Morrow D, Rifai N, Rose LM, McCabe CH, Pfeffer MA, Braunwald E (2005) C-reactive protein levels and outcomes after statin therapy. N Engl J Med 352:20–28 [DOI] [PubMed] [Google Scholar]
- Ridker PM, Cushman M, Stampfer MJ, Tracy RP, Hennekens CH (1997) Inflammation, aspirin, and the risk of cardiovascular disease in apparently healthy men. N Engl J Med 336:973–979 [DOI] [PubMed] [Google Scholar]
- Ridker PM, Hennekens CH, Buring JE, Rifai N (2000) C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med 342:836–843 [DOI] [PubMed] [Google Scholar]
- Russell AI, Cunninghame Graham DS, Shepherd C, Roberton CA, Whittaker J, Meeks J, Powell RJ, Isenberg DA, Walport MJ, Vyse TJ (2004) Polymorphism at the C-reactive protein locus influences gene expression and predisposes to systemic lupus erythematosus. Hum Mol Genet 13:137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine B, de Beer FC, Pepys MB (1981) Solid phase radioimmunoassays for human C-reactive protein. Clin Chim Acta 117:13–23 [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk HJ, Ridker PM, Cook NR, Zee RY (2005) Relation of polymorphism within the C-reactive protein gene and plasma CRP levels. Atherosclerosis 178:139–145 [DOI] [PubMed] [Google Scholar]
- Szalai AJ, McCrory MA, Cooper GS, Wu J, Kimberly RP (2002) Association between baseline levels of C-reactive protein (CRP) and a dinucleotide repeat polymorphism in the intron of the CRP gene. Genes Immun 3:14–19 [DOI] [PubMed] [Google Scholar]
- Szalai AJ, Wu J, Lange EM, McCrory MA, Langefeld CD, Williams A, Zakharkin SO, George V, Allison DB, Cooper GS, Xie F, Fan Z, Edberg JC, Kimberly RP. Single-nucleotide polymorphisms in the C-reactive protein (CRP) gene promoter that affect transcription factor binding, alter transcriptional activity, and associate with differences in baseline serum CRP level. J Mol Med (in press) [DOI] [PubMed] [Google Scholar]
- Tillet W, Francis T (1930) Serologic reactions in pneumonia with a non-protein somatic fraction of pneumococcus. J Exp Med 52:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins LO (2004) Perspectives on coronary heart disease in African Americans. Rev Cardiovasc Med (Suppl 3) 5:S3–S13 [PubMed] [Google Scholar]
- Wener MH, Daum PR, McQuillan GM (2000) The influence of age, sex, and race on the upper reference limit of serum C-reactive protein concentration. J Rheumatol 27:2351–2359 [PubMed] [Google Scholar]
- Wolford JK, Gruber JD, Ossowski VM, Vozarova B, Antonio Tataranni P, Bogardus C, Hanson RL (2003) A C-reactive protein promoter polymorphism is associated with type 2 diabetes mellitus in Pima Indians. Mol Genet Metab 78:136–144 [DOI] [PubMed] [Google Scholar]
- Zee RY, Ridker PM (2002) Polymorphism in the human C-reactive protein (CRP) gene, plasma concentrations of CRP, and the risk of future arterial thrombosis. Atherosclerosis 162:217–219 [DOI] [PubMed] [Google Scholar]