Abstract
The human genome contains numerous blocks of highly homologous duplicated sequence. This higher-order architecture provides a substrate for recombination and recurrent chromosomal rearrangement associated with genomic disease. However, an assessment of the role of segmental duplications in normal variation has not yet been made. On the basis of the duplication architecture of the human genome, we defined a set of 130 potential rearrangement hotspots and constructed a targeted bacterial artificial chromosome (BAC) microarray (with 2,194 BACs) to assess copy-number variation in these regions by array comparative genomic hybridization. Using our segmental duplication BAC microarray, we screened a panel of 47 normal individuals, who represented populations from four continents, and we identified 119 regions of copy-number polymorphism (CNP), 73 of which were previously unreported. We observed an equal frequency of duplications and deletions, as well as a 4-fold enrichment of CNPs within hotspot regions, compared with control BACs (P < .000001), which suggests that segmental duplications are a major catalyst of large-scale variation in the human genome. Importantly, segmental duplications themselves were also significantly enriched >4-fold within regions of CNP. Almost without exception, CNPs were not confined to a single population, suggesting that these either are recurrent events, having occurred independently in multiple founders, or were present in early human populations. Our study demonstrates that segmental duplications define hotspots of chromosomal rearrangement, likely acting as mediators of normal variation as well as genomic disease, and it suggests that the consideration of genomic architecture can significantly improve the ascertainment of large-scale rearrangements. Our specialized segmental duplication BAC microarray and associated database of structural polymorphisms will provide an important resource for the future characterization of human genomic disorders.
Introduction
Segmental duplications (also termed “low-copy repeats”) are blocks of DNA that range from 1 to 400 kb in length, occur at more than one site within the genome, and typically share a high level of (>90%) sequence identity (reviewed by Eichler [2001]). Both in situ hybridization and in silico analyses have shown that ∼5% of the human genome is composed of duplicated sequence (Cheung et al. 2001; Bailey et al. 2002; Cheung et al. 2003; She et al. 2004a), and many studies have noted a significant association between the location of segmental duplications and regions of chromosomal instability or evolutionary rearrangement (Ji et al. 2000; Samonte and Eichler 2002; Armengol et al. 2003; Locke et al. 2003a, 2003b; Bailey et al. 2004). Indeed, segmental duplications have been implicated as the probable mediators of >25 recurrent genomic disorders (reviewed by Stankiewicz and Lupski [2002]). Molecular studies have shown that the presence of large, highly homologous flanking repeats predisposes these regions to recurrent rearrangement by nonallelic homologous recombination, resulting in deletion, duplication, or inversion of the intervening sequence (Chance et al. 1994; Shaw et al. 2002).
A growing body of evidence now suggests that the duplication architecture of the genome may also mediate normal variation. The existence of large genomic polymorphisms, originally termed “heteromorphisms” or “euchromatic variants,” has been recognized since the advent of high-resolution cytogenetic banding techniques (summarized at the Chromosome Anomaly Register Web site). With the use of more-targeted molecular analyses, a number of submicroscopic polymorphic rearrangements between homologous blocks of sequence have been identified in the normal population (Siniscalco et al. 2000; Sprenger et al. 2000; Giglio et al. 2001; Osborne et al. 2001; Gimelli et al. 2003; Skaletsky et al. 2003). Recently, the use of methods such as array comparative genomic hybridization (array CGH) and representational oligonucleotide microarray analysis (ROMA) have revealed the presence of numerous copy-number polymorphisms (CNPs) in the human genome and have suggested an enrichment of segmental duplications associated with these variants (Iafrate et al. 2004; Sebat et al. 2004). However, these studies used arrays with either limited genomic coverage or limited resolution with respect to regions of segmental duplication, and even current tiling-path arrays with >30,000 BAC clones (Ishkanian et al. 2004) do not achieve complete coverage of regions rich in segmental duplications (Z.C. and E.E.E., unpublished data).
Because regions flanked by segmental duplications are susceptible to rearrangement by nonallelic homologous recombination, we hypothesized that these regions represent potential hotspots of genomic instability that are prone to copy-number variation. It has been shown that several factors—including the length, sequence identity, and orientation of and the distance between duplications—influence the probability of meiotic misalignment (Stankiewicz and Lupski 2002). Most of the blocks of duplicated sequence that have been implicated in known genomic disorders are large (10–400 kb in size) and have >96% sequence identity. This level of sequence sharing between intrachromosomal sites provides ample substrate for aberrant recombination, on the basis of the estimated minimal efficient-processing segment length (Waldman and Liskay 1988). In general, the larger and more homologous the block of duplicated sequence is, the more frequently sporadic segmental aneusomy events occur. For example, the most frequently occurring microdeletion syndrome (velocardiofacial and DiGeorge syndromes; frequency 1/3,000) occurs between blocks of duplications that are in excess of 300 kb in length and that share 99.7% sequence identity (Edelmann et al. 1999; Shaikh et al. 2000).
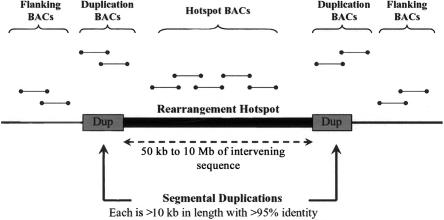
Thus, a review of the recurrent genomic disorders characterized to date suggests a strategy for the identification of novel regions of genomic instability. With a focus on regions flanked by intrachromosomal duplications that are >10 kb in length, share >95% sequence identity, and span from 50 kb to 10 Mb of intervening sequence (Stankiewicz and Lupski 2002), novel sites of genomic variation may be uncovered. On the basis of these criteria, in silico analysis of the human genome defines a map of potential rearrangement hotspots (Bailey et al. 2002). In total, 130 regions—covering 274 Mb, or ∼10% of the entire genome—are flanked by intrachromosomal duplications whose characteristics suggest a potential predisposition to genomic instability. Whereas 25 of these regions are associated with known genomic disorders, the remainder represent novel sites whose genomic architecture is susceptible to either polymorphic or disease-causing rearrangement. We have constructed a custom BAC array, termed the “segmental duplication microarray” (SD microarray), specifically targeted to these rearrangement hotspots, and we used it to investigate copy-number variation in a panel of ethnically diverse normal individuals. We report the discovery of numerous novel CNPs distributed throughout the human population and demonstrate an enrichment of copy-number variation in regions of the genome flanked by segmental duplications.
Material and Methods
A total of 130 nonredundant regions of potential genomic instability (termed “rearrangement hotspots”) were defined by the presence of intrachromosomal duplications >10 kb in length, with >95% similarity and flanking 50 kb to 10 Mb of intervening sequence in the July 2003 build of the human genome (Bailey et al. 2002; She et al. 2004b). A total of 1,986 nonredundant BACs (mean insert size 164 kb) were ultimately selected from the RPCI-11, CTC, and CTD libraries to encompass each rearrangement hotspot and were processed for construction of a microarray in accordance with protocols established elsewhere (Snijders et al. 2004) (table A1 in appendix A [online only]). When possible, three classes of BAC were selected: (1) BACs that were contained entirely within each rearrangement hotspot, (2) BACs overlapping the segmental duplications, and (3) flanking BACs in the peripheral unique sequence (as a local control) (fig. 1). During the design of this microarray, we confirmed the identity and location of all BACs by end-sequencing and by alignment of the end sequences against the human genome reference (builds 33 and 34). During our first-pass analysis, ∼93% of all selected clones were confirmed. A second round of surrogate BAC selection resulted in a total of 1,986 identity-confirmed BACs. Of these, 1,206 overlapped a rearrangement hotspot, and 760 were contained within segmental duplications. As a control, an additional 192 randomly selected single-copy clones that had been extensively tested elsewhere (Snijders et al. 2001) were incorporated into the array. In total, our BAC array consisted of 2,194 confirmed BACs. All BAC DNA was amplified by ligation-mediated PCR (Snijders et al. 2004), and each BAC was printed in triplicate on GAPS II glass slides (Corning), with a spot diameter of 80 μm and 130-μm spacing.
Figure 1.
Targeted design strategy for choosing BACs for array CGH on the basis of genomic architecture. We hypothesized that the presence of large, highly homologous segmental duplications predisposes certain regions of the genome to microdeletion and/or microduplication by nonallelic homologous recombination (Bailey et al. 2002). Termed “rearrangement hotspots,” these are defined by the presence of flanking intrachromosomal duplications >10 kb in length with >95% similarity and separated by 50 kb to 10 Mb of intervening sequence. For each region, we selected BACs that were contained entirely within each rearrangement hotspot, BACs that overlapped the segmental duplications, and BACs in the peripheral unique sequence for microarray manufacture. The SD microarray comprised a total of 2,194 BACs.
DNA was extracted from 47 lymphoblastoid cell lines (NIGMS Cell Repository), representing seven ethnicities from Asia, Europe, sub-Saharan Africa, and South America. This collection comprised 8 Chinese, 4 Japanese, 10 Czech, 2 Druze, 7 Biaka, 9 Mbuti, and 7 American Indian samples (table A1 in appendix A [online only]). All hybridizations were performed in duplicate—once with test and reference DNAs labeled with Cy3 and Cy5, respectively, and a second time with the fluorochrome dyes reversed. All hybridizations used reference DNA isolated from blood lymphocytes from a single anonymous male donor of unknown ethnicity. A single reference genome was analyzed, as opposed to a pool of normal individuals, to increase signal-to-noise ratios over sites of polymorphic variation. In cases in which the reference sample represented the rare allele (i.e., the majority of individuals showed a change in the fluorescence intensity ratio), we classified the CNPs as minor-allele variants.
Test and reference DNAs were labeled with Cy3 or Cy5 (Amersham Biosciences) by random prime labeling (Bioprime DNA Labeling System [Invitrogen]) and were mixed and purified using Sephadex G-50 Spin Columns (Roche). Labeled DNA was coprecipitated with 75 μg of COT DNA (Roche), was resuspended in 50 μl of hybridization mix (50% formamide, 10% dextran sulfate, 2× saline sodium citrate [SSC], and 4% SDS), was denatured, and was prehybridized at 37°C for 1 h to allow blocking of repetitive sequences. Array hybridizations were performed in a humid chamber at 37°C for 40 h in open rubber-cement wells on a gently rocking platform, to allow circulation of hybridization solution over the array (Snijders et al. 2004). Hybridization solution was removed under a stream of buffer (0.1 M Na2HPO4 and 0.1% Nonidet P-40; pH 8.0), and slides were washed for 15 min at 45°C in 50% formamide and 2× SSC, were rinsed in buffer, and were mounted (in 90% glycerol, 10% PBS, and 1 μM 4′,6-diamidino-2-phenylindole) under a glass cover slip before being scanned with a custom-built CCD system.
Array image analysis and normalization were performed using UCSF Spot and Sproc software (Jain et al. 2002). BACs for which only one of the triplicates printed on the array yielded data, or for which the standard deviation of log2 ratio for the triplicates was >0.2, were removed from final analysis. Furthermore, we discarded BACs that failed to yield data in <20% of cases. For each hybridization experiment, we established a threshold log2 ratio of 2 SDs from the mean of all autosomal clones, and BACs that exceeded this threshold in both independent dye-swap experiments were classified as variant.
For comparison, we also hybridized a subset of our samples to a genomewide BAC microarray containing clones selected to minimize segmental-duplication content, spaced at an average resolution of ∼1.4 Mb throughout the genome (Snijders et al. 2001). For these experiments, we used an identical protocol, except for the use of a different reference DNA (from GM15724, a male Czech). Verification of array CGH results by FISH was performed on the lymphoblastoid cell lines derived from the individuals used for array profiling, in accordance with standard procedures, by use of isolated BAC DNA as a probe source (Nucleobond [BD Biosciences]) (Pinkel et al. 1986).
Results
Initial Array Validation
To assess the performance of the SD microarray and to establish appropriate thresholds for the detection of copy-number changes, we performed a series of test hybridizations. First, we analyzed three previously characterized individuals possessing one, four, or six copies of the 15q11-q13 region (Locke et al. 2004). There was a strong correlation (r2=0.945) between copy number and relative fluorescence ratios yielded by BACs within the variant region, which demonstrates the ability of our array to detect bona fide rearrangements and to reliably distinguish deletions, duplications, and triplications. Second, to determine the false-positive rate, we performed four self-versus-self hybridizations. Results indicated that the use of the threshold of log2 hybridization ratios deviating >2 SDs from the mean in both replicates of an experiment yields a false-positive rate of ∼3 per 4,000 clones, or ∼0.08% (table A1 in appendix A [online only]). As has been reported elsewhere (Vissers et al. 2003), we found that, by performing all hybridizations in duplicate and incorporating a dye reversal, we significantly reduced the false-positive rate.
Assessment of CNPs in the Human Population
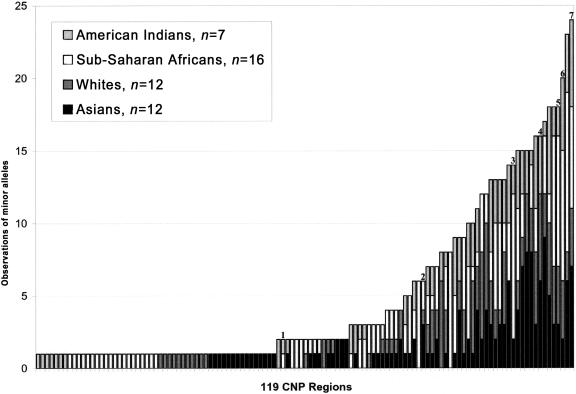
Using the SD microarray, we analyzed a diverse panel of 47 unrelated humans representing four continental groups. On the basis of our conservative criteria, we identified 160 variant BACs among these 47 individuals. We identified 27 regions where neighboring or overlapping clones yielded concordant results, which suggested that some copy-number variations extended over regions of several hundred kilobases. Under the assumption that concordantly variant neighboring clones separated by <250 kb represent the same copy-number variation, the 160 variant clones correspond to 119 nonredundant regions. Whereas 66 of these 119 regions were polymorphic in multiple individuals, 53 were observed in only a single individual (fig. 2 and table A2 in appendix A [online only]). Conversely, 38 regions were variant in >10% of the individuals studied (fig. 2). The proportions of gains and losses were approximately equal (table 1).
Figure 2.
Observations of minor alleles in CNP regions. The 47 samples were categorized as American Indian, sub-Saharan African (Biaka and Mbuti), white (Czech and Druze), and Asian (Chinese and Japanese), and the 119 CNPs identified were plotted (see table A2 in appendix A [online only]). In cases in which the reference sample represented the rare allele (i.e., the majority of individuals showed a change in fluorescence intensity ratio), we classified the CNPs as minor-allele variants. Some previously identified CNPs are indicated by numerals above the bars: 1 = CHRNA7/CHRFAM7A at 15q13.3 (Riley et al. 2002); 2 = IGVH/SLC6A8/CDM pseudogene cluster at 16p11.2 (Barber et al. 1999); 3 = CCL3-L1/CCL4-L1 at 17q12 (Townson et al. 2002); 4 = IGHG1 gene cluster at 14q32.33 (Sasso et al. 1995); 5 = IGL gene cluster at 22q11.22 (van der Burg et al. 2002); 6 = NF1/IGVH/GABRA5 pseudogene amplification at 15q11.2-q13 (Fantes et al. 2002); and 7 = β-defensin gene cluster at 8p23.1 (Hollox et al. 2003).
Table 1.
Summary of CNPs Detected by Array CGH[Note]
|
No. of CNPs |
||||||||
| Ethnic Group | No. of SubjectsAnalyzed | Gains | Losses | Showing BothGain and Loss | Detected inPresent Study | Detected bySebat et al. (2004) | Detected byIafrate et al. (2004) | Novel |
| American Indians | 7 | 9 | 22 | 13 | 44 | 18 | 9 | 22 |
| Sub-Saharan Africans | 16 | 27 | 31 | 17 | 75 | 24 | 14 | 43 |
| Whites | 12 | 17 | 22 | 12 | 51 | 16 | 13 | 25 |
| Asians | 12 | 26 | 27 | 15 | 68 | 21 | 14 | 37 |
| Nonredundant total | 47 | 52 | 50 | 17 | 119 | 22 | 21 | 73 |
Note.— CNPs were scored as either increased or decreased log2 ratios (i.e., gains or losses), relative to the reference DNA. In total, 46 (39%) of 119 of the CNPs we identified have been reported previously, which thus validates the ability of our array to detect known copy-number variations. In our sample population, we identified 22 (71%) of the 31 CNPs detected by Sebat et al. (2004) and 22 (39%) of the 57 CNPs detected by Iafrate et al. (2004) that were represented on the SD microarray.
Validation of CNPs Detected by Array CGH
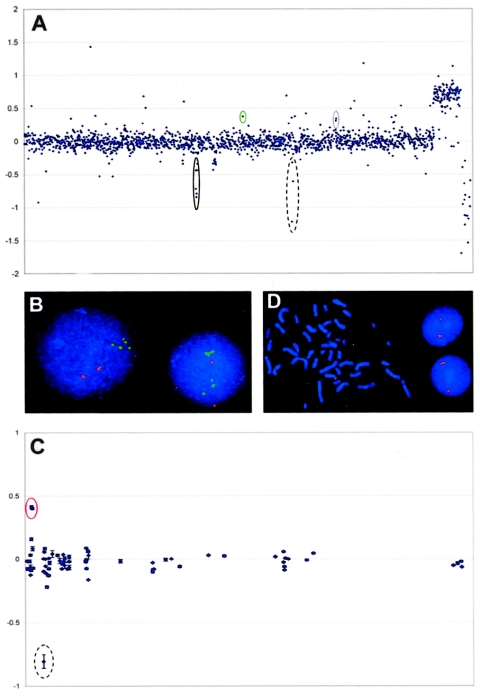
Two analyses were performed to assess the validity of our hybridization results. First, we compared sites of copy-number variation with those identified recently in two other studies (Iafrate et al. 2004; Sebat et al. 2004). Of the sites we detected, 39% (46 of 119) had been reported elsewhere (within a 250-kb overlap), which thus validates the ability of our array to detect known copy-number alterations and which indicates that 73 of the 119 genomic regions we identified in the present study are novel. A comparison of our set of CNPs with those detected in a similar study that used ROMA (Sebat et al. 2004) revealed excellent concordance (table 1). Of the 31 CNPs detected by ROMA that were represented on the SD microarray, we detected 22 (71%). Seven of the nine variations that we did not detect were observed by Sebat et al. (2004) in only one individual, and therefore those variations probably represent rare polymorphisms that were not represented in our sample population. We also identified many other previously reported copy-number changes—for example, the β-defensin gene cluster at 8p23.1 (Hollox et al. 2003), the IGHG1 gene cluster at 14q32.33 (Sasso et al. 1995), and the IGVH/SLC6A8/CDM pseudogene cluster at 16p11.2 (Barber et al. 1999) (fig. 3A). Figure 4 shows an example of the pattern of copy-number variation detected on chromosome 15 by use of the SD microarray. A genomewide map showing all 119 CNPs detected is shown in figure 5.
Figure 3.
CNPs detected by array CGH, and verification by FISH. A, Genomic profile of GM10473A (a Biaka female). After quality filtering, 1,967 BAC clones were ordered sequentially from 1pter to Yqter on the basis of physical location in the July 2003 genome assembly (X-axis). Each data point represents the mean log2 ratio of test:reference intensity of the three replicate spots of each BAC from a single hybridization experiment (Y-axis). Each hybridization was repeated with reverse labeling (dye swap), and clones that yielded log2 ratios deviating >2 SDs from the mean of all autosomal clones in both experiments were classified as variant. The reference DNA is male; thus, clones located on the sex chromosomes show log2 ratios consistent with female:male hybridization. Three previously reported CNPs are circled: the β-defensin gene cluster at 8p23.1 (Hollox et al. 2003) (solid circle), the IGHG1 gene cluster at 14q32.33 (Sasso et al. 1995) (dashed circle), and the IGVH/SLC6A8/CDM pseudogene cluster at 16p11.2 (Barber et al. 1999) (dotted circle). A novel variant locus, RP11-136P13 (chromosome 10:81097351–81263857 [green circle]), yielded log2 ratios of 0.44 and 0.48 in replicate hybridizations. B, FISH confirmation of RP11-136P13, which is composed entirely of segmental-duplication sequence and therefore shows dual signals when used as a FISH probe (green). However, the presence of an additional signal in interphase nuclei shows the polymorphic nature of this locus in GM10473A. A control probe (red) confirms that the cells are diploid. This duplication was also observed in two other subjects. C, Array CGH profile of chromosome 8 in GM10493 (a Biaka). Clones are ordered by physical distance from 8pter in kb (X-axis), with error bars showing the SD of the log2 ratios from the three replicate spots. Two adjacent clones, RP11-159F11 and RP11-46M15 (red circle), yielded log2 ratios of 0.41, indicating the presence of a duplication. A BAC that yielded a log2 ratio of −0.81 in this experiment (dashed circle) was not confirmed in the replicate dye-swapped hybridization; thus, it was classified as nonvariant. D, Use of RP11-159F11 as a FISH probe, showing increased signal intensity on a chromosome 8 homologue in metaphase cells, which resolves to dual signals in interphase cells, confirming the presence of a duplication of this region. An overlapping but nonidentical duplication of 8p23.2 was also observed in GM17051.
Figure 4.
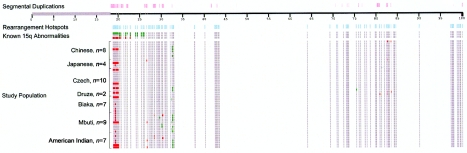
CNPs detected on chromosome 15. For each of the 47 individuals studied, relative duplications (green) and deletions (red) are represented, with invariant BACs shown in gray. The different sizes of the deletions and duplications in the three patients with known copy-number changes in the 15q11-q13 region (Locke et al. 2004) are clearly visible. Note that, as expected, none of the 47 normal individuals (94 chromosomes) in our population survey showed variation over these regions associated with disease. Also shown are the locations of segmental duplications >10 kb in size with >95% identity (pink) and of rearrangement hotspots represented on the array (blue), which illustrates the targeted nature of our array. Each tick mark represents 5 Mb, with gaps in the sequence assembly represented by gray bars.
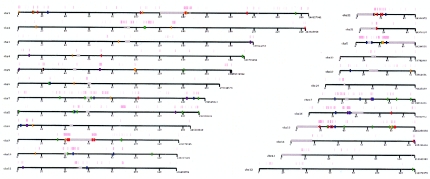
Figure 5.
Genomewide map of sites of copy-number variation detected by array CGH. The location of CNPs are indicated by colored bars on each chromosome. Relative deletions identified in a single individual are shown in orange, and deletions observed in multiple individuals are shown in red. Relative duplications identified in a single individual are shown in green, and duplications observed in multiple individuals are shown in blue. CNPs that showed both relative deletion and relative duplication in different individuals are shown in purple (not to scale). Also shown are the locations of segmental duplications >10 kb in size with >95% identity (pink bars above each chromosome). Each tick mark represents 20 Mb, with gaps in the sequence assembly represented by gray bars.
Second, we selected 11 BACs that showed putative duplication or deletion by array CGH for use as FISH probes (fig. 3 and table A2 in appendix A [online only]). Although multiple signals were evident in some cases, as a result of the presence of segmental duplications in the BAC probe, the results in metaphase and/or interphase nuclei for 7 of the 11 BACs were consistent with copy-number changes at these loci. For two loci, copy number could not be reliably determined because of the high segmental-duplication content; thus, results were ambiguous. For the remaining two loci, FISH results did not support the presence of CNPs. However, at one of these loci (CTD-3185D7), results by array CGH showed copy-number variation in seven of the individuals we tested, and, in addition, this same locus was independently reported in another study (Iafrate et al. 2004). Thus, it seems unlikely that this represents a false-positive result, and this structural variation simply may not be easily resolved by FISH. For the remaining BAC (RP11-325E8), only 1 of the 47 individuals analyzed by array CGH showed a log2 ratio >2 SDs from the mean for this locus, which suggests that this is a potential false-positive result.
Analysis of variation with respect to ethnicity revealed very little population stratification. Almost all of the CNPs detected were present in multiple ethnic groups, with no apparent clustering with respect to geographic origin. Among the 50 regions that were polymorphic in more than two individuals, only two CNPs (in regions 12 and 91) were confined to a single ethnicity, being present specifically in sub-Saharan Africans (table A2 in appendix A [online only]). It is surprising, however, that the African population did not show significantly more variation than any other continental group (table 1 and fig. 2).
Analysis of CNPs
Analysis of the sequence properties of structurally variant regions revealed some important trends. We observed a significant enrichment of copy-number variation within rearrangement hotspots (defined by the presence of flanking intrachromosomal duplications >10 kb in length, with >95% similarity and separated by 50 kb to 10 Mb of intervening sequence [fig. 1]), when those regions were compared with the control loci (Bailey et al. 2002). Of the 718 BACs that were contained within rearrangement-hotspot regions, 113 (15.7%) were variant, compared with 46 (4.1%) of the 1,110 BACs that were located outside of hotspot regions (3.8-fold enrichment; χ2=73.8; P<.000001). There was also an enrichment of rearrangement-hotspot sequence within the more frequent CNPs. The 96 BACs that were variant in multiple individuals contained an average of 76.4% hotspot sequence, compared with an average of 50.8% hotspot sequence in the 64 BACs that were variant in only a single individual. In total, CNPs were identified in 51 (39%) of 130 rearrangement hotspots.
It is interesting that both intrachromosomal and interchromosomal segmental duplications showed an association with copy-number variation. Variant BACs contained an average of 52.1% intrachromosomal duplication, compared with 12.0% in nonvariant BACs (4.3-fold enrichment). Similarly, interchromosomal duplications comprised 28.8% of variant BACs, compared with 5.9% of nonvariant BACs (4.9-fold enrichment). However, despite the accumulation of segmental duplications within pericentromeric and subtelomeric regions (Bailey et al. 2002), we observed no significant bias for copy-number variations within these regions of the genome.
Regions showing evidence of copy-number variation were not particularly gene poor. The 160 BACs exhibiting copy-number variation completely encompass a total of 108 genes and partially overlap a further 33 coding regions (table A3 in appendix A [online only]). This includes numerous gene families, consistent with the observed enrichment of segmental duplications with CNPs.
Hybridizations to a Genomewide Array
A subset of the samples were also hybridized to a second BAC microarray containing clones spaced at an average resolution of ∼1.4 Mb throughout the genome (Snijders et al. 2001). With the use of this nontargeted array, only 8 (0.3%) of 2,460 BACs were variant in the eight individuals analyzed (table A4 in appendix A [online only]), compared with 82 (3.7%) of 2,194 BACs that were analyzed using the SD microarray, which represents an 11.5-fold difference. Two of the eight BACs that were classified as variant on the genomewide array overlapped clones on the SD microarray that were also classified as variant, thus yielding concordant results.
Discussion
We have performed an analysis of copy-number variation in the human genome by using a targeted BAC array. Although many of the CNPs we observed were identified in previous studies—which thus validates the ability of our array to detect bona fide polymorphisms—our targeted experimental approach significantly improves the ascertainment of structural rearrangements. We grouped BACs into those that were contained within each rearrangement hotspot, those that overlapped the segmental duplications, or those that were in the flanking unique sequence (fig. 1). We observed a 4–5-fold enrichment of CNPs within regions that were flanked by or contained large, highly homologous segmental duplications, as compared with control clones. These data indicate that genomic duplication architecture is strongly associated with CNP in the human genome. Presumably, segmental duplications mediate the deletion or duplication of the intervening sequence via nonallelic homologous recombination, supporting the notion that certain regions of the genome are predisposed to rearrangement as a result of their underlying genomic architecture (Stankiewicz and Lupski 2002; Stankiewicz et al. 2003; Bailey et al. 2004). Consistent with this hypothesis, we also observed an enrichment of hotspot sequence in the more common CNPs, suggesting that the presence of flanking duplications renders these sites prone to recurrent rearrangement. In total, we detected CNPs within 51 (39%) of 130 rearrangement hotspots. It is surprising that, in most cases, CNPs were not continuous across entire hotspot regions. Several factors could account for this observation. The highly complex segmental-duplication architecture bracketing these regions means that there are multiple potential sites of rearrangement and that these configurations may differ among individuals. This provides the potential for alternate sites of rearrangement. Such an effect has been observed for the Prader-Willi and Angelman syndromes, for which atypical breakpoints have been mapped to other duplication structures (Amos-Landgraf et al. 1999; Locke et al. 2004). Other possibilities include genomic misassembly of these regions and incorrect mapping of the arrayed BAC clones. The latter possibility, however, seems less likely, because BAC end-sequence confirmation and/or FISH verification was performed for every clone on the SD microarray. Finally, if nonallelic homologous recombination is the underlying mechanism of rearrangement in these regions, then additional repair or recombination events could result in more-complex rearrangements. Nevertheless, this study demonstrates that segmental duplications play an important role in normal variation as well as in genomic disease, defining hotspots of rearrangement that are susceptible to variation among the normal population. We hypothesize that the remainder of hotspots that did not show variation in our sample population represent excellent candidate sites that may be associated with genomic disease, and this survey provides the necessary baseline to begin future studies on disease populations.
Our analysis of different ethnic groups allowed an assessment of population-specific copy-number variants. However, almost without exception, the CNPs were present in multiple populations. Of the 50 regions that were polymorphic in more than two individuals, only 2 were confined to a single ethnic group. One of these CNPs (in region 12) represents a partial deletion of two overlapping BACs at 1q31.3, observed in 6 of the 16 sub-Saharan Africans studied. The region of overlap defined by these two BACs is composed of 70 kb of sequence flanked by a pair of intrachromosomal segmental duplications with 91% identity. We suggest that this CNP likely represents a deletion of this 70-kb segment, mediated by nonallelic homologous recombination between the flanking duplications. Such ethnic predilections for copy-number variation have recently been shown to be an important determinant in disease association studies (Gonzalez et al. 2005).
The occurrence of CNPs across multiple ethnic populations suggests that these structural rearrangements either (1) are evolutionarily ancient, having occurred prior to the separation of these ethnic groups, or (2) are recurrent events that have occurred independently in multiple founders. Distinguishing between these two hypotheses will require the integration of SNP haplotype and array CGH data. If CNPs play a role in phenotypic variation or susceptibility to common diseases (Buckland 2003; Gonzalez et al. 2005), the occurrence of the same CNP on multiple haplotype backgrounds as a result of recurrent rearrangement could confound conventional SNP-based association studies.
We also observed a significant enrichment of segmental duplications within regions of CNP, suggesting that these duplications themselves show marked variation in copy number. This is consistent with previous observations of polymorphic copy-number variation in segmental duplications (Fredman et al. 2004). A striking example of such polymorphism is illustrated by a series of CNPs within the pericentromeric region of chromosome 9, which is composed almost exclusively of large, highly homologous (>99% identity) blocks of intrachromosomal duplication that our data suggest vary in copy number. We hypothesize that many CNPs in the human genome may be due to the presence or absence of evolutionarily recent segmental-duplication events that have not yet become fixed within the population, providing evidence that the process of duplicative transposition is ongoing within the human population. It should be noted, however, that this study likely underestimates the amount of copy-number variation that exists within regions of segmental duplication. By definition, these sequences occur at multiple genomic locations, with some present in >40 copies (Horvath et al. 2003); thus, unlike for unique portions of the genome, the gain or loss of a single duplication will often be below the resolution of array CGH, which undoubtedly biases our results. Despite the sequence complexity of these clones, they provided valuable information. We observed significantly more copy-number variation in BACs that contained segmental duplications than in unique regions of the genome. Indeed, the parallel analysis of samples on both our SD microarray and a second array with BACs spaced at an average resolution of ∼1.4 Mb throughout the genome showed a >10-fold increase in the ascertainment of CNPs with the use of the duplication-targeted array. This suggests that the inclusion of BACs enriched in segmental-duplication content significantly increases the ascertainment of structurally polymorphic regions. We suggest that either the copy number or the sequence composition of these regions must differ dramatically among different individuals to produce significant differences on array CGH. However, until targeted sequencing of the variant regions from multiple individuals occurs, the true nature of the variation at these loci will not be fully ascertained.
A total of 141 genes either completely or partially overlapped variant BACs (table A3 in appendix A [online only]). However, because the majority of CNPs do not encompass complete BACs, and, because their boundaries are difficult to define accurately by use of array CGH, it is likely that only a subset of these genes vary in copy number. Because many of the genes previously confirmed as polymorphic have functions in metabolism and immunity, alterations in copy number of these genes often have profound effects on an individual's metabolic rate or resistance to environmental pathogens (Lackner et al. 1991; McLellan et al. 1997; Dalen et al. 1998; Rao et al. 2000; Sprenger et al. 2000; Townson et al. 2002; van der Burg et al. 2002; Hollox et al. 2003; Gonzalez et al. 2005) and, as such, are likely to be significant susceptibility factors for some common human diseases.
Although results yielded by array CGH only reveal copy-number changes relative to the reference sample used, deletions and duplications were observed in our sample population at approximately equal frequencies (they were observed in 50 and 52 regions, respectively). Of note, a further 17 regions showed both relative deletion and relative duplication in different individuals, which indicates that these loci have multiple variations in copy number. These may represent a variable number of tandem repeats or, alternatively, the product of reciprocal nonhomologous recombination events, as has been observed at sites of known genomic disorders (Bi et al. 2003). Similarly, many other loci exhibited a wide range of hybridization ratios among different individuals, suggesting the presence of multiple alleles at these loci, such as that observed at the AMY1A gene cluster (Iafrate et al. 2004).
As in a previous study by Iafrate et al. [2004], we observed a significant increase in the frequency of genome assembly gaps at sites of copy-number variation. Of the 160 variant BACs, 42 (26%) were located within 100 kb of an assembly gap, compared with 130 (8%) of the 1,663 nonvariant BACs (a 3.4-fold enrichment; χ2=58.0; P<.000001), suggesting that the presence of these polymorphisms may represent a significant impediment to correct genome assembly. However, because there was also an association of segmental duplications with CNPs, which are themselves known to be correlated with gap location (Eichler et al. 2004), it is difficult to draw firm conclusions about the causality of this relationship. The coinciding of CNPs, segmental duplications, and sequence assembly gaps highlights the continued importance of resolving and studying the nature of these biologically relevant regions.
To facilitate the study of structural variation, we have compiled a Web interface, based on the UCSC Human Genome Browser, that displays all CNPs reported both here and in previous studies. Our Structural Variation Database (see Web Resources) includes an additional 297 sites of fine-scale variation (resolution ∼8 kb) identified by fosmid paired-end sequence analysis (Tuzun et al. 2005), including sites of inversion not detectable by array-based approaches, as well as CNPs identified in two other studies (Iafrate et al. 2004; Sebat et al. 2004). Given the importance of this type of variation for genetic disease, a coordinated effort should be made to incorporate results and the underlying raw data into central repositories of data on human variation (such as the dbSNP and the Data Coordinating Center for the International HapMap Consortium).
In conclusion, this study demonstrates that segmental duplications define hotspots of chromosomal rearrangement in the human genome. Our data suggest not only that intrachromosomal segmental duplications frequently mediate polymorphic rearrangement of intervening sequence via nonallelic homologous recombination but also that segmental duplications themselves are often variant in copy number. Thus, the consideration of genomic architecture can significantly enrich the detection of large-scale variation. The array we have constructed includes BACs covering 25 regions associated with known pathogenic microdeletions/duplications (Stankiewicz and Lupski 2002) and includes an additional 105 regions for which the pathological relevance has not yet been determined, thus representing an excellent resource for the study of genomic disease. Although we did not detect CNPs in 79 of the 130 rearrangement hotspots in our sample population of normal individuals, we suggest that these hotspots represent excellent candidate sites of recurrent rearrangement that may be associated with novel genomic disorders (Bailey et al. 2002; Mehan et al. 2004). This survey of normal variation provides the requisite baseline for distinguishing copy-number variation associated with genomic disease.
Supplementary Material
Acknowledgments
We thank Cassy Gulden and Marla Eichler, for technical assistance. This work was supported in part by National Institutes of Health grant HD043569 (to E.E.E.).
Appendix A
Tables A1, A2, and A3 are tab-delimited ASCII files that can be imported into a Microsoft Excel spreadsheet for data manipulation and plotting.
Table A1 is a complete list of reporting BAC clones in 47 individuals and self-versus-self hybridizations.
Table A2 is a summary of the 160 BACs identified as variant by use of the SD microarray. For each individual, results of the duplicate dye-swapped hybridizations are shown. BACs that yielded log2 hybridization ratios concordantly deviating >2 SDs from the mean in both independent dye-swap experiments were classified as variant.
Table A3 is a summary of the 141 genes either completely or partially overlapping variant BACs.
Table A4.
Variant BACs Identified by Use of a Genomewide BAC Microarray with an Average Resolution of 1.4 Mb[Note]
|
Build 34 Position |
Log2 Ratio for Individual |
|||||||||||||||||||
| Clone ID | Chromosome | Overlap with SD Microarray BACs | Start | End | GM10971 | GM10971 | GM10976 | GM10976 | GM10469A | GM10469A | GM17015 | GM17015 | GM15733 | GM15733 | GM11521 | GM11521 | JK1061 | JK1061 | P86GA | P86GA |
| RP11-80G24 | 1 | None | 77139377 | 77212787 | −.03356 | −.074366 | −.019094 | −.026857 | .072919 | .035307 | .017376 | .010557 | .058324 | −.045758 | −.024474 | −.00556 | .353048 | .303538 | .092873 | −.022958 |
| RP11-88L18 | 5 | None | 17465158 | 17646347 | .257222 | .04048 | .460069 | .167617 | 1.156814 | .899637 | .345384 | .237609 | .235486 | .096557 | .514499 | .31079 | .914636 | .700019 | 1.09805 | .851648 |
| RP11-54H13 | 6 | None | 32633341 | 32815611 | −.144892 | −.385706 | .011892 | −.124511 | −.186515 | −.261263 | −.23622 | −.358063 | −.321434 | −.507876 | −.15266 | −.421456 | −.231978 | −.399803 | −.251672 | −.248022 |
| RP11-43B19 | 6 | None | 160895056 | 160989860 | −.088462 | −.194265 | .119669 | .058714 | .024056 | −.02856 | .06629 | .22024 | −.219736 | −.208599 | .044621 | −.15075 | −.068096 | −.216495 | −.30175 | −.421675 |
| RP11-122N11 | 8 | CNP region 45 | 7295548 | 7755862 | .084569 | −.172854 | −.111666 | −.372474 | .502866 | .250299 | .114427 | −.007112 | .069663 | .045114 | .179389 | −.076298 | .672897 | .376702 | .434174 | .176032 |
| RP11-95J4 | 9 | None | 111201223 | 111218764 | −.202168 | −.051224 | −.010057 | −.020496 | −.147954 | .112502 | −.067557 | −.129185 | −.166902 | −.041462 | −.194072 | .035134 | .297673 | .494059 | −.434126 | −.342147 |
| RP11-41D21 | 10 | None | 107840549 | 107948457 | .053775 | .164889 | .063302 | .106675 | −.169527 | .008357 | −.097841 | .124418 | −.438486 | −.305911 | .135821 | .00394 | ||||
| CTB-305D20 | 17 | CNP region 90 | 45231067 | 45431400 | −.292745 | −.127378 | .144236 | .315087 | −.108068 | .169836 | .36981 | .371404 | −.224268 | .041454 | −.126897 | .093954 | .133636 | .270973 | .314351 | .426032 |
| 2 × SD | .218 | .254 | .249 | .205 | .219 | .229 | .18 | .226 | .319 | .264 | .229 | .233 | .217 | .247 | .275 | .22 | ||||
Note.— The genomewide BAC microarray contained clones selected to minimize segmental-duplication content, spaced at an average resolution of ∼1.4 Mb throughout the genome (Snijders et al. 2001). For each individual, results of the duplicate dye-swapped hybridizations are shown. BACs that yielded log2 hybridization ratios concordantly deviating >2 SDs from the mean in both independent dye-swap experiments were classified as variant.
Web Resources
The URLs for data presented herein are as follows:
- Chromosome Anomaly Register, http://www.som.soton.ac.uk/research/geneticsdiv/anomaly%20register/
- Structural Variation Database, http://paralogy.gs.washington.edu/structuralvariation
- UCSC Human Genome Browser, http://genome.ucsc.edu/
References
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol L, Pujana MA, Cheung J, Scherer SW, Estivill X (2003) Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum Mol Genet 12:2201–2208 10.1093/hmg/ddg223 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE (2004) Hotspots of mammalian chromosomal evolution. Genome Biol 5:R23 10.1186/gb-2004-5-4-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297:1003–1007 10.1126/science.1072047 [DOI] [PubMed] [Google Scholar]
- Barber JC, Reed CJ, Dahoun SP, Joyce CA (1999) Amplification of a pseudogene cassette underlies euchromatic variation of 16p at the cytogenetic level. Hum Genet 104:211–218 10.1007/s004390050938 [DOI] [PubMed] [Google Scholar]
- Bi W, Park SS, Shaw CJ, Withers MA, Patel PI, Lupski JR (2003) Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am J Hum Genet 73:1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland PR (2003) Polymorphically duplicated genes: their relevance to phenotypic variation in humans. Ann Med 35:308–315 10.1080/07853890310001276 [DOI] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR (1994) Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3:223–228 [DOI] [PubMed] [Google Scholar]
- Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui LC, Scherer SW (2003) Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol 4:R25 10.1186/gb-2003-4-4-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen XN, Furey TS, et al (2001) Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409:953–958 10.1038/35057192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalen P, Dahl ML, Ruiz ML, Nordin J, Bertilsson L (1998) 10-Hydroxylation of nortriptyline in white persons with 0, 1, 2, 3, and 13 functional CYP2D6 genes. Clin Pharmacol Ther 63:444–452 10.1016/S0009-9236(98)90040-6 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE (1999) Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet 64:1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE (2001) Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 17:661–669 10.1016/S0168-9525(01)02492-1 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Clark RA, She X (2004) An assessment of the sequence gaps: unfinished business in a finished human genome. Nat Rev Genet 5:345–354 10.1038/nrg1322 [DOI] [PubMed] [Google Scholar]
- Fantes JA, Mewborn SK, Lese CM, Hedrick J, Brown RL, Dyomin V, Chaganti RS, Christian SL, Ledbetter DH (2002) Organisation of the pericentromeric region of chromosome 15: at least four partial gene copies are amplified in patients with a proximal duplication of 15q. J Med Genet 39:170–177 10.1136/jmg.39.3.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredman D, White SJ, Potter S, Eichler EE, Den Dunnen JT, Brookes AJ (2004) Complex SNP-related sequence variation in segmental genome duplications. Nat Genet 36:861–866 10.1038/ng1401 [DOI] [PubMed] [Google Scholar]
- Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, Weber JL, Ledbetter DH, Zuffardi O (2001) Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 68:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelli G, Pujana MA, Patricelli MG, Russo S, Giardino D, Larizza L, Cheung J, Armengol L, Schinzel A, Estivill X, Zuffardi O (2003) Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum Mol Genet 12:849–858 10.1093/hmg/ddg101 [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O'Connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK (2005) The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:1434–1440 10.1126/science.1101160 [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Armour JA, Barber JC (2003) Extensive normal copy number variation of a β-defensin antimicrobial-gene cluster. Am J Hum Genet 73:591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath JE, Gulden CL, Bailey JA, Yohn C, McPherson JD, Prescott A, Roe BA, de Jong PJ, Ventura M, Misceo D, Archidiacono N, Zhao S, Schwartz S, Rocchi M, Eichler EE (2003) Using a pericentromeric interspersed repeat to recapitulate the phylogeny and expansion of human centromeric segmental duplications. Mol Biol Evol 20:1463–1479 10.1093/molbev/msg158 [DOI] [PubMed] [Google Scholar]
- Iafrate JA, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, Ling V, MacAulay C, Lam WL (2004) A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet 36:299–303 10.1038/ng1307 [DOI] [PubMed] [Google Scholar]
- Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D (2002) Fully automatic quantification of microarray image data. Genome Res 12:325–332 10.1101/gr.210902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD (2000) Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res 10:597–610 10.1101/gr.10.5.597 [DOI] [PubMed] [Google Scholar]
- Lackner C, Boerwinkle E, Leffert CC, Rahmig T, Hobbs HH (1991) Molecular basis of apolipoprotein (a) isoform size heterogeneity as revealed by pulsed-field gel electrophoresis. J Clin Invest 87:2153–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Archidiacono N, Misceo D, Cardone MF, Deschamps S, Roe B, Rocchi M, Eichler EE (2003a) Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol 4:R50 10.1186/gb-2003-4-8-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE (2003b) Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res 13:347–357 10.1101/gr.1003303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Segraves R, Nicholls RD, Schwartz S, Pinkel D, Albertson DG, Eichler EE (2004) BAC microarray analysis of 15q11-q13 rearrangements and the impact of segmental duplications. J Med Genet 41:175–182 10.1136/jmg.2003.013813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan RA, Oscarson M, Alexandrie A, Seidegård J, Price Evans DA, Rannug A, Ingelman-Sundberg M (1997) Characterization of a human glutathione S-transferase μ cluster containing a duplicated GSTM1 gene that causes ultrarapid enzyme activity. Mol Pharmacol 52:958–965 [DOI] [PubMed] [Google Scholar]
- Mehan MR, Freimer NB, Ophoff RA (2004) A genome-wide survey of segmental duplications that mediate common human genetic variation of chromosomal architecture. Hum Genomics 1:335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW (2001) A 1.5 million–base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 10.1038/ng753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Hoffmann E, Zia M, Bodin L, Zeman M, Sellers EM, Tyndale RF (2000) Duplications and defects in the CYP2A6 gene: identification, genotyping, and in vivo effects on smoking. Mol Pharmacol 58:747–755 [DOI] [PubMed] [Google Scholar]
- Riley B, Williamson M, Collier D, Wilkie H, Makoff A (2002) A 3-Mb map of a large segmental duplication overlapping the α7-nicotinic acetylcholine receptor gene (CHRNA7) at human 15q13-q14. Genomics 79:197–209 10.1006/geno.2002.6694 [DOI] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE (2002) Segmental duplications and the evolution of the primate genome. Nat Rev Genet 3:65–72 10.1038/nrg705 [DOI] [PubMed] [Google Scholar]
- Sasso EH, Buckner JH, Suzuki LA (1995) Ethnic differences of polymorphism of an immunoglobulin VH3 gene. J Clin Invest 96:1591–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528 10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS (2000) Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet 9:489–501 10.1093/hmg/9.4.489 [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Bi W, Lupski JR (2002) Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 71:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Horvath JE, Jiang Z, Liu G, Furey TS, Christ L, Clark R, Graves T, Gulden CL, Alkan C, Bailey JA, Sahinalp C, Rocchi M, Haussler D, Wilson RK, Miller W, Schwartz S, Eichler EE (2004a) The structure and evolution of centromeric transition regions within the human genome. Nature 430:857–864 10.1038/nature02806 [DOI] [PubMed] [Google Scholar]
- She X, Jiang Z, Clark RA, Liu G, Cheng Z, Tuzun E, Church DM, Sutton G, Halpern AL, Eichler EE (2004b) Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431:927–930 10.1038/nature03062 [DOI] [PubMed] [Google Scholar]
- Siniscalco M, Robledo R, Orru S, Contu L, Yadav P, Ren Q, Lai H, Roe B (2000) A plea to search for deletion polymorphism through genome scans in populations. Trends Genet 16:435–437 10.1016/S0168-9525(00)02101-6 [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, et al (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837 10.1038/nature01722 [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG (2001) Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 29:263–264 10.1038/ng754 [DOI] [PubMed] [Google Scholar]
- Snijders AM, Segraves R, Blackwood S, Pinkel D, Albertson DG (2004) BAC microarray-based comparative genomic hybridization. Methods Mol Biol 256:39–56 [DOI] [PubMed] [Google Scholar]
- Sprenger R, Schlagenhaufer R, Kerb R, Bruhn C, Brockmoller J, Roots I, Brinkmann U (2000) Characterization of the glutathione S-transferase GSTT1 deletion: discrimination of all genotypes by polymerase chain reaction indicates a trimodular genotype-phenotype correlation. Pharmacogenetics 10:557–565 10.1097/00008571-200008000-00009 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 10.1016/S0168-9525(02)02592-1 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, Elizondo L, Park SS, Lupski JR (2003) Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet 72:1101–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson JR, Barcellos LF, Nibbs RJB (2002) Gene copy number regulates the production of the human chemokine CCL3-L1. Eur J Immunol 32:3016–3026 [DOI] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE (2005) Fine-scale structural variation of the human genome. Nat Genet, published online May 15 [DOI] [PubMed] [Google Scholar]
- van der Burg M, Barendregt BH, van Gastel-Mol EJ, Tümkaya T, Langerak AW, van Dongen JJM (2002) Unraveling of the polymorphic Cλ2-Cλ3 amplification and the Ke+Oz− polymorphism in the human Iλg locus. J Immunol 169:271–276 [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, Choy CO, Straatman H, van der Vliet W, Huys EH, van Rijk A, Smeets D, van Ravenswaaij-Arts CM, Knoers NV, van der Burgt I, de Jong PJ, Brunner HG, van Kessel AG, Schoenmakers EF, Veltman JA (2003) Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet 73:1261–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman AS, Liskay RM (1988) Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol Cell Biol 8:5350–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.