Abstract
Endometriosis is a common gynecological disease that affects up to 10% of women in their reproductive years. It causes pelvic pain, severe dysmenorrhea, and subfertility. The disease is defined as the presence of tissue resembling endometrium in sites outside the uterus. Its cause remains uncertain despite >50 years of hypothesis-driven research, and thus the therapeutic options are limited. Disease predisposition is inherited as a complex genetic trait, which provides an alternative route to understanding the disease. We seek to identify susceptibility loci, using a positional-cloning approach that starts with linkage analysis to identify genomic regions likely to harbor these genes. We conducted a linkage study of 1,176 families (931 from an Australian group and 245 from a U.K. group), each with at least two members—mainly affected sister pairs—with surgically diagnosed disease. We have identified a region of significant linkage on chromosome 10q26 (maximum LOD score [MLS] of 3.09; genomewide P = .047) and another region of suggestive linkage on chromosome 20p13 (MLS = 2.09). Minor peaks (with MLS > 1.0) were found on chromosomes 2, 6, 7, 8, 12, 14, 15, and 17. This is the first report of linkage to a major locus for endometriosis. The findings will facilitate discovery of novel positional genetic variants that influence the risk of developing this debilitating disease. Greater understanding of the aberrant cellular and molecular mechanisms involved in the etiology and pathophysiology of endometriosis should lead to better diagnostic methods and targeted treatments.
Introduction
Endometriosis (MIM 131200) is a common gynecological disease that causes pelvic pain, severe dysmenorrhea (painful periods), and subfertility. It is defined as the presence of tissue resembling endometrium in sites outside the uterus, most commonly the pelvic peritoneum, ovaries, and rectovaginal septum (Giudice and Kao 2004). The main pathological processes associated with the disease are peritoneal inflammation and fibrosis and the formation of adhesions and ovarian cysts.
The diagnosis is usually made by visual inspection of the pelvis at laparoscopy, because noninvasive diagnostic tools, such as ultrasound scanning, can reliably detect only severe forms of the disease—that is, ovarian endometriotic cysts. Therefore, the population prevalence is difficult to measure. The best estimates indicate that endometriosis affects 8%–10% of women in their reproductive years (Eskenazi and Warner 1997), and it has been suggested that, in North America alone, >5.5 million women are affected (National Institute of Child Health and Human Development 2002). These data are compatible with the prevalence estimate of 7.2% from our own study of a large community sample of Australian twins (Treloar et al. 1999).
The cause of endometriosis remains uncertain despite >50 years of hypothesis-driven research, and thus the therapeutic options are limited. However, there is now convincing evidence that the disease is inherited as a complex genetic trait (Simpson and Bischoff 2002; Kennedy 2003; Giudice and Kao 2004). Genetic factors accounted for 52% of the variation in liability to endometriosis in our Australian twin study (Treloar et al. 1999). Familial aggregation has been reported in humans (Kennedy et al. 1995; Stefansson et al. 2002) and in nonhuman primates with spontaneous disease (Zondervan et al. 2004). In an Icelandic population study, the average kinship coefficient for the women identified with endometriosis was significantly higher than that calculated for 1,000 sets of 750 matched controls (Stefansson et al. 2002). The genetic relative-recurrence risk for sibs (λs) was estimated to be 2.34 in our Australian sample of twins and their families (Treloar et al. 1999).
The most widely accepted theory to explain endometriosis is that viable endometrial cells reach the peritoneal cavity through retrograde menstruation along the fallopian tubes (Sampson 1927). Some cells then adhere to the peritoneal surface and proliferate. However, it is well established that menstrual debris is present in the peritoneal cavity of 90% of menstruating women, suggesting that endometrial cells from only some women are capable of establishing ectopic endometrial implants. There are several possible explanations for such susceptibility, including differences in genetic predisposition, increased exposure to menstrual debris, abnormal eutopic endometrium, altered peritoneal environment, reduced immune surveillance, and increased angiogenic capacity (Healy et al. 1998; Vinatier et al. 2001; Treloar et al. 2002; Varma et al. 2004).
Various functional candidates have been tested for association as disease-susceptibility genes, but many of these case-control studies have lacked power and/or adequate controls; the results have therefore been inconclusive (Zondervan et al. 2001). Several researchers have adopted a positional-cloning approach to identify loci for endometriosis (Kennedy 2003). One unpublished study reported modest linkage to an unidentified chromosome 10 candidate region in 32 Puerto Rican families (Flores et al. 2004). Linkage to two candidate genes—GSTM1 on chromosome 1p13 (Hadfield et al. 2001) and GALT (MIM 606999) on 9p13 (Stefansson et al. 2001)—has been excluded, but no positive-linkage regions from genomewide mapping have been reported to date.
We recruited >1,000 families—mainly affected sister pair (ASP) families—for a positional-cloning approach in our International Endogene Study (Treloar et al. 2002), which resulted from the merger of two independent groups: the Australian Genes Behind Endometriosis Study and the United Kingdom–based, international Oxford Endometriosis Gene (OXEGENE) Study; this ensured that the combined resource had 80% power to detect loci of modest effect (λs⩾1.3) (Treloar et al. 2002), which is consistent with current expectations for most complex diseases. Here, we report results from the genomewide linkage scan in 1,176 families, one of the largest ASP linkage studies conducted for any disease to date.
Methods
Family and Sample Collection
From 1995 to 2002, the Australian and U.K. groups recruited affected families with the use of protocols that have been described in detail elsewhere (Treloar et al. 2002). All women classified as affected had surgically confirmed endometriosis. In both studies, disease severity was assessed retrospectively from medical records by use of the revised American Fertility Society (rAFS) classification system (American Fertility Society 1985), which assigns patients to one of four stages (I–IV) on the basis of the extent of the disease and the associated adhesions present. The study families mainly comprised ASPs, although parents and other affected relatives were also recruited. If one or both parents were unavailable, sibs were recruited to increase the identity-by-descent (IBD) information, but they were assigned “unknown” disease status. There were three or more affected sisters in 104 (9%) of the sibships, which added power for linkage detection. The final data set comprised 1,176 families (931 from the Australia group and 245 from the U.K. group), with 1,242 independent affected sibships (i.e., a sibship of size S is equivalent to S-1 independent sib pairs [Suarez and Hodge 1979]) that included two or more siblings. The mean number of affected family members in both Australian and U.K. kindreds was 2.3. Their ethnic backgrounds were similar, and >95% of subjects were white. Affected women completed a questionnaire about their pain symptoms and fertility history (table 1).
Table 1.
Characteristics of Affected Women in Combined Genome Scan Data[Note]
| Characteristic | Value |
| Age in 2002 (years): | |
| Median | 39 |
| Range | 15–87 |
| No. (%) with disease: | |
| Stage A (rAFS I–II) | 1,550 (62) |
| Stage B (rAFS III–IV) | 957 (38) |
| No. (%) with pelvic pain: | |
| Yes | 2,109 (79) |
| No | 555 (21) |
| Age at symptom onset (years): | |
| Median | 19 |
| Range | 8–51 |
| Age at diagnosis (years): | |
| Median | 28 |
| Range | 10–64 |
| No. (%) with subfertility: | |
| Any problem in conceiving | 1,237 (48) |
| No problem in conceiving | 902 (35) |
| Unknown/untested | 456 (17) |
Note.— In calculation of values, N ranged from 2,516 to 2,696.
Approval for obtainment of medical records and collection of blood for DNA extraction and for all questionnaires and interview schedules was obtained from the Human Research Ethics Committee of the Queensland Institute of Medical Research. In the United Kingdom, the study received approval from the regional Multi-Centre Research Ethics Committee, from local research ethics committees, and from collaborating centers in Leuven and Dublin. All participants gave written informed consent.
Blood Sample Collection and Storage
In both studies, DNA was extracted from peripheral blood lymphocytes (Miller et al. 1988) or from buccal swabs with the use of Microcon Centrifugal Filter Devices (Amicons) and was stored at 4°C at a concentration of 300–600 μg/ml.
Genome Scans
In total, 4,985 individuals were genotyped, including 2,709 women with endometriosis. The genome scans for the Australian families were performed by the Australian Genome Research Facility (Ewen et al. 2000) and by Oxagen, United Kingdom. Both groups used the 400 dinucleotide microsatellite markers from Linkage Mapping Set version 2 (LMSV2 [PE Biosystems]) to provide ∼10-cM coverage of the genome on the autosomes and the X chromosome. To save cost and time, the final 79 Australian families were genotyped using only the 113 markers on chromosomes 9, 10, 11, 19, 20, 21, 22, and X. The U.K. study genotyped a small number of additional microsatellite markers from the ABI Prism LMS +HD5 set (Applied Biosystems).
At both sites, markers were amplified individually and were later combined into appropriate electrophoretic running panels, with up to 20 PCR products combined for one panel of markers (LMSV2 [PE Biosystems]). The pooled PCR products were electrophoresed through polyacrylamide gels on a PE Biosystems 377 (Australia) or 3700 (United Kingdom) Automated Sequencer, with the use of the recommended gel conditions and run protocols. PstI–restriction-digested lambda-phage DNA labeled with 6-carboxy rhodamine (GS500-ROX [PE Biosystems]) was included in each lane as a size standard (Ewen et al. 2000).
Combining Genotype Data from the Two Studies
Linkage analyses used allele frequencies calculated from pedigree founders by use of Sib-Pair (Duffy 2001). One approach to combining the data for common markers in different studies is to pool the genotypes, but doing so for samples typed at different facilities can be problematic because of allele binning and frequency differences. To avoid such difficulties, markers common to both studies were treated as different markers located at almost the same location (0.01 cM between them), which thereby kept the data from the two studies separate by a method we called “merging.” Hence, the separate allele frequencies were maintained—that is, allele frequencies were based on the Australian sample for the Australian pedigrees and were based on the U.K. sample for the U.K. pedigrees. Analyses of the markers for one chromosome with this merging method and with the much more labor-intensive pooling of alleles between studies produced near-identical linkage results, indicating that this is a valid approach.
Data Integrity
In both studies, sibling and family relationships were confirmed using GRR (which gives a graphical representation of relationships) (Abecasis et al. 2001). Data were cleaned prior to analysis of Mendelian inconsistencies with the use of Sib-Pair (Duffy 2001), PedCheck (O’Connell and Weeks 1998), and MERLIN (Abecasis et al. 2002). If a Mendelian inconsistency was detected, data for that marker were dropped for all family members. Two families common to both data sets (which occured because the U.K. group recruited a small number of families in Australia) were removed from the Australian data set prior to analysis of combined data.
Statistical Analysis
For linkage analyses, we used a version of GENEHUNTER (v2.1) (Kruglyak et al. 1996) recompiled to handle the very large numbers of families. The statistic chosen for the analysis was the “possible triangle” maximum LOD score (MLS) that restricts maximization to the set of possible IBD-sharing probabilities for ASPs (Holmans 1993), where sibships of size S were weighted to be equivalent to S-1 independent sib pairs. This approach has been shown to be the most powerful use of available data that maintained the correct type I error rate (Suarez and Hodge 1979; Davis and Weeks 1997). We opted for a nonparametric statistic because (1) there was no a priori reason to assume a particular disease model and (2) the assignment of the status “unaffected” is problematic (because a surgical procedure is required to exclude endometriosis). The MLS statistic was chosen because it has more-consistent power across disease models in ASP studies than do single-parameter statistics such as the nonparametric linkage (NPL) score (Cordell 2004). However, the U.K. data set contained several non-ASP affected relative pairs (ARPs). Huang and Vieland (2001) recently showed that the possible triangle MLS statistic is approximately equivalent to the ordinary parametric heterogeneity LOD (HLOD) score under the assumption of a simple recessive model (Huang and Vieland 2001). Therefore, we also compared results obtained from parametric HLOD scores calculated under a simple (i.e., with no phenocopies) recessive mode of inheritance (which yields HLOD-R scores) (Greenberg et al. 1998), assuming 50% penetrance (Hodge et al. 1997). As recommended by Pal et al. (2001), we specified a disease gene frequency of 0.1. The X chromosome was analyzed using an earlier version of the GENEHUNTER program (X-GENEHUNTER) because the chromosome is not included in later versions. In the final analyses, we used published deCODE map positions for microsatellite markers when available and used Marshfield sex-averaged positions when deCODE positions were not available.
In accordance with the now widely accepted practice (Abecasis et al. 2004a, 2004b; Song et al. 2004) to obtain empirical estimates of genomewide significance levels, simulations were performed using the pedigree structures in the data set and any missing genotypes. These simulations allow us to take account of uneven marker spacing and informativeness (see the work of Kruglyak and Daly [1998] for a discussion of the utility of empirical significance levels in linkage analysis). Simulations were run on the data prior to the addition of the fine markers but after the addition of the last 79 families. Data for 1,000 genome scans were generated using MERLIN (Abecasis et al. 2002), under the assumption of no susceptibility loci, and were analyzed using our modified version of GENEHUNTER. The empirical significance level of an MLS peak was then estimated by counting the proportion of genome scans containing one or more peaks of that size. The cutoff for suggestive linkage (MLS=1.88) was calculated as the mean of the genomewide MLS from each genome scan, which determines the maximum peak size expected once per genome scan by chance alone. The significant linkage threshold (MLS=3.08) was defined as the MLS occurring with probability 0.05 in a genome scan (i.e., 50 peaks of equal or greater size observed in the 1,000 simulations).
Stratified analyses of the combined genome scan data were performed for stage of disease, age at symptom onset, age at surgical diagnosis, presence of pelvic pain, and problems in conceiving (subfertility). The number of affected women per family (zero, one, or more than one) who had the more extreme phenotype was the general criterion used to allocate families to one of three strata. For stage of disease, the three disease-severity strata were defined by the presence of zero, one, or more than one individual per family who had stage B (rAFS stages III–IV) endometriosis. Families were stratified by age at onset and age at diagnosis by use of the thresholds <20 years and <27 years of age, respectively. For subfertility, the relevant subphenotype for defining three strata was the reported lifetime history of any problem in conceiving. Since pelvic pain was so prevalent, we defined only two strata—zero or one family member and two or more family members reporting pelvic pain. To assess the significance of evidence for linkage in a phenotypic subset, compared with that in the entire sample, we randomly chose from the total sample the same number of families (1,000 replicates) in the subset of interest and calculated the MLS under the null hypothesis, which is analogous to the empirical estimation of genomewide significance levels.
Results
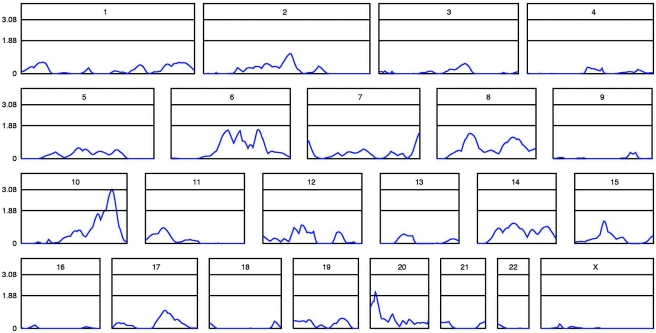
For the final merged Australian and U.K. data, the highest MLSs were on chromosome 10q (3.09 at D10S587) and chromosome 20p (2.09 at D20S889/D20S116). In our simulation study, 47 of the 1,000 replicates had peaks ⩾3.09, and 360 had peaks ⩾2.09, which implied significant linkage on chromosome 10 (genomewide empirical P=.047) and suggestive linkage on chromosome 20. The linkage (MLS) peaks for all chromosomes are shown in figure 1, and table 2 gives the positions and nearest markers for all MLS peaks ⩾1.0.
Figure 1.
Linkage (MLS) curves for all chromosomes in the combined data set, prior to inclusion of additional fine markers. This composite figure shows relative chromosome lengths. The genetic map of each chromosome is shown on the X-axis, and the MLS is shown on the Y-axis. Thresholds for significant (MLS=3.08) and suggestive (MLS=1.88) genomewide linkage are indicated by horizontal lines.
Table 2.
Chromosomal Regions with Multipoint MLS ⩾1 in Genome Scan
| Chromosomea | Position of Peak(cM) | MLS | Closest Marker(s) |
| 2 | 141.79 | 1.14 | D2S2256, D2S112 |
| 6 | 92.11 | 1.63 | D6S460 |
| 6 | 111.49 | 1.59 | D6S434 |
| 6 | 140.80 | 1.65 | D6S292, D6S308 |
| 7 | 7.44 | 1.00 | D7S517 |
| 7 | 182.19 | 1.48 | D7S2423 |
| 8 | 56.60 | 1.44 | D8S1771, D8S505 |
| 8 | 127.84 | 1.22 | D8S514 |
| 10 | 147.57 | 3.09 | D10S587 |
| 12 | 66.12 | 1.06 | D12S368 |
| 14 | 64.11 | 1.16 | D14S63 |
| 15 | 45.69 | 1.29 | D5S987 |
| 17 | 79.63 | 1.03 | D17S787, D17S944 |
| 20 | 11.32 | 2.09 | D20S889, D20S116 |
For chromosomes 10 and 20, 1,176 pedigrees were included. For the other chromosomes listed in this table, 1,097 pedigrees were included.
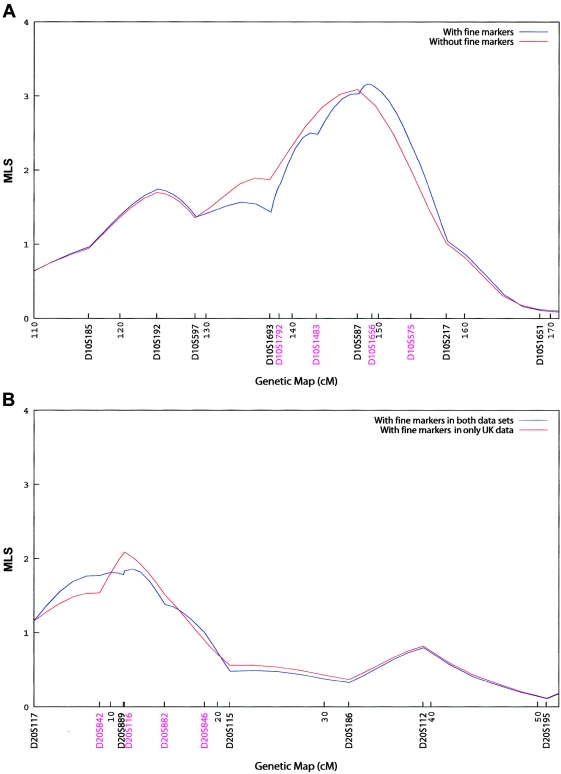
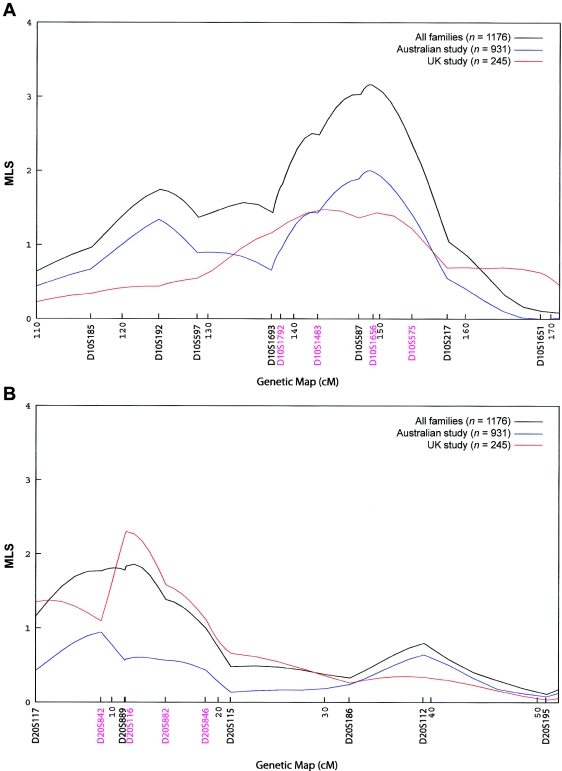
To better define these peaks, we genotyped four additional markers under each of the chromosome 10 and 20 peaks in the Australian families. One of these markers (D20S116) had already been typed in the U.K. families. These markers changed the shape of the peaks but had minimal effects on the peak MLS: it increased from 3.09 to 3.16 for chromosome 10 and decreased from 2.09 to 1.86 for chromosome 20. The MLS curves are shown in figure 2. The summit for the chromosome 20 region moved marginally, from 8.49 cM to 12.09 cM, and the location of the chromosome 10 peak shifted from 145.44 cM to 148.75 cM—that is, from D10S587 toward D10S1656 in the telomeric direction. Both Australian and U.K. families contributed to the chromosome 10 linkage, with respective peaks of MLS = 2.06 and MLS = 1.48 when data were analyzed separately (fig. 3A). Both sets of families also contributed to the chromosome 20 peak (fig. 3B). Additional markers are shown in figures 2 and 3.
Figure 2.
Fine mapping of chromosome 10 peak (A) and chromosome 20 peak (B). Linkage (MLS) curves are presented for the chromosomes before and after the addition of several markers, shown in different colors. Microsatellite markers are shown in genetic map order along the X-axis, with positions given in cM (Haldane). The Y-axis represents the MLS.
Figure 3.
Study of origin strata at chromosome 10 peak (A) and chromosome 20 peak (B). Linkage (MLS) curves that include the fine markers are presented for the Australian and U.K. studies separately and also for the combined data set. Microsatellite markers are shown in genetic map order along the X-axis, with positions given in cM (Haldane). The Y-axis represents the MLS. A, Results show the increase in MLS when the two data sets are combined.
Given the large sample size from which our linkage statistics were calculated, conversion of the distribution of the likelihood-ratio test statistic of linkage versus no linkage into a χ2 distribution will be asymptotically valid (Walling et al. 2000). An MLS of 1.0 (which is a mixture of χ2 distributions with 0, 1, and 2 df) corresponds to an asymptotic (two-sided) χ2 with a P value of .05 (Nyholt 2000). The end points of the 95% CI are calculated by finding the locations on either side of the 148.75 cM peak on chromosome 10 that have an MLS of 2.16 (i.e., 1.0 less than the peak at 3.16). On the basis of our fine-mapping results, the 95% CI for the peak at 148.75 cM spans 15 cM, from 139.49 (10q26.11) to 154.77 cM (10q23.33).
Stratification by Subphenotypes
Families with no stage B disease and those with multiple stage B–affected members contributed to the chromosome 10 peak (table 3). Other results of stratified analyses for chromosomes 10 and 20 are shown in table 3. Except for families with two or more affected members reporting pelvic pain, no subphenotype stratum contributed disproportionally more to the chromosome 10 peak, and no particular strata contributed more to the chromosome 20 peak. More specifically, the original chromosome 10 peak MLS increased to 3.39 in the 817 families with more than one affected woman reporting pelvic pain (see table 3). One thousand permutations using subgroups of 817 families chosen at random produced 94 replicates with an MLS ⩾3.39, suggesting that this observation was most likely a chance finding (P=.094). Indeed, a difference in MLS of 0.3 is not significant at the nominal P<.05 level. Further, very few families had no women reporting pelvic pain (Treloar et al. 2002).
Table 3.
Linkage Results for Phenotypic Strata of Families, Including Fine Marker Data
| MLSb |
|||
| Phenotype and Strataa | No. of Families | Chromosome 10(at 148.75 cM) | Chromosome 20(at 12.09 cM) |
| Stage B (III–IV) disease: | |||
| 0 Members (stage A only) | 436 | 1.43 | 1.23 |
| 1 Member | 401 | .31 | .96 |
| >1 Member | 329 | 1.82 | .07 |
| Age at onset <20 years: | |||
| 0 Members | 392 | .92 | .46 |
| 1 Member | 461 | 1.98 | 1.16 |
| ⩾2 Members | 313 | .47 | .34 |
| Age at diagnosis <27 years: | |||
| 0 Members | 410 | 2.30 | 1.20 |
| 1 Member | 390 | .51 | 1.45 |
| ⩾2 Members | 366 | .71 | .01 |
| Pelvic pain: | |||
| 0 or 1 Member | 349 | .17 | 1.65 |
| ⩾2 Members | 817 | 3.39 | .68 |
| Subfertility: | |||
| 0 Members | 323 | .81 | .86 |
| 1 Member | 487 | 1.91 | .45 |
| ⩾2 Members | 356 | .57 | .80 |
Families were stratified by the no. of family members with the phenotype.
1,166 families were informative at these positions.
Conditional Linkage Results
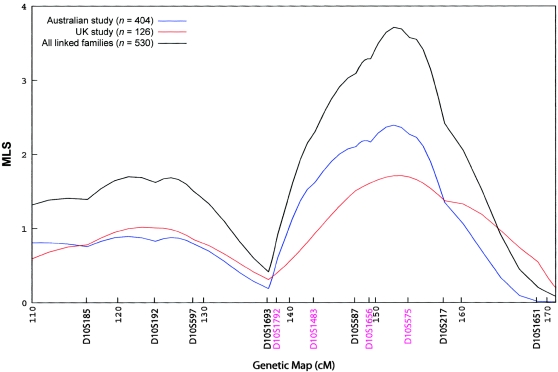
Conditioning on status at the chromosome 10 peak did not increase the evidence for linkage at the chromosome 20 peak. In contrast, conditional analysis of a subset of 530 families with positive allele sharing at the chromosome 20 peak resulted in an increased MLS at the chromosome 10 peak of 3.71, and this was consistent between both the Australian and the U.K. data sets (see fig. 4). Permutations using subgroups of 530 families chosen at random produced only 28 of 1,000 replicates with an MLS ⩾3.71 (P=.028), suggesting that this observation may not be a chance finding. However, this result should be interpreted with caution, given that we investigated a large number of stratifications.
Figure 4.
Linkage in chromosome 10 conditional on linkage to chromosome 20. Chromosome 10 linkage curves are presented for families showing linkage at the chromosome 20 peak. Curves show MLS results for families linked to the chromosome 20 peak in the two studies separately and for all linked families. Microsatellite markers are shown in genetic map order along the X-axis, with positions given in cM (Haldane). The Y-axis represents the MLS.
Discussion
We present results from the first large-scale genomewide scan for loci influencing susceptibility to endometriosis, which is a common disease affecting millions of women worldwide. To the best of our knowledge, this data set is the largest collection of ASPs assembled to study endometriosis and is one of the largest family collections for mapping disease genes for any complex trait. Analysis of our combined set of families identified significant linkage (MLS=3.16) to a novel susceptibility locus on chromosome 10q26.
The magnitude of the linkage scores we report is consistent with λs estimates predicted by our twin study (Treloar et al. 1999) and with those thought to pertain to most complex genetic diseases. Expected MLSs calculated for 1,243 independent ASPs showed higher values for incremental increases in λs than those obtained. For example, with the formula of Risch (1990) and with the assumption of fully informative ASPs, the expected MLS was 5.1 for a locus-specific λs value of 1.3; raising the locus-specific λs value to 1.5 would yield an expected MLS of 10.9. Estimating the locus-specific λs value from the IBD-sharing probabilities at the chromosome 10 peak, we obtained a very modest value of 1.07. However, it is possible that this estimate is biased, and hence unreliable, because of reported problems in estimating locus-specific heritability from the peak of a variance-components genomewide scan (Goring et al. 2001). Our chromosome 10 data are consistent with inclusion of dominance variance (i.e., interaction among alleles at a locus) in the MLS model; the maximum-likelihood estimates of the probability of affected sibs sharing 0, 1, and 2 alleles IBD are 0.233, 0.466, and 0.301, respectively, which suggests some degree of recessivity of the disease allele. Indeed, had we not allowed for dominance variance, we would have had <1% power to detect this locus under a purely additive model (Risch 1990).
The MLS statistic is approximately equivalent to the ordinary HLOD score with the assumption of a simple recessive model (Huang and Vieland 2001), which suggests that the MLS allows for heterogeneity without explicitly modeling for it. In our data, multipoint heterogeneity linkage analysis calculated under a simple (i.e., with no phenocopies) recessive mode of inheritance (Greenberg et al. 1998) with the assumption of 50% penetrance (Hodge et al. 1997) and a gene frequency of 0.1 (as recommended by Pal et al. [2001]), produced an HLOD-R of 3.58 at 148.75 cM (maximum HLOD-R=3.75; 2 cM distal). However, heterogeneity analysis under a simple dominant model with the assumption of 50% penetrance and a gene frequency of 0.01 produced a maximum HLOD-D of 1.05 at 148.75 cM. These analyses add weight to the conclusion that the chromosome 10q locus best fits a recessive mode of inheritance.
The chromosome 10 peak MLS showed a steady and generally consistent incremental increase with the addition of more data during the course of the study. Families with more than two affected members comprised ∼22% of the entire sample and contributed ∼28% of the linkage signal. Thus, their inclusion did not explain the linkage result. Indeed, both the chromosome 10q and the chromosome 20p peaks diminished with the addition of the final 79 Australian families. Prior to their addition, the chromosome 10q MLS was 4.16 and the chromosome 20p peak was 2.13. Although the 79 additional Australian kindreds diminished both the chromosome 10 and the chromosome 20 peaks, no systematic ascertainment bias can be identified. These kindreds were not selected for any particular characteristic or for having multiple family members. The mean number of affected members was 2.3 for the original kindreds and 2.5 for the last 79 families added. Recruitment of many of those 79 kindreds took longer to complete than that of the other kindreds, for a variety of logistic reasons. They therefore fell into the last batch to be sent for genome scanning, and we did not have enough money to complete the scan on all chromosomes. The clinical profile of those families was very similar to that of earlier recruits with regard to pelvic pain (80% vs. 77%), but they had somewhat lower proportions of women with stage B disease (28% vs. 34%) and women with a history of subfertility (39% vs. 49%) than did the previous combined family collection.
It clearly would be desirable to have our finding independently confirmed, but other groups may not have the resources to achieve the sample size of the present study for replication purposes. Although we had sufficient power to detect linkage to loci with effect sizes as low as λs=1.3, it is interesting that only one peak achieved genomewide significance (P=.047) in this large data set. To obtain one MLS of this magnitude from 1,176 families suggests substantial genetic heterogeneity and/or that more than one disease entity exists, as suggested elsewhere (Nisolle and Donnez 1997). However, analysis based on alternative phenotypic information is problematic if the criteria for stratification are unknown, and it must be conducted cautiously, since power can be lost through multiple testing.
Chromosome 10q26 had already been implicated in a candidate gene study that reported aberrant endometrial EMX2 (MIM 600035) expression in women with endometriosis (Daftary and Taylor 2004). EMX2 (10q26.1) is the gene for a transcription factor required for reproductive-tract development, which maps to a region of allelic deletion corresponding to a putative endometrial tumor suppressor at 10q26 (Peiffer et al. 1995; Peiffer-Schneider et al. 1998). Another candidate gene in the region is PTEN (MIM 601728) (10q23.31), which has been implicated in the malignant transformation of ovarian endometriosis (Bischoff and Simpson 2000; Sato et al. 2000). Although outside the reported support interval, both EMX2 and PTEN fall within our 99.9% CI. We are therefore planning to test for association in large numbers of cases and controls with the use of SNPs in these and other positional-candidate genes. In the first instance, cases will be drawn from ASPs who share two alleles or one allele, which are identical by descent, under the peak (Fingerlin et al. 2004; Wicks et al. 2004), to enrich for genetic predisposition at this locus and to remove genetic heterogeneity, and we will then conduct replication studies with independent case-parents triads. Although the gene or genes in this region are likely to comprise only a subset of all genes influencing susceptibility to endometriosis, discovery of even one should lead to a clearer understanding of the aberrant cellular and molecular mechanisms and to better diagnosis and more-targeted treatment.
Acknowledgments
We thank the women with endometriosis and their families, for their participation and support of our research. We also thank the many hospital directors and staff, gynecologists, general practitioners, and pathology services in the United Kingdom, the United States, Australia, and New Zealand who provided assistance with confirmation of diagnoses. The Endometriosis Associations of the United States, the United Kingdom, Queensland, Victoria, Western Australia, and Tasmania and the Endometriosis Foundation of New Zealand supported and assisted with recruitment for the study. We acknowledge Barbara Haddon and Alison McKenzie, Australian project coordinators at the Queensland Institute of Medical Research (QIMR), for their contributions; the team of Rebekah Cicero, Shona Holloway, Alana Goldman, Carmel Cassar, Cassie Grace, Lynnette Kennedy, Linda McInnes, Jan Callanan, Sue McCoombe, Stephanie Brook, Heather Park, Lorraine Kelpie, Sally Von Bibra, Maureen Norris, Lyn Barnes, and Kim Eldridge, for recruitment and phenotyping; Olivia Zheng, David Smyth, and Xiaping Lin, for computing and database management; and Anjali Henders, Patricia Keith, Leanne McNeill, and Renée Mayne, for blood processing and DNA extraction. We wish to acknowledge the contribution of many pathology services for pro bono blood collection and, in particular, Sullivan Nicolaides Pathology and QML Pathology (Brisbane, Australia), for also contributing courier return of blood samples to the QIMR.
We thank Ann Lambert, Louise Cotton, Lesley Pope, Gillian Spencer-Webb, Gail Farmer, Stephen Golding, and Xiao-Hui Liao (University of Oxford). We also thank Philippe Koninckx (Leuven, Belgium), Christopher Sutton (Guildford, United Kingdom), Mary Wingfield (Dublin), Enda McVeigh (Oxford), Leila Adamyan (Moscow), David Adamson (Palo Alto, CA), Ronald Batt (Buffalo, NY), Agneta Bergqvist (Huddinge, Sweden), Mette Moen (Trondheim, Norway), Matthew Peterson (Salt Lake City), and Martin Sillem (Heidelberg). We also thank Emma Jones, Oxagen’s Database Manager for Endometriosis, for managing data and quality checking prior to statistical analysis.
The two studies that formed the International Endogene Study were funded independently. The U.K. study was funded by Oxagen (Abingdon, United Kingdom) and the Australian study was funded by the Cooperative Research Centre for Discovery of Genes for Common Human Diseases (CRC), by Cerylid Biosciences (Melbourne), and by donations from Neville and Shirley Hawkins, the Western Australian Endometriosis Support Association, and the Endometriosis Association. We thank Mark Edwards and Diana Schuette (Oxagen), Nick Gough, Brandon Wainwright, and Andrea Douglas (CRC), and Matthijs Smith (Cerylid), for their encouragement and backing of this project over the past seven years.
Web Resources
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for endometriosis, EMX2, GALT, and PTEN)
References
- Abecasis GR, Burt RA, Hall D, Bochum S, Doheny KF, Lundy SL, Torrington M, Roos JL, Gogos JA, Karayiorgou M (2004a) Genomewide scan in families with schizophrenia from the founder population of Afrikaners reveals evidence for linkage and uniparental disomy on chromosome 1. Am J Hum Genet 74:403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 [DOI] [PubMed] [Google Scholar]
- ——— (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Yashar BM, Zhao Y, Ghiasvand NM, Zareparsi S, Branham KE, Reddick AC, Trager EH, Yoshida S, Bahling J, Filippova E, Elner S, Johnson MW, Vine AK, Sieving PA, Jacobson SG, Richards JE, Swaroop A (2004b) Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet 74:482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Fertility Society (1985) Revised American Fertility Society classification of endometriosis: 1985. Fertil Steril 43:351–352 [DOI] [PubMed] [Google Scholar]
- Bischoff FZ, Simpson JL (2000) Heritability and molecular genetic studies of endometriosis. Hum Reprod Update 6:37–44 [DOI] [PubMed] [Google Scholar]
- Cordell HJ (2004) Bias toward the null hypothesis in model-free linkage analysis is highly dependent on the test statistic used. Am J Hum Genet 74:1294–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftary GS, Taylor HS (2004) EMX2 gene expression in the female reproductive tract and aberrant expression in the endometrium of patients with endometriosis. J Clin Endocrinol Metab 89:2390–2396 [DOI] [PubMed] [Google Scholar]
- Davis S, Weeks DE (1997) Comparison of nonparametric statistics for detection of linkage in nuclear families: single-marker evaluation. Am J Hum Genet 61:1431–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL (2001) Sib-Pair release 0.99.9, Brisbane, Australia [Google Scholar]
- Eskenazi B, Warner ML (1997) Epidemiology of endometriosis. Obstet Gynecol Clin North Am 24:235–258 [DOI] [PubMed] [Google Scholar]
- Ewen KR, Bahlo M, Treloar SA, Levinson DF, Mowry B, Barlow JW, Foote SJ (2000) Identification and analysis of error types in high-throughput genotyping. Am J Hum Genet 67:727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerlin TE, Boehnke M, Abecasis GR (2004) Increasing the power and efficiency of disease-marker case-control association studies through use of allele-sharing information. Am J Hum Genet 74:432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores I, Mandal DM, Bailey-Wilson JE (2004) Search for endometriosis susceptibility genes in Puerto Rico [program number 1905]. Presented at the American Society for Human Genetics 2004 Annual Meeting, Toronto, Canada, October 26–30 [Google Scholar]
- Giudice LC, Kao LC (2004) Endometriosis. Lancet 364:1789–1799 [DOI] [PubMed] [Google Scholar]
- Goring HH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield RM, Manek S, Weeks DE, Mardon HJ, Barlow DH, Kennedy SH (2001) Linkage and association studies of the relationship between endometriosis and genes encoding the detoxification enzymes GSTM1, GSTT1 and CYP1A1. Mol Hum Reprod 7:1073–1078 [DOI] [PubMed] [Google Scholar]
- Healy DL, Rogers PA, Hii L, Wingfield M (1998) Angiogenesis: a new theory for endometriosis. Hum Reprod Update 4:736–740 [DOI] [PubMed] [Google Scholar]
- Hodge S, Abreu P, Greenberg D (1997) Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60:217–227 [PMC free article] [PubMed] [Google Scholar]
- Holmans P (1993) Asymptotic properties of affected–sib-pair linkage analysis. Am J Hum Genet 52:362–374 [PMC free article] [PubMed] [Google Scholar]
- Huang J, Vieland VJ (2001) Comparison of “model-free” and “model-based” linkage statistics in the presence of locus heterogeneity: single data set and multiple data set applications. Hum Hered 51:217–225 [DOI] [PubMed] [Google Scholar]
- Kennedy S (2003) Genetics of endometriosis: a review of the positional cloning approaches. Semin Reprod Med 21:111–118 [DOI] [PubMed] [Google Scholar]
- Kennedy S, Mardon H, Barlow D (1995) Familial endometriosis. J Assist Reprod Genet 12:32–34 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly M (1998) Linkage thresholds for two-stage genome scans. Am J Hum Genet 62:994–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Child Health and Human Development (2002) Endometriosis. U.S. Department of Health and Human Services, National Institutes of Health, Rockville, MD (available at: http://www.nichd.nih.gov/publications/pubs/endometriosis.pdf; accessed July 18, 2005)
- Nisolle M, Donnez J (1997) Peritoneal endometriosis, ovarian endometriosis, and adenomyotic nodules of the rectovaginal septum are three different entities. Fertil Steril 68:585–596 [DOI] [PubMed] [Google Scholar]
- Nyholt DR (2000) All LODs are not created equal: invited editorial. Am J Hum Genet 67:282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal D, Durner M, Greenberg D (2001) Effect of misspecification of gene frequency on the two-point LOD score. Eur J Hum Genet 9:855–859 [DOI] [PubMed] [Google Scholar]
- Peiffer SL, Herzog TJ, Tribune DJ, Mutch DG, Gersell DJ, Goodfellow PJ (1995) Allelic loss of sequences from the long arm of chromosome 10 and replication errors in endometrial cancers. Cancer Res 55:1922–1926 [PubMed] [Google Scholar]
- Peiffer-Schneider S, Noonan FC, Mutch DG, Simpkins SB, Herzog T, Rader J, Elbendary A, Gersell DJ, Call K, Goodfellow PJ (1998) Mapping an endometrial cancer tumor suppressor gene at 10q25 and development of a bacterial clone contig for the consensus deletion interval. Genomics 52:9–16 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- Sampson JA (1927) Peritoneal endometriosis due to the menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gynecol 14:422–469 [Google Scholar]
- Sato N, Tsunoda H, Nishida M, Morishita Y, Takimoto Y, Kubo T, Noguchi M (2000) Loss of heterozygosity on 10q23.3 and mutation of the tumor suppressor gene PTEN in benign endometrial cyst of the ovary: possible sequence progression from benign endometrial cyst to endometrioid carcinoma and clear cell carcinoma of the ovary. Cancer Res 60:7052–7056 [PubMed] [Google Scholar]
- Simpson JL, Bischoff FZ (2002) Heritability and molecular genetic studies of endometriosis. Ann N Y Acad Sci 955:239–251 [DOI] [PubMed] [Google Scholar]
- Song K, Weeks D, Sobel E, Feingold E (2004) Efficient simulation of P values for linkage analysis. Genet Epidemiol 26:88–96 [DOI] [PubMed] [Google Scholar]
- Stefansson H, Einarsdottir A, Geirsson RT, Jonsdottir K, Sverrisdottir G, Gudnadottir VG, Gunnarsdottir S, Manolescu A, Gulcher J, Stefansson K (2001) Endometriosis is not associated with or linked to the GALT gene. Fertil Steril 76:1019–1022 [DOI] [PubMed] [Google Scholar]
- Stefansson H, Geirsson RT, Steinthorsdottir V, Jonsson H, Manolescu A, Kong A, Ingadottir G, Gulcher J, Stefansson K (2002) Genetic factors contribute to the risk of developing endometriosis. Hum Reprod 17:555–559 [DOI] [PubMed] [Google Scholar]
- Suarez B, Hodge S (1979) A simple method to detect linkage for rare recessive diseases: an application to juvenile diabetes. Clin Genet 15:126–136 [DOI] [PubMed] [Google Scholar]
- Treloar S, Hadfield R, Montgomery G, Lambert A, Wicks J, Barlow DH, O’Connor DT, Kennedy S (2002) The International Endogene Study: a collection of families for genetic research in endometriosis. Fertil Steril 78:679–685 [DOI] [PubMed] [Google Scholar]
- Treloar SA, O’Connor DT, O’Connor VM, Martin NG (1999) Genetic influences on endometriosis in an Australian twin sample. Fertil Steril 71:701–710 [DOI] [PubMed] [Google Scholar]
- Varma R, Rollason T, Gupta JK, Maher ER (2004) Endometriosis and the neoplastic process. Reproduction 127:293–304 [DOI] [PubMed] [Google Scholar]
- Vinatier D, Orazi G, Cosson M, Dufour P (2001) Theories of endometriosis. Eur J Obstet Gynecol Reprod Biol 96:21–34 [DOI] [PubMed] [Google Scholar]
- Walling GA, Visscher PM, Andersson L, Rothschild MF, Wang L, Moser G, Groenen MA, Bidanel JP, Cepica S, Archibald AL, Geldermann H, de Koning DJ, Milan D, Haley CS (2000) Combined analyses of data from quantitative trait loci mapping studies: chromosome 4 effects on porcine growth and fatness. Genetics 155:1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks J, Treloar SA, Martin NG (2004) Using identity-by-descent information in affected sib pairs to increase the efficiency of genetic association studies. Twin Res 7:211–216 [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR, Kennedy SH (2001) The genetic basis of endometriosis. Curr Opin Obstet Gynecol 13:309–314 [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Weeks DE, Colman R, Cardon LR, Hadfield R, Schleffler J, Trainor AG, Coe CL, Kemnitz JW, Kennedy SH (2004) Familial aggregation of endometriosis in a large pedigree of rhesus macaques. Hum Reprod 19:448–455 [DOI] [PubMed] [Google Scholar]