Abstract
On the basis of genomewide linkage studies of families affected with age-related maculopathy (ARM), we previously identified a significant linkage peak on 10q26, which has been independently replicated by several groups. We performed a focused SNP genotyping study of our families and an additional control cohort. We identified a strong association signal overlying three genes, PLEKHA1, LOC387715, and PRSS11. All nonsynonymous SNPs in this critical region were genotyped, yielding a highly significant association (P < .00001) between PLEKHA1/LOC387715 and ARM. Although it is difficult to determine statistically which of these two genes is most important, SNPs in PLEKHA1 are more likely to account for the linkage signal in this region than are SNPs in LOC387715; thus, this gene and its alleles are implicated as an important risk factor for ARM. We also found weaker evidence supporting the possible involvement of the GRK5/RGS10 locus in ARM. These associations appear to be independent of the association of ARM with the Y402H allele of complement factor H, which has previously been reported as a major susceptibility factor for ARM. The combination of our analyses strongly implicates PLEKHA1/LOC387715 as primarily responsible for the evidence of linkage of ARM to the 10q26 locus and as a major contributor to ARM susceptibility. The association of either a single or a double copy of the high-risk allele within the PLEKHA1/LOC387715 locus accounts for an odds ratio of 5.0 (95% confidence interval 3.2–7.9) for ARM and a population attributable risk as high as 57%.
Introduction
Age-related maculopathy (ARM), or age-related macular degeneration (ARMD-1 [MIM 603075]), is a leading cause of central blindness in the elderly population, and numerous studies support a strong underlying genetic component to this complex disorder. Genomewide linkage scans performed using large pedigrees, affected sib pairs, and, more recently, discordant sib pairs have identified a number of potential susceptibility loci (Klein et al. 1998; Weeks et al. 2000; Majewski et al. 2003; Schick et al. 2003; Seddon et al. 2003; Abecasis et al. 2004; Iyengar et al. 2004; Kenealy et al. 2004; Schmidt et al. 2004; Weeks et al. 2004; Santangelo et al. 2005). Our genomewide linkage screen strongly implicated the 10q26 region as likely to contain an ARM gene (Weeks et al. 2004); this region has also been supported by many other studies and was the top-ranked region in a recent meta-analysis (Fisher et al. 2005). Recently, three studies (Edwards et al. 2005; Haines et al. 2005; Klein et al. 2005) identified an allelic variant in the complement factor H gene (CFH [MIM 134370]) as responsible for the linkage signal seen on chromosome 1 and as the variant accounting for a significant attributable risk (AR) of ARM in both familial and sporadic cases. We and others have confirmed these findings (Conley et al. 2005; Hageman et al. 2005; Zareparsi et al. 2005a). CFH has previously been suspected of playing a role in ARM, as a result of the work of Hageman and Anderson (Hageman and Mullins 1999; Johnson et al. 2000, 2001; Mullins et al. 2000; Hageman et al. 2001), who have shown that the subretinal deposits (drusen) that are observed in many patients with ARM contain complement factors. However, until other genes that contribute to ARM are identified, CFH remains an isolated piece of the puzzle, implicating the alternative pathway and inflammation as part of the ARM pathogenesis but failing to fully account for the unique pathology that is observed in the eye.
We have expanded our family linkage studies and have also undertaken a case-control association study, using a high-density SNP panel in two regions of linkage on 1q31 and 10q26 that we had previously reported. Our SNP linkage and association results for chromosome 1q31 yielded the same findings as others, confirming that the peak of linkage and the strongest associations with ARM were localized over the CFH gene. We have analyzed both our family data and the case-control data on chromosome 10q26 to identify the next major ARM susceptibility–related gene.
Material and Methods
Families and Case-Control Cohort
A total of 612 ARM-affected families and 184 unrelated controls were sent to the Center for Inherited Disease Research (CIDR) for genotyping. Because of possible population substructure, we restricted our analysis to the subset of data from white subjects; we were not able to analyze the set of data from nonwhites separately, because it was too small. The white subset had 594 ARM-affected families, containing 1,443 genotyped individuals, and 179 unrelated controls. The white families contained 430 genotyped affected sib pairs, 38 genotyped affected avuncular pairs, and 52 genotyped affected first-cousin pairs.
A total of 323 white families, 117 unrelated controls, and 196 unrelated cases were also genotyped locally for additional SNPs. The local subset contained 824 genotyped individuals, 298 genotyped affected sib pairs, 23 genotyped affected avuncular pairs, and 38 genotyped affected first-cousin pairs. We used PedStats from the Merlin package (Abecasis et al. 2000) to easily get summary counts on the family data.
Affection-Status Models
We have defined three classification models (types A, B, and C) for the severity of ARM status (Weeks et al. 2004). For simplicity, we have restricted our attention here to individuals affected with “type A” ARM, our most stringent and conservative diagnosis. We used only unrelated controls who were unaffected under all three diagnostic models. Unaffected individuals were those for whom eye-care records and/or fundus photographs indicated either no evidence of any macular changes (including drusen) or a small number (<10) of hard drusen (⩽50 μm in diameter) without any other retinal pigment epithelial (RPE) changes. Individuals with evidence of large numbers of extramacular drusen were not coded as unaffected.
In our efforts to examine specific ARM subphenotypes, we chose to look at only patients with end-stage disease, either those with evidence of choroidal neovascular membrane (CNV) in either eye or those with geographic atrophy (GA) in either eye. There are a significant number of individuals who have been described as having both GA and CNV, though this is problematic, since, in these cases, it is often difficult to determine whether the GA is secondary to the damage from the CNV or is from the treatment given to limit the CNV growth (i.e., laser, surgery, or photodynamic therapy). Because it is often difficult to discern from photographs or records whether a person had GA in an eye prior to the development of CNV, we included the patients who had both pathologies in the CNV group. However, we allowed only a subset of this overlapping group to be included in the GA group, specifically those who reportedly had GA in one eye that did not have evidence of CNV. Table 1 shows the numbers of individuals in each of our three sets. This approach may have excluded a small proportion of individuals from the GA group who had asymmetric GA prior to the development of CNV in the same eye or who may have had bilateral GA but developed CNV in both eyes.
Table 1.
Distribution of Subphenotypes in Patients with Advanced ARM[Note]
|
No. of Patients fromCIDR Familiesa |
No. of Patients fromLocal Familiesa |
No. of LocalUnrelated Patients |
||||
| Subphenotype | With GA | Without GA | With GA | Without GA | With GA | Without GA |
| With CNV | 220 (76) | 187 | 130 (45) | 106 | 71 (17) | 59 |
| Without CNV | 108 | 62 | 57 | 28 | 40 | 26 |
Note.— The numbers in parentheses are the numbers of individuals with both CNV and GA who were also included in GA group (see text for selection criteria) for OR and AR estimation and association tests.
Counts are based on the set of unrelated cases generated by selecting one type A–affected person from each family (see section “Part III: Interaction and OR Analysis: Unrelated cases”).
Pedigree and Genotyping Errors and Data Handling
We used the program PedCheck (O’Connell and Weeks 1998) to check for Mendelian inconsistencies. Since it can be extremely difficult to determine which genotypes within small families are erroneous (Mukhopadhyay et al. 2004), we set all genotypes at each problematic marker to missing within each family containing a Mendelian inconsistency. Mega2 (Mukhopadhyay et al. 2005; see Division of Statistical Genetics Web site) was used to set up files for linkage analysis and for allele-frequency estimation by gene counting.
Allele Frequencies and Hardy-Weinberg Equilibrium
The allele frequencies used in the linkage analyses were estimated, by direct counting, from the unrelated and unaffected controls. All controls were unaffected under all three affection status models. Genotyped spouses who had no children or who had children who were not yet part of the study were combined with the controls for this study. The exact test of Hardy-Weinberg equilibrium (HWE), implemented in Mega2 (Mukhopadhyay et al. 2005), was performed on our SNPs.
We also used Mendel (version 5) (Lange et al. 2001) to estimate allele frequencies directly from the family data, because Mendel properly accounts for relatedness of the subjects while estimating the allele frequencies. Since the majority of the genotyped family members were affected, these estimates were quite close to estimates obtained using our unrelated affected cases.
Genetic Map
We used linear interpolation on the Rutgers combined linkage-physical map (version 2.0) (Kong et al. 2004) to predict the genetic position of the SNPs that were not already present in the Rutgers map. Since the distribution of our SNPs was very dense in the regions of interest, the estimated recombination between several SNPs was zero; for these, we set the recombination to 0.000001. We obtained the physical positions for all our SNPs from the National Center for Biotechnology Information (NCBI) dbSNP database (human build 35).
LD Structure
Ignoring high linkage disequilibrium (LD) between SNPs when performing linkage analysis can result in false-positive findings (Schaid et al. 2002; Huang et al. 2004). Our efforts to take high SNP-SNP LD into account included the following measures. (1) We used the H-clust method (Rinaldo et al. 2005), which is implemented in R (R Development Core Team 2004; see R Project for Statistical Computing Web site), to determine haplotype-tagging SNPs (htSNPs) for linkage analysis. The method uses hierarchical clustering to cluster highly correlated SNPs. After the clustering, the H-clust method chooses a htSNP from each cluster; the htSNP chosen is the SNP that is most correlated with all other SNPs in the cluster. We chose to cluster the SNPs so that each SNP had a correlation coefficient (r2) >0.5 with at least one htSNP; we used HaploView (Barrett et al. 2005) to get a graphical view of SNP-SNP LD along both chromosomes, and we compared LD estimates of htSNPs with SNPs omitted by H-clust. (2) We performed haplotype-based association analyses using two- and three-SNP moving windows (see “Association Analysis” section).
Linkage Analysis
Two-point analysis
As in our previous study (Weeks et al. 2004), we computed LOD scores under a single simple dominant model (with disease-allele frequency of 0.0001 and penetrance vector of [0.01, 0.90, 0.90]). Because of the complexities and late onset of the ARM phenotype, only two disease phenotypes were used: “affected under model A” (i.e., “type A–affected”) and “unknown.” Parametric LOD scores were computed under heterogeneity (HLOD), whereas model-free LOD scores were computed with the linear Sall statistic. Both scores were computed using Allegro (Gudbjartsson et al. 2000).
Multipoint analysis ignoring LD
Since intermarker distances are often very small, LD between SNPs can be high and thus violate the assumption of no LD made by most linkage analysis programs. Multipoint analyses ignoring LD were performed using Allegro (Gudbjartsson et al. 2000). Both HLOD scores and Sall statistics were computed. Our main goal in estimating the multipoint linkage curve without properly accounting for LD was not to predict the position of ARM-associated loci but to compare the results with those from analyses in which LD was taken into account.
Multipoint analysis using htSNPs
When only htSNPs were used for LOD score calculation, the number of SNPs decreased from 679 to 533 on chromosome 1 and from 196 to 159 on chromosome 10. Multipoint linkage analyses were done as described elsewhere (Weeks et al. 2004). The SNPs that were omitted fit well into the SNP-SNP LD structure estimated by HaploView (Barrett et al. 2005).
Association Analysis
To incorporate all cases from the families, we used the new CCREL program (Browning et al. 2005), which permits testing for association with the use of related cases and unrelated controls simultaneously. CCREL was used to analyze SNPs under the linkage peak on chromosomes 1 and 10, to test for association. The CCREL test accounts for biologically related subjects by calculating the effective number of cases and controls. For these analyses, type A–affected family members were assigned the phenotype “affected,” unrelated controls were assigned the phenotype “normal,” and family members that were not affected with type A ARM were assigned the phenotype “unknown.” (The CCREL approach has not yet been extended to permit the simultaneous use of both related cases and related controls.) The effective number of controls for each SNP used for association testing is therefore the number of controls genotyped for that SNP. An allelic test, a haplotype test with a two-SNP sliding window, a haplotype test with a three-SNP sliding window, and a genotype test were performed. We used the CCREL R package for analysis, as provided by Browning et al. (2005).
GIST Analysis
To explore which allele/SNP contributes the most to the linkage signal, we performed the genotype–identity by descent (IBD) sharing test (GIST) using our locally genotyped SNPs and significant SNPs from the CCREL test that are located around the linkage peaks on chromosomes 1 and 10. GIST determines whether an allele, or another allele in LD with it, accounts in part for the observed linkage signal (Li et al. 2004). Weights were computed for each affected sibship under three different disease models (recessive, dominant, and additive); these weights are unbiased under the null hypothesis of no disease-marker association. The correlation between the family weight variable and the nonparametric linkage (NPL) score is the basis of the test statistic. Since the GIST method is currently applicable only to affected sib pair families, we split our families into their component nuclear families before computing the NPL scores. Since we do not know the underlying disease model, we performed tests using three different disease models (recessive, dominant, and additive) and then took the maximum result, using a P value that was adjusted for multiple testing over the three models.
Tripartite Analyses
Our analyses were performed in three sequential steps. First, we analyzed the set of data that had been genotyped at CIDR. Second, after locally genotyping eight additional SNPs in the PLEKHA1/LOC387715/PRSS11 region on chromosome 10, we then analyzed the locally genotyped data set. Note that all of the known nonsynonymous SNPs in the region from PLEKHA1 (MIM 607772) through PRSS11 (MIM 602194) were investigated. Because these two data sets differ in size and composition, it is most straightforward to analyze them separately (table 2). Allele-frequency estimation, CCREL association testing, and GIST analysis were performed on both of these (overlapping) data sets, as described above. Third, we tested for interaction between the chromosome 1 and chromosome 10 regions and examined whether or not the risk differed as a function of the presence of either GA or CNV.
Table 2.
Summary of Statistical Analyses and Sample Sizes in Parts I–III
| Part and Analysis | Set of SNPs, Method, and Sample Used | Results Shown in |
| I: | ||
| htSNP selection | CIDR SNPs, 179 controls | … |
| SNP-SNP LD | CIDR SNPs, 179 controls | Figs. 4 and 6 |
| Linkage | CIDR SNPs and htSNPs, 594 ARM-affected families | Figs. 3 and 5 |
| Allele frequencies | Mendel v5 for 594 ARM-affected families; counting for 179 controls | Table 5 |
| CCREL | CIDR SNPs, 594 ARM-affected families and 179 controls | Table 5 |
| GIST | 594 ARM-affected families split into 734 typed nuclear families | Table 5 |
| II: | ||
| Allele frequencies | All SNPs (CIDR and local); Mendel v5 for 323 ARM-affected families; counting for 117 controls | Table 6 |
| CCREL | CIDR SNPs and local SNPs, 323 families and 117 controls | Table 6 |
| GIST | 323 ARM-affected families split into 407 typed nuclear families | Table 6 |
| SNP-SNP LD | CIDR and local SNPs, 117 unrelated controls | Fig. 2 |
| III: | ||
| Interaction by GIST | See GIST in I and II above | Tables 5 and 6 |
| Logistic regression | CIDR SNPs, 577 cases and 179 controls | Table 7 |
| OR and AR | CIDR SNPs, 577 cases and 179 controls; local SNPs, 517 cases (321 familial, 196 sporadic) and 117 controls | Table 8 |
| OR and AR of subtypes: | ||
| CIDR SNPs | For CNV, 407 cases and 179 controls; for GA, 184 cases and 179 controls | Table 9 |
| Local SNPs | For CNV, 366 cases and 117 controls; for GA, 159 and 117 controls | Table 9 |
Part I: Analysis of CIDR SNPs
To identify the responsible gene on chromosome 10q26, the CIDR performed high-density custom SNP genotyping of 612 ARM-affected families and 184 unrelated controls with the use of 199 SNPs spanning 13.4 Mbp (26.7 cM), from rs7080289 through rs6597818 (nucleotide position: 115094788–128517320 bp), which spans our region of interest. For our analysis, we used 196 SNPs; 3 were skipped because of a lack of polymorphism in the controls (when this was checked within the family data, the less common allele was extremely rare and was only present in heterozygotes). In addition, 684 SNPs spanning 45.7 Mbp (47.1 cM) on chromosome 1q31, from rs723858 through rs653734 (nucleotide position: 169749920–215409007 bp), were also genotyped; 5 SNPs were skipped because of a lack of polymorphism in the controls—the less common allele was either not present or very rare and, in the family data, was only present in heterozygotes. Table 3 shows the correspondence between our allele labels and the actual alleles, and, for nonsynonymous SNPs, the amino acid change.
Table 3.
Allele Labeling
|
Allele Frequency inb |
|||||
| SNP and Allele | Label | Amino Acida | CIDRControls | LocalControls | HWEP Valuec |
| rs6658788: | |||||
| A | 1 | .511 | .483 | ||
| G | 2 | .489 | .517 | .58 | |
| rs1538687: | |||||
| A | 1 | .693 | .658 | ||
| G | 2 | .307 | .342 | .41 | |
| rs1416962: | |||||
| T | 1 | .648 | .607 | ||
| C | 2 | .352 | .393 | .44 | |
| rs946755: | |||||
| T | 1 | .656 | .620 | ||
| C | 2 | .344 | .380 | .70 | |
| rs6428352: | |||||
| T | 1 | .997 | .996 | ||
| C | 2 | .003 | .004 | 1.00 | |
| rs800292: | |||||
| A | 1 = Ile | .232 | .269 | ||
| G | 2 = Val | Ile62Val | .768 | .731 | .82 |
| rs1061170: | |||||
| T | 1 = Tyr | .690 | |||
| C | 2 = His | Tyr402His | .310 | .26 | |
| rs10922093: | |||||
| G | 1 | .295 | |||
| A | 2 | .705 | .66 | ||
| rs70620: | |||||
| T | 1 | .173 | .150 | ||
| C | 2 | .827 | .850 | .28 | |
| rs1853883: | |||||
| G | 1 | .511 | .568 | ||
| C | 2 | .489 | .432 | .45 | |
| rs1360558: | |||||
| A | 1 | .397 | .389 | ||
| G | 2 | .603 | .611 | .70 | |
| rs955927: | |||||
| T | 1 | .609 | .615 | ||
| A | 2 | .391 | .385 | .85 | |
| rs4350226: | |||||
| A | 1 | .905 | .897 | ||
| G | 2 | .095 | .103 | .34 | |
| rs4752266: | |||||
| A | 1 | .777 | .774 | ||
| G | 2 | .223 | .226 | .18 | |
| rs915394: | |||||
| T | 1 | .813 | .791 | ||
| A | 2 | .187 | .209 | 1.00 | |
| rs1268947: | |||||
| G | 1 | .883 | .885 | ||
| C | 2 | .117 | .115 | .65 | |
| rs1537576: | |||||
| G | 1 | .567 | .581 | ||
| C | 2 | .433 | .419 | .35 | |
| rs2039488: | |||||
| T | 1 | .885 | .885 | ||
| C | 2 | .115 | .115 | .01 | |
| rs1467813: | |||||
| T | 1 | .293 | .295 | ||
| C | 2 | .707 | .705 | .66 | |
| rs927427: | |||||
| A | 1 | .464 | .487 | ||
| G | 2 | .536 | .513 | .10 | |
| rs4146894: | |||||
| A | 1 | .466 | .474 | ||
| G | 2 | .534 | .526 | 1.00 | |
| rs12258692: | |||||
| C | 1 = Pro | 1.000 | |||
| G | 2 = Arg | Pro233Arg | .000 | … | |
| rs4405249: | |||||
| T | 1 | .158 | |||
| C | 2 | .842 | 1.00 | ||
| rs1045216: | |||||
| G | 1 = Ala | .573 | |||
| A | 2 = Thr | Ala320Thr | .427 | .46 | |
| rs1882907: | |||||
| A | 1 | .813 | .816 | ||
| G | 2 | .187 | .184 | .76 | |
| rs10490923: | |||||
| G | 1 = Arg | .859 | |||
| A | 2 = His | His3Arg | .141 | .39 | |
| rs2736911: | |||||
| C | 1 = Arg | .881 | |||
| T | 2 = Ter | Arg38Ter | .119 | 1.00 | |
| rs10490924: | |||||
| G | 1 = Ala | .807 | |||
| T | 2 = Ser | Ser69Ala | .193 | .21 | |
| rs11538141: | |||||
| A | 1 = Glu | .995 | |||
| G | 2 = Gly | Gly54Glu | .005 | 1.00 | |
| rs760336: | |||||
| T | 1 | .520 | .526 | ||
| C | 2 | .480 | .474 | .58 | |
| rs763720: | |||||
| A | 1 | .212 | .226 | ||
| G | 2 | .788 | .774 | .79 | |
| rs1803403: | |||||
| T | 1 = Cys | .030 | |||
| G | 2 = Gly | Cys384Gly | .970 | 1.00 | |
Amino acid change of nonsynonymous SNPs.
CIDR controls (n=179) and local controls (117 overlap CIDR controls).
HWE P value of the exact test.
Part II: Analysis of Locally Genotyped SNPs
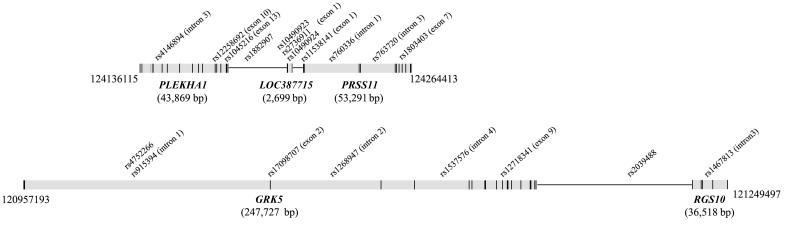
We genotyped eight additional SNPs on chromosome 10 that overlie three susceptibility genes, PLEKHA1 (rs12258692, rs4405249, and rs1045216), LOC387715 (rs10490923, rs2736911, and rs10490924), and PRSS11 (rs11538141 and rs1803403). This genotyping effort included all of the nonsynonymous SNPs that have been reported for these genes in the NCBI databases (see fig. 1). As part of another study (Conley et al. 2005), we genotyped two CFH variants (rs10922093 and rs1061170), which we have used here as well. Genotyping of additional SNPs under the GRK5/RGS10 (MIM 600870/MIM 602856) locus is in process. Genotype data for rs12258692, rs1803403, and the newly characterized SNP rs4405249 (which is 1 base 3′ of rs12258692) were collected by sequencing (Rexagen) and were analyzed using Sequencher software (Gene Codes). Genotype data for rs11538141, rs2736911, rs10490923, and rs10490924 were collected using RFLP. The primers, amplification conditions, and restriction endonucleases, where appropriate, for SNPs that were genotyped by sequencing or RFLP can be found in table 4. Genotype data for rs1045216 were collected using a 5′ exonuclease Assay-on-Demand TaqMan assay (Applied Biosystems). Amplification and genotype assignments were conducted using the ABI 7000 and SDS 2.0 software (Applied Biosystems). Two unrelated CEPH samples were genotyped for each variant and were included on each gel and in each TaqMan tray, to assure internal consistency in genotype calls. Additionally, double-masked genotyping assignments were made for each variant and were compared, and each discrepancy was addressed using raw data or regenotyping. Table 3 shows the correspondence between our allele labels and the actual alleles and, for nonsynonymous SNPs, the amino acid change.
Figure 1.
Location of CIDR SNPs and locally genotyped SNPs with respect to candidate genes. Positions, distances, and nucleotide positions along chromosome 10 are derived from NCBI Entrez Gene and dSNP databases.
Table 4.
Primers, Annealing Conditions, and Restriction Endonucleases Used for Genotype Data Collection[Note]
|
Primer Sequences |
||||
| Variant | Forward | Reverse | Annealing Temperature(°C) | Restriction Enzyme |
| rs11538141 | CAG AGT CGC CAT GCA GAT CC | CCC GAA GGG CAC CAC GCA CT | 58 | MnlI |
| rs2736911 | GCA CCT TTG TCA CCA CAT TA | GCC TGA TCA TCT GCA TTT CT | 54 | DraIII |
| rs10490923 | GCA CCT TTG TCA CCA CAT TA | GCC TGA TCA TCT GCA TTT CT | 54 | HhaI |
| rs10490924 | GCA CCT TTG TCA CCA CAT TA | GCC TGA TCA TCT GCA TTT CT | 54 | PvuII |
| rs1803403 | TGC TGT CCC TTT GTT GTC TC | AGA CAC AGA CAC GCA TCC TG | 55 | NA |
| rs12258692 (and rs4405249) | GCC AGG AAA AGG AAC CTC | GCC AGG CAT CAA GTC AGA | 54 | NA |
Note.— NA = not applicable.
Part III: Interaction and OR Analysis
Unrelated cases
No unrelated cases were genotyped by CIDR, but 196 unrelated cases were genotyped locally for our additional SNPs. For computation of odds ratios (ORs) and for interaction analyses (see below), we chose to generate a set of unrelated cases by drawing one type A–affected person from each family. A total of 321 locally genotyped families had at least one type A–affected person. If a family had more than one type A–affected person, we chose the person who had the most complete genotyping at the Y402H variant (rs1061170) and three CIDR SNPs representative of CFH, GRK5, and PLEKHA1: rs800292 (CFH), rs1537576 (GRK5), and rs4146894 (PLEKHA1; rs4146894 also represents LOC387715, because of high LD with rs10490924) (see fig. 2). If they could not be distinguished by the number of genotyped SNPs, we chose the person who developed the disease at the youngest age, or, if more than one shared the earliest age at onset, we selected one type A–affected individual at random from those with the most SNPs genotyped and the earliest age at onset. A total of 577 CIDR families had at least one type A–affected person; 321 of these families were also genotyped locally, and the type A–affected person was the same one chosen for the local set. For the remaining 256 families, we based our selection on the same criteria described above, except that only rs800292 (CFH), rs1537576 (GRK5), and rs4146894 (PLEKHA1) were used to identify the person with the most complete genotyping.
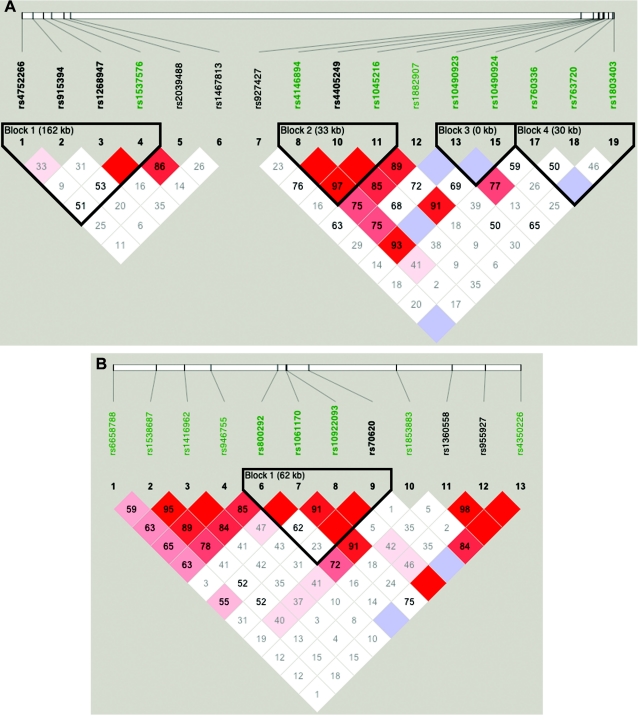
Figure 2.
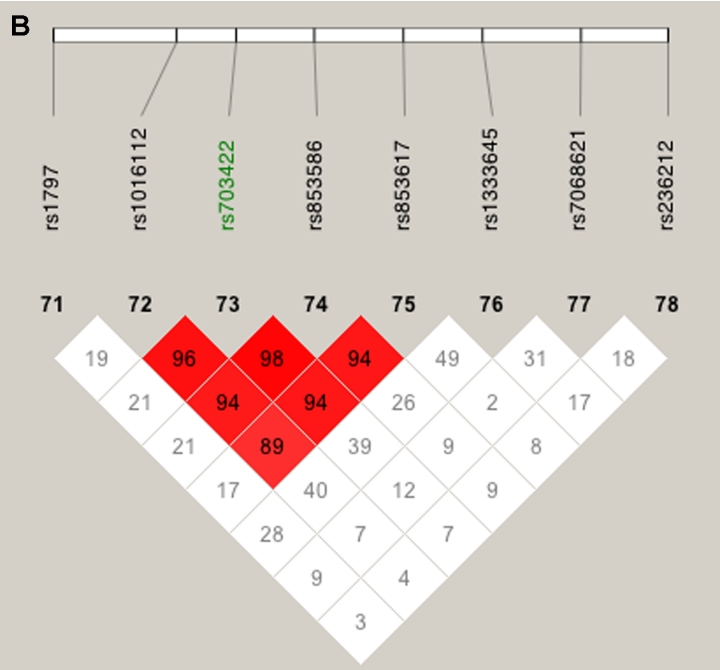
A, LD patterns in GRK5 (Block 1), RGS10 (SNP 6), PLEKHA1 (Block 2), LOC387715 (Block 3), and PRSS11 (Block 4). B, LD patterns in CFH (Block 1). Squares shaded pink or red indicate significant LD between SNP pairs (bright red indicates pairwise D′=1), white squares indicate no evidence of significant LD, and blue squares indicate pairwise D′=1 without statistical significance. Significant SNPs from the CCREL allele test are highlighted in green (see table 6). Three SNPs (rs6428352, rs12258692, and rs11538141) were not included, because of very low heterozygosity, and one SNP (rs2736911) was not included, because it was uninformative. Note that the blocks were drawn to show clearly the position of the genes and do not represent haplotype blocks.
Analysis of interaction with CFH
We investigated possible interaction between CFH on chromosome 1 and the genes on chromosome 10 by using GIST to test whether SNPs in CFH are associated with the linkage signal on chromosome 10 and whether SNPs on chromosome 10 are associated with the linkage signal on chromosome 1. We did this by using weights from SNPs on one chromosome and family-based NPLs from the other.
We also used logistic regression to evaluate different interaction models and to test for interaction by use of the approach described by North et al. (2005). In this approach, many different possible models of the interactions, allowing simultaneously for additive and dominant effects at both of the loci, are fit, and relative likelihoods of the different models are compared to draw inferences about the most likely and parsimonious model. As described elsewhere (North et al. 2005), the fitted models include a MEAN model, in which only the mean term is estimated; ADD1, ADD2, and ADD models, which assume an additive effect at one or both loci; DOM1, DOM2, and DOM models, which additionally incorporate dominance effects; and three further models, ADDINT, ADDDOM, and DOMINT, which allow for interactive effects (for more details, see the work of North et al. [2005]). Since some pairs of these models are not nested, we compared them by using the Akaike information criteria (AIC); in this approach, the model with the lowest AIC is considered to be the best fitting and the most parsimonious. For these analyses, we used the program provided by North and colleagues (2005), after some bugs that we discovered had been fixed; we double-checked our results with our own R program. To maximize the sample size, we chose CIDR SNPs in high LD with a highly significant nonsynonymous SNP within each gene. The CIDR SNP rs800292 was chosen to represent rs10611710 (the Y402H variant of CFH), and the CIDR SNP rs4146894 represented rs1045216 in PLEKHA1. Similarly, we also selected a representative CIDR SNP in GRK5, RGS10, and PRSS11.
Magnitude of association
We calculated crude ORs and estimated ARs for SNPs in each gene. The allele that was least frequent in the controls was considered to be the risk allele. AR was estimated using the formula AR=100×P×(OR-1)/[1+P×(OR-1)], where OR is the OR and P is the frequency of the risk factor (genotype) in the population, as estimated from the controls. We did this by comparing type A–affected subjects with controls, comparing subjects who had CNV with controls, and comparing subjects who had GA with controls. To have the maximum possible sample size, we used different but overlapping samples for CIDR and locally typed SNPs. A total of 577 cases selected from the families and 179 unrelated cases were used for calculating OR and AR of CIDR SNPs, but 517 cases (of those, 321 are within the 577 CIDR SNP cases) and 117 controls (all within the 179 CIDR SNP controls) were used for calculating the OR and AR on the locally genotyped SNPs.
Multiple-testing issues
Since we have very strong evidence from previous studies that there is an ARM-susceptibility locus in the chromosome 10q26 region, the analyses performed here were aimed at estimating the location of the susceptibility gene, rather than testing a hypothesis. Multiple-testing issues are most crucial and relevant in the context of hypothesis testing. In estimation, we are simply interested in determining where the signal is strongest. In any event, any correction for multiple testing would not alter the rank order of the results. A Bonferroni correction, which does not account for any correlation between tests due to LD, for 196 tests at the 0.05 level would lead to a significance threshold of 0.05/196=0.00026; correlations due to LD would lead to a larger threshold.
Results
Our analyses were performed in three sequential steps. First, we analyzed the set of data that had been genotyped at CIDR. Second, after locally genotyping eight additional SNPs in the PLEKHA1/LOC387715/PRSS11 region on chromosome 10, we then analyzed the locally genotyped data set. Allele-frequency estimation, testing for HWE (table 3), CCREL association testing, and GIST testing were performed on both of these (overlapping) data sets, as described above. Third, we tested for interaction between the chromosome 1 and chromosome 10 regions and examined whether or not the risk differed as a function of the presence of GA or CNV.
Part I: Analysis of CIDR SNPs
CIDR linkage results
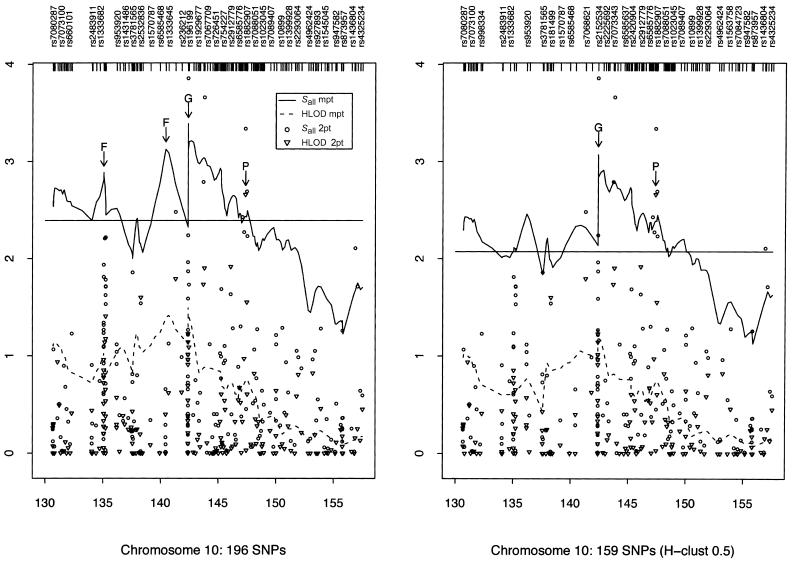
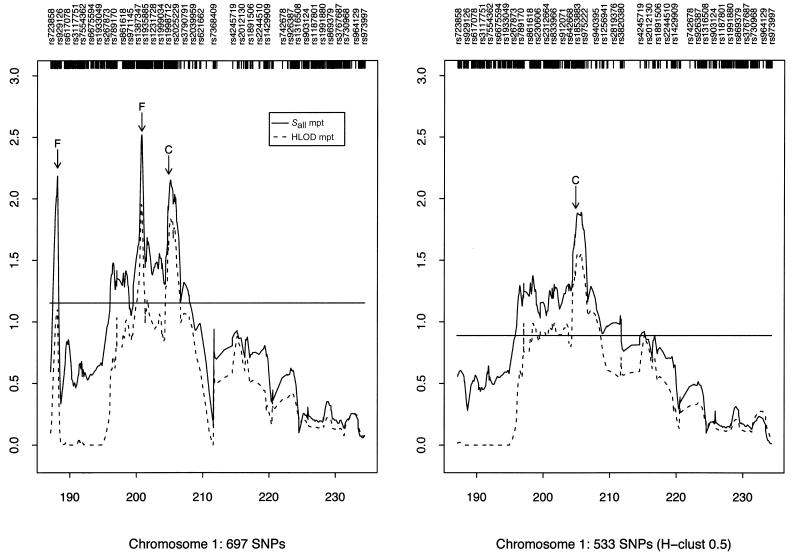
The narrow peak of our Sall linkage curve obtained using the 159 htSNPs on chromosome 10 suggests that there might be an ARM gene in the GRK5 region (marked “G” in fig. 3, right panel); rs1537576 in GRK5 had a two-point Sall of 1.87, whereas the largest (across our whole region) two-point Sall of 3.86 occurred at rs555938, 206 kb centromeric of GRK5. Several elevated two-point nonparametric Sall LOD scores and our highest HLOD score drew attention to the PLEKHA1/LOC387715/PRSS11 region (marked “P” in fig. 3). In this region, SNP rs4146894 in PLEKHA1 had a two-point Sall of 3.34 and the highest two-point HLOD of 2.66, whereas SNPs rs760336 and rs763720 in PRSS11 had two-point Sall values of 2.69 and 2.23, respectively. However, the 1-unit support interval is large (10.06 cM) (fig. 3), and so localization from the linkage analyses alone is rather imprecise.
Figure 3.
Two-point (2pt) and multipoint (mpt) linkage results on chromosome 10. The panel on the left summarizes the results when all SNPs were used for analysis. The panel on the right summarizes the results when only htSNPs were used. The peaks marked “F” represent likely false peaks due to high SNP-SNP LD, whereas the peaks marked “G” and “P” correspond to the loci containing GRK5 and PLEKHA1, respectively. The horizontal lines indicate the 1-unit support interval of multipoint Sall (i.e., maximum Sall-1).
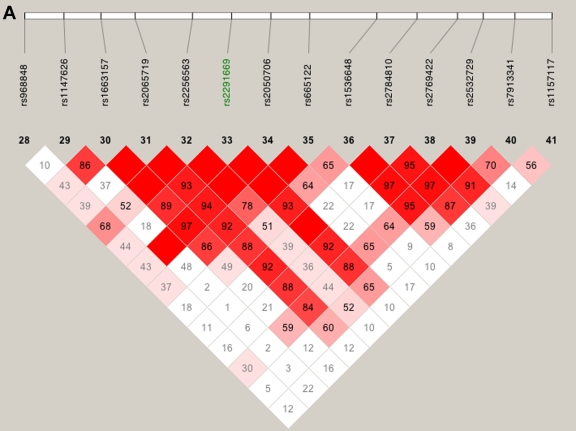
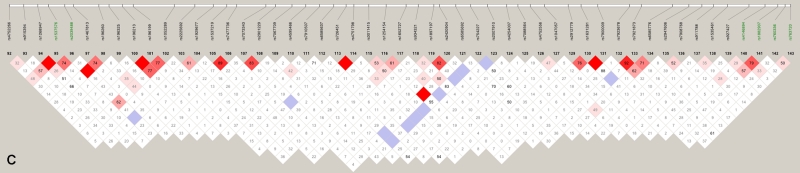
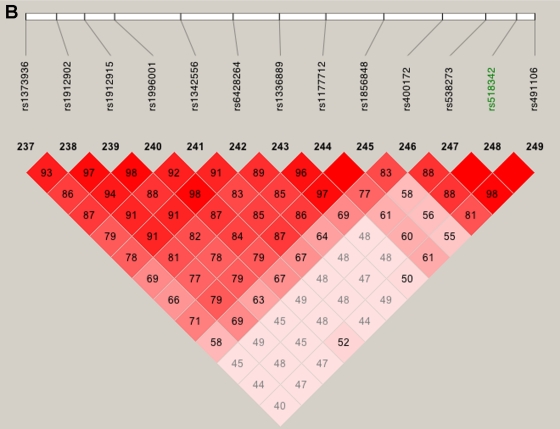
We also explored the effect of failing to take SNP-SNP LD into account, by comparing the multipoint scores computed using all SNPs (fig. 3, left panel) with those computed using only the htSNPs (fig. 3, right panel). Two of the peaks found using all SNPs (referred to as “false peaks”; marked “F” in fig. 3, left panel) almost vanish completely when using only htSNPs; interestingly, these two peaks lie within haplotype blocks (fig. 4A and 4B), whereas the LD around our highest multi- and two-point LOD scores is low (fig. 4C), indicating the importance of taking LD into account when performing linkage analysis.
Figure 4.
LD patterns on chromosome 10 based on analysis of 196 CIDR SNPs and 179 unrelated controls. A, The false peak at 135 cM (see fig. 3); the SNP with the largest Sall in the peak is highlighted in green. B, The false peak at 142 cM (see fig. 3); the SNP with the largest Sall in the peak is highlighted in green. C, Linkage peak. Significant SNPs, from CCREL (table 5), that overlie the five genes (GRK5, RGS10, PLEKHA1, LOC387715, and PRSS11) are highlighted in green. Squares shaded pink or red indicate significant LD between SNP pairs (bright red indicates pairwise D′=1), white squares indicate no evidence of significant LD, and blue squares indicate pairwise D′=1 without statistical significance. LD is measured using D′, and the values within the squares give pairwise LD in D′/100.
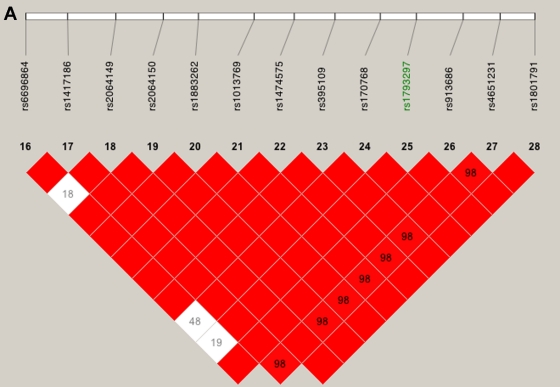
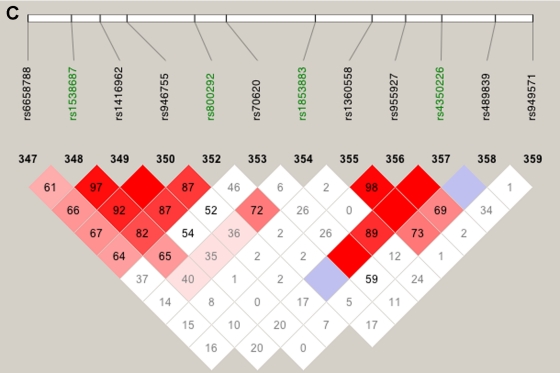
Our linkage results on chromosome 1 gave three peaks with Sall>2, and only one of those peaks was observed when we restricted our analysis to htSNPs (fig. 5). This remaining peak overlies CFH and includes two SNPs with very high two-point Sall and HLOD scores: rs800292, a nonsynonymous SNP in CFH, had an Sall of 1.53 and an HLOD of 2.11, whereas SNP rs1853883, 165 kb telomeric of CFH, had an Sall of 4.06 and an HLOD of 3.49. These results strongly support earlier findings of CFH’s involvement in ARM (Conley et al. 2005; Edwards et al. 2005; Hageman et al. 2005; Haines et al. 2005; Klein et al. 2005; Zareparsi et al. 2005a). The vanishing peaks (marked “F” in fig. 5, left panel) that we saw when we used all of our SNPs in the linkage analysis are located within strong haplotype blocks (fig. 6A and 6B), whereas the LD under the CFH peak is relatively low (fig. 6C).
Figure 5.
Multipoint linkage results for chromosome 1. The panel on the left summarizes results when all SNPs were used for analysis, and the panel on the right summarizes results when only htSNPs were used. The peaks marked “F” represent likely false peaks due to high SNP-SNP LD, whereas the peak marked “C” corresponds to the CFH gene. The horizontal lines indicate the 1-unit support interval of multipoint Sall (i.e., maximum Sall over CFH-1).
Figure 6.
LD patterns on chromosome 1 based on analysis of 679 CIDR SNPs and 179 unrelated controls. A, The false peak at 188 cM (see fig. 5); the SNP with the largest Sall in the peak is highlighted in green. B, The false peak at 202 cM (see fig. 5); the SNP with the largest Sall in the peak is highlighted in green. C, Linkage peak. Significant SNPs, from CCREL (table 5), that overlie CFH are highlighted in green. Squares shaded pink or red indicate significant LD between SNP pairs (bright red indicates pairwise D′=1), white squares indicate no evidence of significant LD, and blue squares indicate pairwise D′=1 without statistical significance. LD is measured using D′, and the values within the squares give pairwise LD in D′/100.
CIDR association results
For finer localization than can be obtained by linkage, we turned to association analyses (which were very successful in discovering CFH on chromosome 1). Here, we performed association analyses using the CCREL approach (Browning et al. 2005), which permitted the simultaneous use of our unrelated controls and all of our related familial cases by appropriately adjusting for the relatedness of the cases. In the CIDR sample on chromosome 10, within our linkage peak, we found a cluster of four adjacent SNPs with very small P values (rs4146894, rs1882907, rs760336, and rs763720) that overlies three genes: PLEKHA1, LOC387715, and PRSS11. Our strongest CCREL results on chromosome 10 were for SNP rs4146894 in PLEKHA1 (table 5). The moving-window haplotype analyses using three SNPs at a time resulted in very small P values across the whole PLEKHA1 to PRSS11 region (table 5). The association testing also generated some moderately small P values in the GRK5 region, which is where our highest evidence of linkage occurred.
Table 5.
CCREL, GIST, and Allele-Frequency Estimation for Families and Controls Typed at CIDR[Note]
|
P Value for Test |
|||||||||
|
Frequency in |
Moving-WindowHaplotype Test |
GIST |
|||||||
| SNP | Gene | Families(n=594) | Controls(n=179) | Allele Test | With 2 SNPs | With 3 SNPs | Genotype Test | NPL 10 | NPL 1 |
| rs6658788 | .460 | .489 | .37312 | .01616 | .00778 | .44415 | .106 | .055 | |
| rs1538687 | .234 | .307 | .00178 | .00206 | .00674 | .0054 | .781 | .129 | |
| rs1416962 | .321 | .352 | .16378 | .39256 | .4157 | .38009 | .566 | .019 | |
| rs946755 | .317 | .344 | .20073 | .20147 | <.00001 |
.37434 | .513 | .012 | |
| rs6428352 | .001 | .003 | … | <.00001 |
<.00001 |
… | … | … | |
| rs800292 | CFH | .132 | .232 | <.00001 |
<.00001 |
<.00001 |
<.00001 |
.437 | .001 |
| rs70620 | CFH | .147 | .173 | .15602 | <.00001 |
<.00001 |
.33122 | .893 | .333 |
| rs1853883 | .630 | .489 | <.00001 |
<.00001 |
<.00001 |
<.00001 |
.521 | <.001 |
|
| rs1360558 | .425 | .397 | .34842 | .60377 | .01118 | .63012 | .183 | .296 | |
| rs955927 | .416 | .391 | .36201 | .00833 | … | .65613 | .065 | .145 | |
| rs4350226 | .055 | .095 | .00182 | … | … | .00183 | .171 | .242 | |
| rs4752266 | GRK5 | .220 | .223 | .84131 | .23223 | .28973 | .03802 | .088 | .475 |
| rs915394 | GRK5 | .214 | .187 | .15214 | .19235 | .00309 | .35594 | .028 | .643 |
| rs1268947 | GRK5 | .112 | .117 | .97426 | .00969 | .01031 | .97976 | .052 | .345 |
| rs1537576 | GRK5 | .507 | .433 | .01881 | .01354 | .03257 | .0295 | .006 | .251 |
| rs2039488 | .078 | .115 | .01339 | .07877 | … | .05075 | .004 | .609 | |
| rs1467813 | RGS10 | .286 | .293 | .63177 | … | … | .71857 | .539 | .582 |
| rs927427 | .514 | .464 | .06936 | .00003 |
.00002 |
.05976 | .198 | .577 | |
| rs4146894 | PLEKHA1 | .598 | .466 | <.00001 |
<.00001 |
.00001 |
<.00001 |
.008 | .802 |
| rs1882907 | .127 | .187 | .00261 | .00013 |
.00006 |
.00521 | .169 | .172 | |
| rs760336 | PRSS11 | .395 | .480 | .00469 | .00126 | … | .02036 | .232 | .581 |
| rs763720 | PRSS11 | .295 | .212 | .00053 |
… | … | .00290 | .198 | .021 |
Note.— The minor-allele frequency is reported for controls (estimated by counting) and families (estimated by Mendel, version 5). Allele frequencies that differed between controls and families by >0.1 are in bold italics. For the allele test, the two-SNP and three-SNP moving-window haplotype tests, and the genotype test from the CCREL, P values ⩽.05 are in bold italics and P values ⩽.001 are underlined. The moving-window haplotype P values correspond to the SNPs in the same row as the P value and the next one or two SNPs for the two- and three-SNP moving window, respectively. For GIST, with the use of NPL scores from chromosome 1 (NPL 1) and chromosome 10 (NPL 10), P values ⩽.05 are in bold italics and P values ⩽.001 are underlined. Blank spaces separate the three chromosomal regions corresponding to SNPs in and around CFH, GRK5/RGS10, and PLEKHA1/LOC687715/PRSS11.
We performed the CCREL on 56 SNPs spanning the linkage peak on chromosome 1 and found two highly significant SNPs (rs800292 and rs1853883) that overlie CFH (table 5). The moving-window haplotype analyses, performed using two and three SNPs at a time, resulted in extremely low P values across the whole CFH gene (table 5), which supports earlier findings of strong association between CFH and ARM.
CIDR GIST results
When GIST was performed on the CIDR data set, the two smallest P values in chromosome 10q26 (.006 and .004) occurred in the GRK5/RGS10 region, whereas the third smallest P value (.008) occurred in PLEKHA1 (table 5). All four SNPs in the GRK5 gene have small GIST P values. The GIST results suggest that both GRK5 and PLEKHA1 contribute significantly to the linkage signal on chromosome 10 and that CFH contributes to the linkage signal on chromosome 1. Neither of the two SNPs in PRSS11 contributes significantly to the linkage signal on chromosome 10. There was no evidence that the genes on chromosome 10 were related to the linkage signal seen on chromosome 1.
PART II: Analysis of Locally Genotyped SNPs
Local association results
After additional SNPs were typed locally, the allele and genotype test generated extremely small P values for each of the three genes PLEKHA1, LOC387715, and PRSS11 (table 6). The moving-window haplotype analyses with three SNPs resulted in very small P values across the entire PLEKHA1/LOC387715/PRSS11 region (table 6). Thus, although association implicates the PLEKHA1/LOC387715/PRSS11 region, it does not distinguish between these genes.
Table 6.
CCREL, GIST, and Allele-Frequency Estimation for Locally Typed Families and Controls[Note]
|
P Value for Test |
|||||||||
|
Frequency in |
Moving-WindowHaplotype Test |
GIST |
|||||||
| SNP | Gene | Families(n=323) | Controls(n=117) | Allele Test | With 2 SNPs | With 3 SNPs | Genotype Test | NPL 10 | NPL 1 |
| rs6658788 | .563 | .483 | .02200 | .00052 |
.00162 | .04920 | .319 | .244 | |
| rs1538687 | .213 | .342 | .00004 |
.00043 |
.00066 |
.00014 |
.652 | .302 | |
| rs1416962 | .299 | .393 | .00597 | .02623 | .02051 | .01819 | .442 | .041 | |
| rs946755 | .295 | .380 | .01234 | .01243 | <.00001 |
.04531 | .409 | .040 | |
| rs6428352 | .001 | .004 | … | <.00001 |
<.00001 |
… | … | … | |
| rs800292 | CFH | .120 | .269 | <.00001 |
<.00001 |
<.00001 |
<.00001 |
.315 | .014 |
| rs1061170 | CFH | .609 | .310 | <.00001 |
<.00001 |
<.00001 |
<.00001 |
.895 | .132 |
| rs10922093 | CFH | .210 | .295 | .00693 | .00175 | <.00001 |
.01723 | .360 | .327 |
| rs70620 | CFH | .148 | .150 | .91163 | <.00001 |
<.00001 |
.56770 | .737 | .356 |
| rs1853883 | .633 | .432 | <.00001 |
<.00001 |
<.00001 |
<.00001 |
.776 | .011 | |
| rs1360558 | .437 | .389 | .18014 | .43576 | .02079 | .37993 | .975 | .488 | |
| rs955927 | .433 | .385 | .15343 | .01037 | … | .36087 | .017 | .585 | |
| rs4350226 | .050 | .103 | .00312 | … | … | .00373 | .228 | .174 | |
| rs4752266 | GRK5 | .223 | .226 | .81772 | .27748 | .64917 | .08279 | .107 | .453 |
| rs915394 | GRK5 | .228 | .209 | .34489 | .83219 | .05560 | .62183 | .049 | .320 |
| rs1268947 | GRK5 | .117 | .115 | .81975 | .02748 | .02192 | .78965 | .049 | .689 |
| rs1537576 | GRK5 | .497 | .419 | .02604 | .02232 | .05636 | .06334 | .012 | .023 |
| rs2039488 | .083 | .115 | .11177 | .42428 | … | .42399 | .025 | .358 | |
| rs1467813 | RGS10 | .293 | .295 | .86608 | … | … | .85954 | .506 | .492 |
| rs927427 | .506 | .487 | .56710 | .00056 |
.00083 |
.42264 | .306 | .625 | |
| rs4146894 | PLEKHA1 | .611 | .474 | .00004 |
.00012 |
.00053 |
.00024 |
.006 | .737 |
| rs12258692 | PLEKHA1 | .008 | .000 | … | .54750 | .00018 |
… | … | … |
| rs4405249 | PLEKHA1 | .139 | .158 | .39378 | .00026 |
.00280 | .33118 | .003 | .345 |
| rs1045216 | PLEKHA1 | .289 | .427 | .00004 |
.00036 |
.00001 |
.00026 |
.068 | .825 |
| rs1882907 | .131 | .184 | .01761 | .00140 | .01099 | .04401 | .017 | .372 | |
| rs10490923 | LOC387715 | .089 | .141 | .02112 | .05024 | <.00001 |
.03415 | .086 | .251 |
| rs2736911 | LOC387715 | .121 | .119 | .71668 | <.00001 |
<.00001 |
.64230 | .312 | .968 |
| rs10490924 | LOC387715 | .475 | .193 | <.00001 |
<.00001 |
<.00001 |
<.00001 |
.018 | .327 |
| rs11538141 | PRSS11 | .004 | .005 | … | .00726 | .01676 | … | … | … |
| rs760336 | PRSS11 | .373 | .474 | .00527 | .01386 | .00036 |
.01396 | .479 | .683 |
| rs763720 | PRSS11 | .296 | .226 | .01645 | .00016 |
… | .03899 | .305 | .451 |
| rs1803403 | PRSS11 | .118 | .030 | .00009 |
… | … | .00022 |
.714 | .778 |
Note.— The minor-allele frequency is reported for controls (estimated by counting) and families (estimated by Mendel, version 5). Allele frequencies that differed between controls and families by >0.1 are in bold italics. For the allele test, the two-SNP and three-SNP moving-window haplotype tests, and the genotype test from the CCREL, P values ⩽.05 are in bold italics and P values ⩽.001 are underlined. The moving-window haplotype P values correspond to the SNPs in the same row as the P value and the next one or two SNPs for the two- and three-SNP moving window, respectively. For GIST, with the use of NPL scores from chromosome 1 (NPL 1) and chromosome 10 (NPL 10), P values ⩽.05 are in bold italics and P values ⩽.001 are underlined. Locally typed SNPs are in bold italics. Blank spaces separate the three chromosomal regions corresponding to SNPs in and around CFH, GRK5/RGS10, and PLEKHA1/LOC687715/PRSS11.
Local GIST results
Of the three genes PLEKHA1, LOC387715, and PRSS11, GIST most strongly implicated PLEKHA1 (table 6). It also generated a small P value for rs10490924 in LOC387715, but this SNP is in high LD with the PLEKHA1 SNPs (see fig. 2A). When the locally typed data set was used, GIST did not generate any significant results for PRSS11, similar to the nonsignificant results observed in the larger CIDR sample. This implies that PLEKHA1 (or a locus in strong LD with it) is the most likely to be involved in ARM, and therefore LOC387715 remains a possible candidate locus.
For a fair assessment of which SNP accounts for the linkage signal across the region, the NPLs were computed using only the locally genotyped families. This permitted us to compare the PLEKHA1/LOC387715/PRSS11 results (table 6) directly with the GRK5/RGS10 results. For the locally typed data set, the GIST results for GRK5 are also interesting, with modest P values of the same magnitude as the P values we got from applying GIST to CFH (table 6). However, note that the P values are not as small as those seen when the CIDR data set was analyzed. Since all of the SNPs in the GRK5 region are CIDR SNPs, this difference is solely a function of sample size, because the locally typed data set is smaller than the CIDR data set (see table 2).
Part III: Interaction and OR Analyses
GIST results
We did not see any strong evidence of an interaction between the chromosome 1 and chromosome 10 regions, by use of GIST. When the CIDR data set was used to test whether SNPs on chromosome 10 contribute to the linkage signal on chromosome 1 (see GIST, NPL 1, in table 5), only rs763720 in PRSS1 gave a P value <.05; however, rs763720 does not contribute significantly to the linkage signal on chromosome 10, which makes this P value less convincing. When we used the local data set, one GRK5 variant (rs1537576), which was not significant in the larger CIDR data set, gave a P value <.05. Similarly, we did not see evidence that SNPs within CFH contribute to the linkage signal on chromosome 10; only one SNP (rs955927) gave a P value <.05—this SNP, however, is not in the CFH gene and is not in strong LD (see fig. 2B) with any SNPs in CFH.
Logistic regression results
The logistic regression results (table 7) suggest that an additive model including the variants from CFH and PLEKHA1 is the best model for predicting case-control status; this indicates that both genes are important to the ARM phenotype. The AIC criteria also suggest that an additive model including an additive interaction term is the next best model (table 7); however, the interaction term is not significant (P=.71). We obtain similar results for interaction between CFH and PRSS11, where the additive model including both variants appears to be the best model. Within the GRK5/RGS10 region, a model with the CFH SNP alone is the best-fitting model, which suggests that the prediction of case-control status with CFH genotype does not improve by the addition of either the GRK5 or RGS10 variant to the model.
Table 7.
Results of Fitting Two-Locus Models by Logistic Regression
| Locus 2 and Modela | AIC | AIC Differenceb |
| rs1537576 (GRK5): | ||
| MEAN | 822.5 | 23.65 |
| ADD1 | 798.8 | 0 |
| ADD2 | 821.2 | 22.35 |
| ADD | 799.1 | .26 |
| DOM1 | 799.7 | .91 |
| DOM2 | 820.1 | 21.24 |
| DOM | 799.2 | .37 |
| ADDINT | 800.9 | 2.07 |
| ADDDOM | 802.1 | 3.25 |
| DOMINT | 803.9 | 5.07 |
| rs1467813 (RGS10): | ||
| MEAN | 821.9 | 23.53 |
| ADD1 | 798.4 | 0 |
| ADD2 | 823.6 | 25.25 |
| ADD | 800.3 | 1.92 |
| DOM1 | 799.3 | .91 |
| DOM2 | 825.2 | 26.79 |
| DOM | 802.6 | 4.23 |
| ADDINT | 801.3 | 2.93 |
| ADDDOM | 804.9 | 6.54 |
| DOMINT | 805.2 | 6.83 |
| rs4146894 (PLEKHA1): | ||
| MEAN | 823.02 | 49.26 |
| ADD1 | 799.24 | 25.49 |
| ADD2 | 801.47 | 27.71 |
| ADD | 773.76 | 0 |
| DOM1 | 800.16 | 26.41 |
| DOM2 | 803.44 | 29.68 |
| DOM | 776.44 | 2.68 |
| ADDINT | 775.62 | 1.87 |
| ADDDOM | 779.85 | 6.09 |
| DOMINT | 778.26 | 4.5 |
| rs760336 (PRSS11): | ||
| MEAN | 821.9 | 27.32 |
| ADD1 | 798.4 | 3.78 |
| ADD2 | 817.1 | 22.54 |
| ADD | 794.6 | 0 |
| DOM1 | 799.3 | 4.69 |
| DOM2 | 819 | 24.37 |
| DOM | 796.7 | 2.14 |
| ADDINT | 796 | 1.43 |
| ADDDOM | 802.1 | 7.46 |
| DOMINT | 803.4 | 8.75 |
Locus 1 is rs800292 (CFH). Model definitions are in the text.
Difference of the AIC from the best-fitting model.
OR and AR
We estimated the magnitude of association by calculating OR and AR values; the significant associations we saw (table 8) are, not surprisingly, consistent with the results from the CCREL tests in parts I and II. Our two most significant SNPs in the PLEKHA1/LOC387715 region are SNPs rs4146894 (PLEKHA1) and rs10490924 (LOC387715); the two tests are highly correlated because the LD between those SNPs is very high (D′=0.93) (see fig. 2A). The third most significant SNP (rs1045216) in the chromosome 10 region is a nonsynonymous SNP in PLEKHA1 and in high LD with both rs4146894 (D′=0.97) and rs10490924 (D′=0.91).
Table 8.
ORs, ARs, and Simulated P Values from χ2 Test with 10,000 Replicates[Note]
|
Dominant([RR+RN] vs. NN) |
Heterozygotes(RR vs. NN) |
Recessive(RR vs. [RN+NN]) |
Homozygotes(RR vs. NN) |
||||||||||
| SNP (Allele) | Gene | OR | 95% CI | AR | P | OR | AR | OR | 95% CI | AR | P | OR | AR |
| rs6658788 (2) | .83 | .57–1.22 | −14.04 | .3909 | 1.09 | 2.69 | 1.01 | .68–1.5 | .21 | 1 | .88 | −5.92 | |
| rs1538687 (2) | .68 | .49–.95 | −19.38 | .023 | .5 | −11.74 | .42 | .23–.78 | −6.52 | .0068 | .38 | −12.42 | |
| rs1416962 (2) | .84 | .6–1.18 | −10.02 | .3418 | .89 | −2.57 | .82 | .49–1.38 | −2.31 | .5002 | .77 | −5.74 | |
| rs946755 (2) | .8 | .57–1.13 | −12.52 | .232 | 1 | .04 | .9 | .53–1.52 | −1.24 | .7816 | .81 | −4.34 | |
| rs6428352 (2) | … | … | … | … | … | … | … | … | … | … | … | … | |
| rs800292 (1) | CFH | .43 |
.3–.62 | −30.01 |
<.0001 | .48 |
−23.85 |
.15 |
.05–.45 | −4.98 |
.0001 | .12 |
−8.19 |
| rs1061170 (2) | CFH | 5.29 |
3.35–8.35 | 68.2 |
<.0001 | 2.66 | 28.55 | 4.57 |
2.48–8.42 | 30.06 |
<.0001 | 10.05 |
63.72 |
| rs10922093 (1) | CFH | .59 | .39–.88 | −25.61 | .0111 | .63 | −19.65 | .5 | .24–1.04 | −4.98 | .0736 | .41 | −10.14 |
| rs70620 (1) | CFH | .83 | .57–1.19 | −5.64 | .3366 | .85 | −4.29 | .67 | .27–1.68 | −1.3 | .4525 | .64 | −1.93 |
| rs1853883 (2) | 2.67 |
1.78–4.01 | 54.41 |
<.0001 | 1.65 | 19.21 | 2.08 |
1.43–3.02 | 22.06 |
.0003 | 3.55 |
55.04 |
|
| rs1360558 (1) | 1.16 | .82–1.65 | 9.12 | .414 | 1.1 | 5.39 | 1.25 | .8–1.96 | 3.94 | .3774 | 1.32 | 9.01 | |
| rs955927 (2) | 1.13 | .79–1.6 | 7.5 | .5303 | 1.28 | 6.35 | 1.31 | .83–2.08 | 4.53 | .2588 | 1.36 | 9.38 | |
| rs4350226 (2) | .51 | .32–.81 | −9.68 | .0038 | .27 | −4.76 | .16 | .01–1.74 | −.95 | .142 | .14 | −1.16 | |
| rs4752266 (2) | GRK5 | .88 | .62–1.23 | −5.57 | .4325 | 3.27 | 10.68 | 2.81 | .98–8.04 | 3.89 | .0457 | 2.56 | 5.51 |
| rs915394 (2) | GRK5 | 1.28 | .9–1.82 | 8.91 | .1543 | 1.35 | 2.73 | 1.56 | .58–4.14 | 1.53 | .3892 | 1.68 | 2.72 |
| rs1268947 (2) | GRK5 | 1.05 | .7–1.57 | 1.06 | .841 | 1.24 | 1.82 | 1.27 | .35–4.55 | .45 | .7761 | 1.28 | .58 |
| rs1537576 (2) | GRK5 | 1.59 | 1.11–2.29 | 27.95 | .0109 | .89 | −3.74 | 1.08 | .71–1.62 | 1.59 | .7579 | 1.47 | 15.14 |
| rs2039488 (2) | .7 | .45–1.07 | −6.5 | .1067 | .23 | −11.98 | .19 | .04–.79 | −2.33 | .0242 | .18 | −2.85 | |
| rs1467813 (1) | RGS10 | .96 | .69–1.35 | −1.84 | .8645 | 1.01 | .42 | .77 | .42–1.38 | −2.27 | .4265 | .77 | −3.76 |
| rs927427 (1) | 1.09 | .74–1.62 | 6.57 | .6172 | .94 | −4.66 | 1.67 | 1.09–2.56 | 10.73 | .0201 | 1.6 | 19.91 | |
| rs4146894 (1) | PLEKHA1 | 2.22 |
1.49–3.31 | 46.78 |
.0002 | 1.77 | 33.08 | 2.21 |
1.49–3.29 | 20.46 |
<.0001 | 3.31 |
49.88 |
| rs12258692 (2) | PLEKHA1 | … | … | … | … | … | … | … | … | … | … | … | … |
| rs4405249 (1) | PLEKHA1 | .62 | .33–1.15 | −12.96 | .1692 | .61 | −12.69 | .87 | .1–7.56 | −.23 | 1 | .77 | −.57 |
| rs1045216 (2) | PLEKHA1 | .48 |
.32–.74 | −51.23 |
.0005 | .49 | −18.27 | .37 |
.21–.65 | −14.3 |
.0003 | .28 |
−35.68 |
| rs1882907 (2) | .58 | .4–.84 | −16.73 | .0026 | .44 | −5.79 | .31 | .1–.97 | −2.37 | .0438 | .27 | −3.65 | |
| rs10490923 (2) | LOC387715 | .53 | .31–.9 | −13.27 | .0239 | .34 | −9.01 | .22 | .04–1.09 | −2.51 | .0809 | .2 | −3.32 |
| rs2736911 (2) | LOC387715 | .72 | .42–1.21 | −6.92 | .2552 | 1.47 | 1.99 | 1.1 | .13–9.53 | .1 | 1 | 1.03 | .04 |
| rs10490924 (2) | LOC387715 | 5.03 |
3.2–7.91 | 57.11 |
<.0001 | 2.72 | 22.76 | 5.75 |
2.46–13.46 | 21.2 |
<.0001 | 10.57 |
42.71 |
| rs11538141 (2) | PRSS11 | … | … | … | … | … | … | … | … | … | … | … | … |
| rs760336 (2) | PRSS11 | .64 | .44–.93 | −35.37 | .013 | .8 | −6.95 | .69 | .46–1.03 | −7.95 | .0773 | .55 | −26.43 |
| rs763720 (1) | PRSS11 | 1.69 | 1.2–2.38 | 21.24 | .0018 | 1.55 | 16.95 | 2.63 | 1.1–6.25 | 5.17 | .0277 | 3.16 | 10.14 |
| rs1803403 (1) | PRSS11 | 2.98 | 1.25–7.06 | 10.51 | .0093 | 2.98 | 10.51 | … | … | … | … | … | … |
Note.— Type A–affected individuals are compared with controls. OR and AR values are underlined if corresponding P values are ⩽.001 and are in bold italics if P values are ⩽.05. Allele denotes the risk allele (minor allele in controls). RR = homozygotes for the risk allele; RN = heterozygotes for the risk allele; NN = homozygotes for the normal allele. Locally typed SNPs are in bold italics. Blank spaces separate the three chromosomal regions corresponding to SNPs in and around CFH, GRK5/RGS10, and PLEKHA1/LOC687715/PRSS11.
We obtained results and OR and AR values (table 8) similar to those that others have reported for the CFH gene. The three most significant SNPs were rs1061170 (Y402H variant), rs800292 (in CFH), and rs1853883 (in strong LD with rs1061170; D′=0.91).
The magnitude of the association we saw within PLEKHA1/LOC387715 is very similar to the level of association seen between CFH and ARM; both loci result in extremely low P values (P<.0001). The OR and AR values were also similar—the dominant OR was 5.29 (95% CI 3.35–8.35) within CFH and 5.03 (95% CI 3.2–7.91) within PLEKHA1/LOC387715, and the dominant AR for CFH and PLEKHA1/LOC387715 was 68% and 57%, respectively.
Subphenotype analyses
We estimated ORs and ARs for patients with exudative disease versus controls and for patients with GA versus controls (table 9). ORs and corresponding P values yielded similar findings to those of the allele test of CCREL (tables 5 and 6). We found no major differences between the ORs for the presence of either GA or CNV.
Table 9.
ORs and ARs from Analysis of ARM Subtypes[Note]
|
Dominant([RR+RN] vs. NN) |
Heterozygotes(RR vs. NN) |
Recessive(RR vs. [RN+NN]) |
Homozygotes(RR vs. NN) |
|||||||||||
| SNP (Allele) | Gene | Subtype | OR | 95% CI | AR | P | OR | AR | OR | 95% CI | AR | P | OR | AR |
| rs6658788 (2) | CNV | .84 | .56–1.25 | −13.41 | .36706 | 1.21 | 6.19 | 1.11 | .73–1.68 | 2.5 | .68123 | .95 | −2.26 | |
| rs6658788 (2) | GA | .88 | .55–1.4 | −9.92 | .63064 | 1.07 | 2.18 | 1.01 | .62–1.66 | .35 | 1.00000 | .92 | −3.97 | |
| rs1538687 (2) | CNV | .71 | .5–1.02 | −17.04 | .07499 | .54 | −10.68 | .47 | .25–.9 | −5.97 | .02020 | .42 | −11.38 | |
| rs1538687 (2) | GA | .62 | .41–.94 | −23.86 | .03170 | .56 | −10.14 | .45 | .2–1.01 | −6.25 | .07239 | .38 | −12.32 | |
| rs1416962 (2) | CNV | .88 | .61–1.25 | −7.7 | .41676 | 1.02 | .46 | .95 | .56–1.62 | −.62 | .89111 | .89 | −2.63 | |
| rs1416962 (2) | GA | .77 | .51–1.17 | −15.12 | .24708 | .69 | −7.53 | .62 | .31–1.24 | −5.07 | .22948 | .57 | −11.11 | |
| rs946755 (2) | CNV | .84 | .59–1.2 | −9.81 | .37326 | 1.14 | 2.86 | 1.03 | .6–1.78 | .37 | 1.00000 | .94 | −1.39 | |
| rs946755 (2) | GA | .73 | .48–1.11 | −18.18 | .17258 | .79 | −4.46 | .69 | .34–1.38 | −3.8 | .37606 | .6 | −9.26 | |
| rs6428352 (2) | CNV | … | … | … | … | … | … | … | … | … | … | … | … | |
| rs6428352 (2) | GA | … | … | … | … | … | … | … | … | … | … | … | … | |
| rs800292 (1) | CFH | CNV | .48 |
.33–.7 | −26.97 |
.00020 | .53 | −21.4 | .21 | .07–.64 | −4.59 | .00530 | .18 | −7.64 |
| rs800292 (1) | CFH | GA | .39 |
.25–.62 | −33.02 |
.00020 | .44 | −26.29 | .09 | .01–.75 | −5.33 | .01130 | .08 | −8.66 |
| rs1061170 (2) | CFH | CNV | 5.25 |
3.22–8.55 | 68 |
<.0001 | 2.37 | 24.74 | 4.11 |
2.2–7.69 | 27.24 |
<.0001 | 9.35 |
61.82 |
| rs1061170 (2) | CFH | GA | 5.76 |
3.17–10.47 | 70.42 |
<.0001 | 3.31 | 35.78 | 5.66 |
2.9–11.04 | 35.95 |
<.0001 | 12.26 |
68.61 |
| rs10922093 (1) | CFH | CNV | .56 | .37–.85 | −28.05 | .00830 | .61 | −20.84 | .4 | .18–.91 | −5.96 | .03270 | .33 | −11.72 |
| rs10922093 (1) | CFH | GA | .51 | .31–.84 | −32.2 | .00890 | .58 | −23.06 | .26 | .08–.85 | −7.43 | .03200 | .21 | −14.08 |
| rs70620 (1) | CFH | CNV | .77 | .52–1.14 | −7.45 | .23338 | .8 | −5.9 | .63 | .24–1.69 | −1.46 | .42256 | .6 | −2.2 |
| rs70620 (1) | CFH | GA | .72 | .45–1.15 | −9.48 | .18978 | .78 | −6.4 | .28 | .06–1.36 | −2.9 | .17068 | .26 | −4.1 |
| rs1853883 (2) | CNV | 2.52 |
1.64–3.89 | 52.14 |
.00020 | 1.5 | 15.44 | 1.88 | 1.28–2.78 | 18.84 | .00140 | 3.2 |
51.28 |
|
| rs1853883 (2) | GA | 3.54 |
1.97–6.36 | 64.51 |
<.0001 | 1.95 | 25.93 | 2.57 |
1.65–4 | 29.15 |
.00030 | 5.12 |
66.42 |
|
| rs1360558 (1) | CNV | 1.1 | .76–1.59 | 5.96 | .64364 | 1.04 | 2.29 | 1.24 | .78–1.98 | 3.75 | .41376 | 1.27 | 7.61 | |
| rs1360558 (1) | GA | 1.16 | .75–1.79 | 9.09 | .57904 | 1.13 | 6.6 | 1.17 | .68–2.02 | 2.67 | .67873 | 1.25 | 7.14 | |
| rs955927 (2) | CNV | 1.12 | .78–1.63 | 7.31 | .51105 | 1.32 | 7.01 | 1.34 | .83–2.17 | 4.9 | .20048 | 1.38 | 9.84 | |
| rs955927 (2) | GA | 1.08 | .7–1.67 | 5 | .74163 | 1.18 | 4.02 | 1.2 | .68–2.1 | 2.86 | .57564 | 1.22 | 6.06 | |
| rs4350226 (2) | CNV | .55 | .34–.91 | −8.65 | .02090 | … | … | … | … | … | … | … | … | |
| rs4350226 (2) | GA | .52 | .28–.96 | −9.46 | .04620 | … | … | … | … | … | … | … | … | |
| rs4752266 (2) | GRK5 | CNV | .93 | .65–1.34 | −2.87 | .71243 | 3.13 | 10.08 | 2.82 | .96–8.24 | 3.9 | .06229 | 2.63 | 5.74 |
| rs4752266 (2) | GRK5 | GA | .78 | .51–1.19 | −10.33 | .27667 | 3.67 | 12.31 | 2.88 | .9–9.23 | 4.04 | .06909 | 2.51 | 5.33 |
| rs915394 (2) | GRK5 | CNV | 1.39 | .96–2.01 | 11.91 | .08469 | 1.28 | 2.23 | 1.56 | .57–4.3 | 1.54 | .48645 | 1.74 | 2.96 |
| rs915394 (2) | GRK5 | GA | 1.09 | .7–1.67 | 2.88 | .74493 | 1.38 | 2.94 | 1.42 | .44–4.58 | 1.17 | .57824 | 1.45 | 1.81 |
| rs1268947 (2) | GRK5 | CNV | 1.15 | .75–1.75 | 3.15 | .52415 | 1.23 | 1.72 | 1.35 | .36–5.05 | .58 | .76382 | 1.39 | .8 |
| rs1268947 (2) | GRK5 | GA | .78 | .46–1.32 | −5 | .42146 | 1.24 | 1.82 | 1 | .2–5.02 | 0 | 1.00000 | .95 | −.1 |
| rs1537576 (2) | GRK5 | CNV | 1.57 | 1.07–2.3 | 27.1 | .02110 | .88 | −4.02 | 1.06 | .69–1.63 | 1.3 | .83192 | 1.44 | 14.26 |
| rs1537576 (2) | GRK5 | GA | 1.84 | 1.15–2.94 | 35.48 | .01430 | 1.17 | 5.17 | 1.44 | .89–2.34 | 8.51 | .17778 | 2.04 | 28.32 |
| rs2039488 (2) | CNV | .76 | .48–1.2 | −5.05 | .28877 | .2 | −12.5 | .18 | .03–.91 | −2.36 | .03180 | .17 | −2.88 | |
| rs2039488 (2) | GA | .62 | .35–1.09 | −8.3 | .11599 | .27 | −11.29 | .2 | .02–1.7 | −2.3 | .21758 | .19 | −2.83 | |
| rs1467813 (1) | RGS10 | CNV | .95 | .67–1.36 | −2.31 | .85551 | .98 | −.68 | .83 | .45–1.54 | −1.61 | .63004 | .83 | −2.79 |
| rs1467813 (1) | RGS10 | GA | .85 | .56–1.29 | −7.69 | .52905 | .88 | −5.73 | .81 | .39–1.69 | −1.85 | .70453 | .76 | −3.85 |
| rs927427 (1) | CNV | 1.08 | .72–1.63 | 5.82 | .75722 | .91 | −6.99 | 1.76 | 1.13–2.74 | 11.97 | .01070 | 1.65 | 21.15 | |
| rs927427 (1) | GA | 1.1 | .68–1.78 | 6.67 | .81152 | .98 | −1.63 | 1.5 | .9–2.5 | 8.16 | .15618 | 1.47 | 16.43 | |
| rs4146894 (1) | PLEKHA1 | CNV | 2.53 |
1.64–3.91 | 52.45 |
<.0001 | 1.94 | 37.72 | 2.46 |
1.63–3.71 | 23.64 |
<.0001 | 3.95 |
56 |
| rs4146894 (1) | PLEKHA1 | GA | 2.09 | 1.24–3.51 | 44 | .00690 | 1.77 | 33.08 | 1.92 | 1.2–3.08 | 16.31 | .00840 | 2.87 |
44.63 |
| rs12258692 (2) | PLEKHA1 | CNV | … | … | … | … | … | … | … | … | … | … | … | … |
| rs12258692 (1) | PLEKHA1 | GA | … | … | … | … | … | … | … | … | … | … | … | … |
| rs4405249 (1) | PLEKHA1 | CNV | .53 | .27–1.02 | −16.43 | .06989 | .51 | −16.36 | .97 | .11–8.85 | −.05 | 1.00000 | .83 | −.41 |
| rs4405249 (1) | PLEKHA1 | GA | .63 | .3–1.33 | −12.27 | .17898 | .64 | −11.43 | .57 | .04–9.31 | −.76 | 1.00000 | .51 | −1.2 |
| rs1045216 (2) | PLEKHA1 | CNV | .5 | .32–.78 | −48.35 | .00260 | .4 | −22.18 | .31 |
.17–.58 | −15.72 |
.00020 | .25 |
−38.01 |
| rs1045216 (2) | PLEKHA1 | GA | .44 |
.26–.72 | −58.72 |
.00100 | .45 | −19.85 | .32 | .15–.7 | −15.46 | .00170 | .24 |
−38.67 |
| rs1882907 (2) | CNV | .52 | .35–.77 | −19.38 | .00240 | .7 | −3.08 | .44 | .14–1.38 | −1.91 | .20808 | .38 | −3.11 | |
| rs1882907 (2) | GA | .6 | .38–.95 | −15.9 | .03500 | .22 | −8.27 | .16 | .02–1.37 | −2.89 | .12039 | .14 | −4.32 | |
| rs10490923 (2) | LOC387715 | CNV | .48 | .28–.85 | −14.84 | .01140 | .17 | −11.63 | .1 | .01–.98 | −2.89 | .04130 | .09 | −3.78 |
| rs10490923 (2) | LOC387715 | GA | .74 | .39–1.38 | −7.07 | .41496 | .58 | −5.49 | .48 | .08–2.91 | −1.66 | .65244 | .45 | −2.24 |
| rs2736911 (2) | LOC387715 | CNV | .71 | .41–1.22 | −7.12 | .24548 | 1.22 | .96 | .92 | .09–8.92 | −.08 | 1.00000 | .86 | −.18 |
| rs2736911 (2) | LOC387715 | GA | .62 | .32–1.19 | −9.43 | .13179 | 2.2 | 4.96 | 1.43 | .13–15.97 | .42 | 1.00000 | 1.3 | .38 |
| rs10490924 (2) | LOC387715 | CNV | 5.64 |
3.52–9.06 | 60.52 |
<.0001 | 2.81 | 23.7 | 6.18 |
2.62–14.59 | 22.67 |
<.0001 | 12.11 |
46.39 |
| rs10490924 (2) | LOC387715 | GA | 3.43 |
2.02–5.84 | 44.55 |
<.0001 | 2.63 | 21.83 | 4.74 |
1.9–11.84 | 17.47 |
.00030 | 7.05 |
32.05 |
| rs11538141 (2) | PRSS11 | CNV | … | … | … | … | … | … | … | … | … | … | … | … |
| rs11538141 (2) | PRSS11 | GA | … | … | … | … | … | … | … | … | … | … | … | … |
| rs760336 (2) | PRSS11 | CNV | .63 | .43–.92 | −37.33 | .01780 | .71 | −10.35 | .61 | .39–.95 | −10.03 | .03480 | .49 | −30.78 |
| rs760336 (2) | PRSS11 | GA | .63 | .4–.98 | −36.73 | .03220 | .84 | −5.52 | .71 | .42–1.19 | −7.3 | .23778 | .56 | −25.69 |
| rs763720 (1) | PRSS11 | CNV | 1.77 | 1.24–2.54 | 23.25 | .00310 | 1.69 | 20.43 | 2.1 | .85–5.18 | 3.55 | .12829 | 2.64 | 7.87 |
| rs763720 (1) | PRSS11 | GA | 1.74 | 1.14–2.65 | 22.5 | .01070 | 1.4 | 12.86 | 4.71 |
1.88–11.79 | 11.06 |
.00010 | 5.41 |
18.69 |
| rs1803403 (1) | PRSS11 | CNV | 3.33 | 1.39–8.02 | 12.17 | .00550 | 3.33 | 12.17 | … | … | … | … | … | … |
| rs1803403 (1) | PRSS11 | GA | 3.85 | 1.53–9.72 | 14.49 | .00390 | 3.85 | 14.49 | … | … | … | … | … | … |
Note.— OR and AR values are underlined if corresponding P values (by χ2 test; P values simulated using 10,000 replicates) are ⩽.001, and are in bold italics if P values are ⩽.05. Allele denotes the risk allele (minor allele in controls). RR = homozygotes for the risk allele; RN = heterozygotes for the risk allele; NN = homozygotes for the normal allele. Locally typed SNPs are in bold italics. Blank spaces separate the three chromosomal regions corresponding to SNPs in and around CFH, GRK5/RGS10, and PLEKHA1/LOC687715/PRSS11.
Discussion
Our linkage studies of families with ARM have consistently identified the chromosome 1q31 and chromosome 10q26 loci, in addition to several other loci. Multiple linkage studies have replicated this finding; thus, we undertook a focused SNP analysis of both regions, using ARM-affected families as well as unrelated affected individuals and controls. We confirmed the strong association of chromosome 1q31 with CFH that has been reported by others (see also Conley et al. [2005]), and we have shown, for the first time, that SNPs in CFH significantly account for the linkage signal. Interestingly, our smallest GIST P value (<.001) was for rs1853883 (which has a high D′ of 0.91 with the Y402H variant) and not for the presumed “disease-associated” Y402H variant itself. This raises the possibility that we may still have to consider other possible ARM-related variants within the CFH gene and that these may be in high LD with Y402H.
Our studies of chromosome 10q26 have implicated two potential loci: (1) a very strongly implicated locus that includes three tightly linked genes, PLEKHA1, LOC387715, and PRSS11, and (2) a less strongly implicated locus comprising two genes, GRK5 and RGS10 (fig. 1). The GIST analysis does not support PRSS11 as the ARM-related gene, but it does not completely exclude it as a potential candidate. PLEKHA1 has the lowest GIST-derived P values, whereas LOC387715 harbors the SNP with the strongest association signal and the highest ORs. With the high LD between SNPs in LOC387715 and PLEKHA1, one cannot clearly distinguish between these genes by statistical analyses alone. However, it is clear that the magnitude of the impact of the PLEKHA1/LOC387715 locus on ARM is comparable to that which has been observed for the CFH locus. As in recent studies (Edwards et al. 2005; Haines et al. 2005; Klein et al. 2005), we have found, in our case-control population, that the CFH allele (either heterozygous or homozygous) accounts for an OR of 5.3 (95% CI 3.4–8.4) and a significant population AR of 68%. In the same fashion, the high-risk allele within the PLEKHA1/LOC387715 locus accounts for an OR of 5.0 (95% CI 3.2–7.9) and an AR of 57% when both heterozygous and homozygous individuals are considered. As noted by Klein et al. (2005), unless the disease is very rare, the OR determined from a case-control study will usually overestimate the equivalent relative risk. Estimates of AR based on ORs for common genetic disorders can misrepresent the extent to which a variant accounts for the population AR. However, if this caution is kept in mind, it is still useful for us to present AR values to allow for relative comparisons and to allow the reader to appreciate that the potential impact of the CFH Y402H variant on ARM is comparable to that of the variants observed in the PLEKHA1/LOC387715 locus.
In the case of CFH on chromosome 1, the association data were extremely compelling for a single gene, even though CFH is within a region of related genes. In addition to the association data found by multiple independent groups, there is additional biological data to implicate CFH, including localization of the protein within drusen deposits of patients with ARM. Thus, we also must consider the biological relevance of the potential ARM-susceptibility genes identified by our studies of chromosome 10q26.
As noted above, the GIST analysis most strongly implicated PLEKHA1, particularly when we included the additional nonsynonymous SNPs that we genotyped locally. PLEKHA1 encodes the protein TAPP1, which is a 404-aa protein with a putative phosphatidylinositol 3,4,5-trisphosphate-binding motif (PPBM), as well as two plectstrin homology (PH) domains. The last three C-terminal amino acids have been predicted to interact with one or more of the 13 PDZ domains of MUPP1 (similar to the PDZ domain within PRSS11). Dowler and colleagues (2000) have shown that the entire TAPP1 protein, as well as the C-terminal PH domain, interacts specifically with phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2) but not with any other phosphoinositides. TAPP1, which has 58% identity with the first 300 aa of TAPP2, shows a fivefold higher affinity for PtdIns(3,4)P2 than does TAPP2, and this binding is nearly eliminated by mutation of the conserved arginine 212 to leucine within the PPBM region (which is part of the second PH domain). The most well-defined role for TAPP1 (and its relatives, Bam32 and TAPP2) has been as an activator of lymphocytes. PtdIns(3,4)P2 is preferentially recruited to cell membranes when lipid phosphatase (SHIP) is activated along with PI3K (phosphatidyl inositol 3-kinase). SHIP is responsible for the dephosphorylation of PIP3 to PtdIns(3,4)P2. SHIP is a negative regulator of lymphocyte activation, and thus TAPP1 and TAPP2 may be crucial negative regulators of mitogenic signaling and of the PI3K signaling pathway. Thus, one can envision a role in the eye for PLEKHA1 and its protein, TAPP1, in modifying local lymphocyte activation, consistent with the hypothesis that ARM is closely linked to an inflammatory process.
However, we need to still consider the biological plausibility of the other two candidate genes within this locus, LOC387715 and PRSS11. Little is known regarding the biology of LOC387715, except that its expression appears to be limited to the placenta. Our own reverse transcription experiments with human retinal RNA have confirmed the expression of PLEKHA1 and PRSS11, but we have not detected LOC387715 transcripts in the retina under standard conditions, even though we confirmed its expression with placental RNA (data not shown). However, we cannot exclude the possibility that LOC387715 is expressed at very low levels in the retina or retinal pigment epithelium or that its expression in nonocular tissues, such as dendritic cells or migrating macrophages, could be a factor in the pathogenesis of ARM.
PRSS11 is one of the genes of the mammalian high temperature requirement A (HtrA) serine protease family, which has a highly conserved C-terminal PDZ domain (Oka et al. 2004). These secretory proteases were initially identified because of their homologies to bacterial forms that are required for survival at high temperatures and molecular chaperone activity at low temperatures. The ATP-independent serine protease activity is thought to degrade misfolded proteins at high temperatures. The mammalian form, HtrA1, has been shown to be selectively stimulated by type III collagen alpha 1 C propeptide, in contrast to HtrA2 (Murwantoko et al. 2004). Type III collagen is a major constituent (35%–39% of the total collagen) in Bruch membrane and is also present in small amounts in the retinal microvascular basement membranes. Developmental studies have reported ubiquitous expression of HtrA1, but with temporal and spatial specificities that coincide with those regions in which Tgfβ proteins play a regulatory role (De Luca et al. 2004). Oka and colleagues (2004) have shown that HtrA1 is capable of inhibiting the signaling of a number of Tgfβ family proteins, including Bmp4, Bmp2, and Tgfβ1, presumably by preventing receptor activation with a requirement for protease activity of the HtrA1 molecule. One clue as to the potential importance of these relationships for ARM comes from the studies of Hollborn et al. (2004), who found that human RPE cells in vitro experienced reduced proliferation in the presence of Tgfβ1 and Tgfβ2 and an increase in levels of collagen III and collagen IV transcripts. Normally, a rise in collagen III would activate HtrA1 and would lead to secondary inhibition of the effects of Tgfβ1. However, if the serine protease is less effective (because of either reduced synthesis or a nonfunctional mutation), then this regulatory pathway would be disrupted, leading to an overall reduction in the proliferation potential of the RPE cells, perhaps contributing to RPE atrophy or further changes that could lead to the development of ARM. The gradual reduction in solubility of type III collagen in Bruch membrane that has been observed with aging could also, in part, account for a general reduction in HtrA1 activity as an individual ages.
Both PRSS11 and PLEKHA1 are expressed in the retina, and a SAGE analysis of central and peripheral retina (Gene Expression Omnibus [GEO] expression data) indicates higher levels of transcripts of both genes in the central macula (more so for PLEKHA1 than for PRSS11). Multiple studies (reported in GEO profiles) have shown that PLEKHA1 expression is significantly induced in a variety of cell types in response to exposure to specific inflammatory cytokines. PRSS11 has also been investigated as part of a microarray expression analysis of dermal fibroblasts that have been oxidatively challenged, in a comparison between normal individuals and patients with ARM. In that study, half of the ARM samples (9 of 18) had lower Htra1 expression levels than any of the normal samples. The lower levels of Htra1 in nonocular tissues of patients with ARM would suggest that this is an intrinsic difference in the biology of these patients, compared with that of normal individuals, and is not a consequence of degenerative changes in the eye.
Several lines of evidence support the GRK5/RGS10 locus. The peak of our Sall multipoint curve is directly over GRK5, and our largest two-point Sall=3.86 (rs555938) is only 206 kb centromeric of GRK5. The P values for the GIST analysis of the GRK5/RGS10 CIDR data were .004 and .006, which are even smaller than the P value for the SNP within PLEKHA1 (P=.008). By use of our locally genotyped sample, the GIST P value for the GRK5 locus was .012, which is comparable to the P value that we found for the Y402H variant in CFH (P=.011). However, the CCREL analyses were not very significant for the GRK5 SNPs, and the ORs were mostly nonsignificant.
On the basis of biological evidence, GRK5 is a reasonable ARM candidate gene, given its role in modulating neutrophil responsiveness to chemoattractants and its interactions with the Toll 4 receptor (Haribabu and Snyderman 1993; Fan and Malik 2003), which has also been implicated in ARM (Zareparsi et al. 2005b). The retinal or RPE expression of GRK5 is not especially relevant to the argument of causality, because it would be the expression and function of GRK5 in migrating lymphocytes and macrophages that would be crucial to its role in the immune and/or inflammatory pathways that may be pathogenic in ARM. The strongest GIST results occur at rs2039488, which is located between GRK5 and RGS10, 3′ of the ends of both genes. Several other SNPs within GRK5 also have small GIST P values, whereas the RGS10 SNP has a nonsignificant GIST P value. However, we cannot completely exclude the possibility that there is a SNP within RGS10 that is in strong LD with rs2039488.
RGS10 is one of a family of G protein–coupled receptors that has been implicated in chemokine-induced lymphocyte migration (Moratz et al. 2004) and whose expression in dendritic cells (which have been identified in ARM-related drusen deposits) is modified by the Toll-like signaling pathway (Shi et al. 2004). RGS10 and GRK5 expression in the same microarray study of oxidatively stressed dermal fibroblasts in patients with ARM and control subjects showed minor fluctuations among the samples but no clear differences between the controls and affected individuals. This does not necessarily lower the potential for these genes being involved in ARM, since the dermal fibroblasts lack the cell populations that would be expected to have modulation of RGS10- and/or GRK5-related proteins.
We have attempted to look at potential interactions between the high-risk alleles within the PLEKHA1/LOC387715 and GRK5/RGS10 loci with respect to CFH on chromosome 1. This is perhaps the first report to use GIST to examine these interactions, and we found no evidence that the NPL data on chromosome 1 could be accounted for by the SNP data on chromosome 10. Conversely, we found no such associations between the NPL data on chromosome 10 and the SNP data from the CFH alleles. Logistic regression analysis also failed to identify an interaction, and it appears that a simple additive risk model is the most parsimonious. We have performed some initial logistic analyses that include exposure to smoking. These analyses were initiated because of the previous suggestion of an interaction between smoking and the biology of complement factor H (Esparza-Gordillo et al. 2004) and because of our prior studies, which found an interaction between smoking and the locus on chromosome 10q26 (Weeks et al. 2004). To date, we have found no strong interaction between smoking and either CFH or PLEKHA1/LOC387715, but we are still exploring a possible interaction with the GRK5/RGS10 locus and different modeling strategies.
We also examined the associations of ARM subphenotypes with the SNPs on chromosomes 1 and 10 (table 9). We found no major differences in the ORs for the presence of either GA or CNV, which suggests that these ARM loci contribute to a common pathogenic pathway that can give rise to either end-stage form of the disease. This does not exclude the possibility that there are other as-yet-undescribed genetic loci that may confer specific risk of GA or CNV development separately.
In summary, these SNP-based linkage and association studies illustrate both the power and the limitation of such methods to identify the causative alleles and genes underlying ARM susceptibility. These genetic approaches allow us to consider genes and their variants that may contribute to disease, whether or not there is tissue-specific expression. Through high-density SNP genotyping, we have narrowed the list of candidate genes within the linkage peak found on chromosome 10q26, from hundreds of genes to primarily GRK5 and PLEKHA1, but we cannot completely exclude possible roles for RGS10 and/or PRSS11 and LOC387715. Additional genotyping of nonsynonymous 3′ SNPs within the GRK5 gene may help to further discriminate between GRK5 and RGS10, but it may not establish a definitive assignment of causality. Replication by other studies (as done in the case of CFH) may allow the attention to be focused on a single gene in future studies of ARM pathology, but there is also the distinct possibility that we will be unable to achieve further resolution with association studies or to clearly establish whether there are more than two genes responsible for ARM susceptibility on chromosome 10q26. However, it is now well within the capabilities of molecular biologists to investigate the potential role of each of these candidate genes in mouse models of ARM and to address the issue of a causal role in disease pathogenesis. Association studies are an incredibly powerful means of testing hypotheses of genetic contributions to disease, but, except in the most extreme cases, they cannot provide definitive answers, even when there are impressive P values.
Acknowledgments
We gratefully acknowledge the many individuals and family members who participated as research subjects, as well as their eye care providers for this study. We also wish to express our thanks to our current and past clinical research team: E. Adams, C. Baic, L. R. Barnes, F. Bivins, M. Blagodatny, A. Burchis, I. Cantillo, J. Green, B. Gur, K. Hendricks, C. Klump, S. Pope, M. Shaffer-Gordon, H. Sheth, S. Ward, and B. Zhu, and to our laboratory research staff: Y. Demirci (who tested for retinal expression of the three candidate genes), H. Brockway, S. Deslouches, and B. Rigatti. Illumina-based SNP genotyping services were provided by the Center for Inherited Disease Research, which is fully funded through federal contract N01-HG-65403 from the National Institutes of Health to The Johns Hopkins University. This study was supported by National Eye Institute grant R01EY009859 (to M.B.G.); The Steinbach Foundation, New York (to M.B.G.); Research to Prevent Blindness, New York (Senior Scientist Investigator Award to M.B.G.); and the Eye and Ear Foundation of Pittsburgh (to M.B.G.).
Web Resources
The URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ARMD-1, CFH, PLEKHA1, PRSS11, GRK5, and RGS10)
- Division of Statistical Genetics, http://watson.hgen.pitt.edu/ (for Mega2)
- The R Project for Statistical Computing, http://www.r-project.org/ (for R statistical software)
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Yashar BM, Zhao Y, Ghiasvand NM, Zareparsi S, Branham KE, Reddick AC, Trager EH, Yoshida S, Bahling J, Filippova E, Elner S, Johnson MW, Vine AK, Sieving PA, Jacobson SG, Richards JE, Swaroop A (2004) Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet 74:482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- Browning SR, Briley JD, Briley LP, Chandra G, Charnecki JH, Ehm MG, Johansson KA, Jones BJ, Karter AJ, Yarnall DP, Wagner MJ (2005) Case-control single-marker and haplotypic association analysis of pedigree data. Genet Epidemiol 28:110–122 [DOI] [PubMed] [Google Scholar]
- Conley YP, Thalamuthu A, Jakobsdottir J, Weeks DE, Mah T, Ferrell RE, Gorin MB (2005) Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum Mol Genet 14:1991–2002 [DOI] [PubMed] [Google Scholar]
- De Luca A, De Falco M, De Luca L, Penta R, Shridhar V, Baldi F, Campioni M, Paggi MG, Baldi A (2004) Pattern of expression of HtrA1 during mouse development. J Histochem Cytochem 52:1609–1617 [DOI] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR (2000) Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J 351:19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, Ritter R III, Abel KJ, Manning A, Panhuysen C, Farrer LA (2005) Complement factor H polymorphism and age-related macular degeneration. Science 308:421–424 [DOI] [PubMed] [Google Scholar]
- Esparza-Gordillo J, Soria JM, Buil A, Almasy L, Blangero J, Fontcuberta J, Rodriguez de Cordoba S (2004) Genetic and environmental factors influencing the human factor H plasma levels. Immunogenetics 56:77–82 [DOI] [PubMed] [Google Scholar]
- Fan J, Malik AB (2003) Toll-like receptor-4 (TLR4) signaling augments chemokine-induced neutrophil migration by modulating cell surface expression of chemokine receptors. Nat Med 9:315–321 [DOI] [PubMed] [Google Scholar]
- Fisher SA, Abecasis GR, Yashar BM, Zareparsi S, Swaroop A, Iyengar SK, Klein BE, Klein R, Lee KE, Majewski J, Schultz DW, Klein ML, Seddon JM, Santangelo SL, Weeks DE, Conley YP, Mah TS, Schmidt S, Haines JL, Pericak-Vance MA, Gorin MB, Schulz HL, Pardi F, Lewis CM, Weber BH (2005) Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet 14:2257–2264 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, et al (2005) A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA 102:7227–7232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF (2001) An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res 20:705–732 [DOI] [PubMed] [Google Scholar]
- Hageman GS, Mullins RF (1999) Molecular composition of drusen as related to substructural phenotype. Mol Vis 5:28 [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA (2005) Complement factor H variant increases the risk of age-related macular degeneration. Science 308:419–421 [DOI] [PubMed] [Google Scholar]
- Haribabu B, Snyderman R (1993) Identification of additional members of human G-protein-coupled receptor kinase multigene family. Proc Natl Acad Sci USA 90:9398–9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollborn M, Reichenbach A, Wiedemann P, Kohen L (2004) Contrary effects of cytokines on mRNAs of cell cycle- and ECM-related proteins in hRPE cells in vitro. Curr Eye Res 28:215–223 [DOI] [PubMed] [Google Scholar]
- Huang Q, Shete S, Amos CI (2004) Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet 75:1106–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar SK, Song D, Klein BE, Klein R, Schick JH, Humphrey J, Millard C, Liptak R, Russo K, Jun G, Lee KE, Fijal B, Elston RC (2004) Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am J Hum Genet 74:20–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Leitner WP, Staples MK, Anderson DH (2001) Complement activation and inflammatory processes in drusen formation and age related macular degeneration. Exp Eye Res 73:887–896 [DOI] [PubMed] [Google Scholar]
- Johnson LV, Ozaki S, Staples MK, Erickson PA, Anderson DH (2000) A potential role for immune complex pathogenesis in drusen formation. Exp Eye Res 70:441–449 [DOI] [PubMed] [Google Scholar]
- Kenealy SJ, Schmidt S, Agarwal A, Postel EA, De La Paz MA, Pericak-Vance MA, Haines JL (2004) Linkage analysis for age-related macular degeneration supports a gene on chromosome 10q26. Mol Vis 10:57–61 [PubMed] [Google Scholar]
- Klein ML, Schultz DW, Edwards A, Matise TC, Rust K, Berselli CB, Trzupek K, Weleber RG, Ott J, Wirtz MK, Acott TS (1998) Age-related macular degeneration: clinical features in a large family and linkage to chromosome 1q. Arch Ophthalmol 116:1082–1088 [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Richard S, Sackler RS, Chad Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Murphy K, Raj T, He C, White PS, Matise TC (2004) A combined linkage-physical map of the human genome. Am J Hum Genet 75:1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Cantor RM, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E (2001) MENDEL version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet Suppl 69:A1886 [Google Scholar]
- Li C, Scott LJ, Boehnke M (2004) Assessing whether an allele can account in part for a linkage signal: the Genotype-IBD Sharing Test (GIST). Am J Hum Genet 74:418–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Schultz DW, Weleber RG, Schain MB, Edwards AO, Matise TC, Acott TS, Ott J, Klein ML (2003) Age-related macular degeneration—a genome scan in extended families. Am J Hum Genet 73:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moratz C, Harrison K, Kehrl JH (2004) Regulation of chemokine-induced lymphocyte migration by RGS proteins. Methods Enzymol 389:15–32 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE (2005) Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics 21:2556–2557 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay N, Buxbaum SG, Weeks DE (2004) Comparative study of multipoint methods for genotype error detection. Hum Hered 58:175–189 [DOI] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS (2000) Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J 14:835–846 [PubMed] [Google Scholar]
- Murwantoko, Yano M, Ueta Y, Murasaki A, Kanda H, Oka C, Kawaichi M (2004) Binding of proteins to the PDZ domain regulates proteolytic activity of HtrA1 serine protease. Biochem J 381:895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BV, Curtis D, Sham PC (2005) Application of logistic regression to case-control association studies involving two causative loci. Hum Hered 59:79–87 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka C, Tsujimoto R, Kajikawa M, Koshiba-Takeuchi K, Ina J, Yano M, Tsuchiya A, Ueta Y, Soma A, Kanda H, Matsumoto M, Kawaichi M (2004) HtrA1 serine protease inhibits signaling mediated by Tgfβ family proteins. Development 131:1041–1053 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2004) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rinaldo A, Bacanu SA, Devlin B, Sonpar V, Wasserman L, Roeder K (2005) Characterization of multilocus linkage disequilibrium. Genet Epidemiol 28:193–206 [DOI] [PubMed] [Google Scholar]
- Santangelo SL, Yen C-H, Haddad S, Fagerness J, Huang C, Seddon JM (2005) A discordant sib-pair linkage analysis of age-related macular degeneration. Ophthalmic Genet 26:61–67 [DOI] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Wang L, Cunningham JM, Thibodeau SN (2002) Caution on pedigree haplotype inference with software that assumes linkage equilibrium. Am J Hum Genet 71:992–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick JH, Iyengar SK, Klein BE, Klein R, Reading K, Liptak R, Millard C, Lee KE, Tomany SC, Moore EL, Fijal BA, Elston RC (2003) A whole-genome screen of a quantitative trait of age-related maculopathy in sibships from the Beaver Dam Eye Study. Am J Hum Genet 72:1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Scott WK, Postel EA, Agarwal A, Hauser ER, De La Paz MA, Gilbert JR, Weeks DE, Gorin MB, Haines JL, Pericak-Vance MA (2004) Ordered subset linkage analysis supports a susceptibility locus for age-related macular degeneration on chromosome 16p12. BMC Genet 5:18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Santangelo SL, Book K, Chong S, Cote J (2003) A genomewide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am J Hum Genet 73:780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi GX, Harrison K, Han SB, Moratz C, Kehrl JH (2004) Toll-like receptor signaling alters the expression of regulator of G protein signaling proteins in dendritic cells: implications for G protein-coupled receptor signaling. J Immunol 172:5175–5184 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, Gorin MB (2000) A full genome scan for age-related maculopathy. Hum Mol Genet 9:1329–1349 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Schmidt S, Postel EA, Agarwal A, Haines JL, Pericak-Vance MA, Rosenfeld PJ, Paul TO, Eller AW, Morse LS, Dailey JP, Ferrell RE, Gorin MB (2004) Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. Am J Hum Genet 75:174–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareparsi S, Branham KE, Li M, Shah S, Klein RJ, Ott J, Hoh J, Abecasis GR, Swaroop A (2005a) Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet 77:149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareparsi S, Buraczynska M, Branham KE, Shah S, Eng D, Li M, Pawar H, Yashar BM, Moroi SE, Lichter PR, Petty HR, Richards JE, Abecasis GR, Elner VM, Swaroop A (2005b) Toll-like receptor 4 variant D299G is associated with susceptibility to age-related macular degeneration. Hum Mol Genet 14:1449–1455 [DOI] [PubMed] [Google Scholar]