Abstract
To provide a definitive linkage map for multiple sclerosis, we have genotyped the Illumina BeadArray linkage mapping panel (version 4) in a data set of 730 multiplex families of Northern European descent. After the application of stringent quality thresholds, data from 4,506 markers in 2,692 individuals were included in the analysis. Multipoint nonparametric linkage analysis revealed highly significant linkage in the major histocompatibility complex (MHC) on chromosome 6p21 (maximum LOD score [MLS] 11.66) and suggestive linkage on chromosomes 17q23 (MLS 2.45) and 5q33 (MLS 2.18). This set of markers achieved a mean information extraction of 79.3% across the genome, with a Mendelian inconsistency rate of only 0.002%. Stratification based on carriage of the multiple sclerosis–associated DRB1*1501 allele failed to identify any other region of linkage with genomewide significance. However, ordered-subset analysis suggested that there may be an additional locus on chromosome 19p13 that acts independent of the main MHC locus. These data illustrate the substantial increase in power that can be achieved with use of the latest tools emerging from the Human Genome Project and indicate that future attempts to systematically identify susceptibility genes for multiple sclerosis will have to involve large sample sizes and an association-based methodology.
Introduction
Familial clustering of multiple sclerosis (MIM 126200) has been recognized for more than a century (Eichorst 1896) and has been carefully measured in several large population-based studies (Sadovnick et al. 1988; Robertson et al. 1996; Carton et al. 1997). The increased risk seen in the siblings of affected individuals compared with the general population is a useful measure of the degree of familial clustering, known as “λs” (Risch 1990). In multiple sclerosis, this risk ratio has a value of ∼20, which indicates that the lifetime risk of developing the disease is 20 times greater for a sibling of an affected individual than for an individual from the general population. Supplementary epidemiological studies of twins (Mumford et al. 1994; Willer et al. 2003), adoptees (Ebers et al. 1995), conjugal pairs (Robertson et al. 1997), and half siblings (Ebers et al. 2004) indicate that shared genetic factors are at least partly responsible for the observed familial clustering, whereas studies of migrants (Dean et al. 1976) and concerning the month of birth (Willer et al. 2005) confirm that environmental factors are also important in the pathogenesis of the disease (Hogancamp et al. 1997).
Many candidate genes have been investigated, but, to date, the only region of the genome that has clearly and consistently shown evidence of association with the disease is the major histocompatibility complex (MHC) on chromosome 6p21, where, in Northern Europeans, association with the DR15 human leukocyte antigen (HLA) haplotype (DRB1*1501-DQB1*0602) is a constant finding (Jersild et al. 1972; Winchester et al. 1975; Compston et al. 1976; Olerup and Hillert 1991; Stewart et al. 1997; Haines et al. 1998). In the mid-1990s, it became possible to complete systematic whole-genome screens for linkage; to date, 11 such studies have been published about multiple sclerosis (Ebers et al. 1996; Haines et al. 1996; Sawcer et al. 1996; Kuokkanen et al. 1997; Broadley et al. 2001; Coraddu et al. 2001; Åkesson et al. 2002; Ban et al. 2002; Eraksoy et al. 2003; Hensiek et al. 2003; Kenealy et al. 2004). These screens have all been based on microsatellite marker sets with a density of approximately one marker every 10 cM. This design has long been accepted as the optimal approach (Hauser et al. 1996). Although all of these screens have identified regions of interest, none has demonstrated linkage with genomewide significance, although each has shown more allele sharing among affected individuals than would have been expected by chance alone. A recent meta-analysis of the available raw genotypes demonstrated linkage that just reached genomewide significance in the MHC region but found no significant linkage outside the region, despite the inclusion of >700 multiplex families (GAMES and Transatlantic Multiple Sclerosis Genetics Cooperative 2003). Close inspection of the quality of the data included in this meta-analysis revealed worrying inadequacies: the average genotyping success rate was just 80%, and the average information extraction was only 44%. Although the accuracy of genotyping was not reported in the majority of these studies, others have observed that error rates as low as 0.5%–1% are likely to hold only for good-quality microsatellites (Lathrop et al. 1983) and that higher rates would be expected for many markers (Brzustowicz et al. 1993). This level of inaccuracy is predicted to profoundly limit the expected LOD score and thereby seriously reduce the power to detect linkage (Abecasis et al. 2001a). In light of these inadequacies, we compared existing data from five affected-sib-pair families included in the original 1996 British genome screen with those generated using a more extensively engineered higher density set of microsatellite markers (Applied Biosystems High Density Linkage Mapping Set) and two high-density SNP-based mapping sets (Illumina BeadArray linkage mapping panel [version 3] and the Affymetrix GeneChip Human Mapping 10K array). This pilot experiment indicated that the SNP-based systems offer considerable advantages for genotyping success rate, information extraction, and—most importantly—genotyping accuracy (International Multiple Sclerosis Genetics Consortium 2004). The value of an accurate linkage map cannot be overstated. Demonstration of robust linkage has enabled the identification of susceptibility genes in several complex diseases (Horikawa et al. 2000; Hugot et al. 2001; Haines et al. 2005) and thus makes a definitive linkage study desirable.
On the basis of concerns about the quality of the genotyping data included in previous linkage screens (which are no better or worse than those performed for other complex diseases) and the encouraging results from the pilot study, we rescreened available families from four of the populations (Australian, Scandinavian, British, and American) with the Illumina BeadArray linkage mapping panel (version 4).
Material and Methods
Markers and Maps
The Illumina BeadArray linkage mapping panel (version 4) includes 5,858 markers (table 1). In our samples, 5,773 (98.5%) of these markers generated potentially usable data that satisfied minimum Illumina quality standards. Of these markers, 17 were uninformative for linkage: 5 were monomorphic in our samples and 12 were Y-linked (included in the panel to confirm the sex of typed samples). A further 474 markers were excluded, since <98% of genotypes generated by these markers had GenCall (as defined in the “Genotyping” section) scores >0.7 among those samples considered to be of acceptable quality (see the “Samples and Families” section). None of the remaining 5,282 quality markers showed deviation from Hardy-Weinberg equilibrium (HWE) (the genotyping success rate and heterozygosity for each of these 5,282 markers are provided in data file 1 [online only]). Among these 5,282 markers, there are 5,812 pairs of markers separated by <500 kb. Among the 5,812 pairs, there were 1,168 in which the linkage disequilibrium (LD) was found to have r2⩾0.16. Since simulation studies (Boyles et al., in press) have shown that there is little if any inflation of linkage evidence when the LD between markers had r2<0.16, this threshold was applied to our data, and selected markers were removed until all pairs of markers separated by <500 kb, consecutive and nonconsecutive, had r2<0.16. In this exclusion process, we aimed to remove the minimum number of markers possible. In regions of high LD, this invariably meant that all but one of a cluster of markers showing LD with each other needed to be excluded. In each instance, markers showing the lowest heterozygosity were excluded first, except when the difference in heterozygosity between a pair of markers was <10% and the more heterozygous marker included more than three Mendelian inconsistencies, in which case, the genotyping error rate was considered to “trump” the heterozygosity, and the more error-prone marker was removed. In total, 776 markers were excluded from the 5,282 quality typed markers. Multipoint linkage analysis was thus based on a total of 4,506 markers; all these markers had an average genotyping success rate (for quality genotypes with GenCall scores >0.7) of 99.7% and an average heterozygosity of 44.6% (only 23 markers have a heterozygosity of <5%). When consecutive marker pairs separated by <2 Mb were considered, only six additional pairs had r2>0.16, and all these had r2<0.44. It is therefore unlikely that residual LD has contributed significantly to the background-allele sharing.
Table 1.
Number of Markers per Chromosome
|
Number of Markersper Chromosome by Sample |
||||
| Chromosome | Fulla | Datab | Qualityc | Finald |
| 1 | 478 | 472 | 444 | 396 |
| 2 | 475 | 471 | 454 | 377 |
| 3 | 398 | 395 | 377 | 319 |
| 4 | 296 | 291 | 273 | 246 |
| 5 | 312 | 309 | 296 | 252 |
| 6 | 410 | 408 | 390 | 334 |
| 7 | 285 | 284 | 272 | 230 |
| 8 | 260 | 259 | 246 | 212 |
| 9 | 205 | 204 | 195 | 176 |
| 10 | 248 | 246 | 228 | 196 |
| 11 | 229 | 223 | 203 | 177 |
| 12 | 285 | 281 | 261 | 222 |
| 13 | 187 | 184 | 163 | 145 |
| 14 | 210 | 206 | 189 | 158 |
| 15 | 201 | 199 | 190 | 150 |
| 16 | 197 | 195 | 175 | 145 |
| 17 | 159 | 154 | 134 | 113 |
| 18 | 170 | 168 | 161 | 142 |
| 19 | 149 | 143 | 129 | 102 |
| 20 | 130 | 127 | 121 | 102 |
| 21 | 103 | 103 | 97 | 73 |
| 22 | 121 | 121 | 106 | 96 |
| Pseudo-Xp | 20 | 20 | 17 | 8 |
| X | 304 | 290 | 154 | 132 |
| Pseudo-Xq | 8 | 8 | 7 | 3 |
| Y | 18 |
12 |
0 |
0 |
| Total | 5,858 | 5,773 | 5,282 | 4,506 |
The complete Illumina linkage panel (version 4).
Markers that generated data in samples.
Markers that generated quality data (GQ>0.7) in >98% of samples.
Final set of markers (all pairs having r2<0.16).
All the markers included in the linkage-mapping panel (version 4) have been ordered and placed on the physical map (build 34) and have had genetic coordinates interpolated from the deCODE map (Kong et al. 2002). Markers unresolved on the genetic map were given an arbitrary separation of 0.01 cM. Among the 4,506 markers included in the multipoint linkage analysis, the average interval between consecutive markers is 0.82 cM, there are no intervals of >10 cM, and 71% of intervals are <1 cM.
Samples and Families
At the inception of this project, 2,923 individuals from 780 families were identified and considered. To monitor and assure experimental variables, such as plate orientation, and to provide the opportunity to test genotyping reproducibility, inter- and intraplate duplicate genotyping was performed. Further duplicate genotyping was also performed to obtain data from as many samples as possible when the amount of available DNA was minimal (see below). In total, genotyping of the linkage panel markers was attempted for 3,417 samples. Of these samples, 13% (446) failed to genotype, with most (404) of these failures occurring in whole-genome amplification (WGA)–derived DNA (see the “WGA” section). The included duplicate genotyping ensured that only 83 individuals had no sample that yielded genotypes. However, since some of the individuals who failed to genotype were affected, 41 families became uninformative for linkage (because they had fewer than two successfully typed affected individuals). Among the remaining 739 families, comprising 2,754 typed individuals, 21 individuals yielded <98% quality genotypes among the 5,282 good-quality markers and were therefore also excluded. After the exclusion of these lower-quality samples, a further nine families became uninformative for linkage, and eight families were reduced in size but were still informative. From the five largest multigenerational families, 24 typed individuals had to be excluded to ensure that the “bit” parameter for these families did not exceed 19, the largest value we were able to successfully analyze with this version of MERLIN (Center for Statistical Genetics) (Abecasis et al. 2002).
In conclusion, 2,692 individuals (from 730 multiplex families) were ultimately included in the analysis reported here—1,595 affected individuals (513 males; 1,082 females), 618 unaffected founders (258 males; 360 females), and 479 other unaffected relatives (230 males; 249 females). Families were drawn from four white populations (Australian, Scandinavian, British, and American), each of which had been previously screened for linkage and either is Northern European or has Northern European ancestry (table 2). The included families overlap substantially (70%) with those previously studied, but 216 new families not previously screened are also included. A breakdown of the demographic features of the affected individuals is shown in table 3. Together, the 730 families provide 1,002 affected-relative pairs, including 830 sib pairs, 14 half-sib pairs, 54 cousin pairs, 57 parent-child pairs, and 47 avuncular pairs; a breakdown by population is shown in table 4.
Table 2.
Population-Specific Breakdown of Families and Individuals
| Population | No. ofFamilies | No. ofIndividualsa | AffectedIndividuals(M:F) | UnaffectedFounders(M:F) | UnaffectedNonfounders(M:F) |
| Australian | 97 | 330 | 59:142 | 33:37 | 26:33 |
| Scandinavian | 165 | 475 | 140:215 | 26:54 | 18:22 |
| British | 298 | 1,023 | 186:428 | 107:159 | 65:78 |
| American | 170 | 864 | 128:297 | 92:110 | 121:116 |
Numbers of individuals for whom DNA was available and typed
Table 3.
Demographic Features of Affected Individuals
|
Averagea |
||||||
| Population | Age(years) | Disease Duration(years) | EDSS | RR(%) | SP(%) | PP(%) |
| Australian | 53.6 | 15.8 | 3.5b | 64 | 24 | 12 |
| Scandinavian | 49.5 | 19.1 | 4.7 | 49 | 32 | 19 |
| British | 45.0 | 15.7 | 4.5 | 57 | 28 | 15 |
| American | 45.3 | 15.4 | 5.5 | 62 | 34 | 4 |
EDSS = Expanded Disability Status Scale; RR = Relapse Remitting; SP = Secondary Progressive; PP = Primary Progressive.
The EDSS was not determined for these patients; instead, the average “disease steps” measure is shown (Hohol et al. 1999).
Table 4.
Breakdown of Affected Relative Pairs by Population
|
No. of Affected-Relative Pairs |
|||||
| Population | Sibling | HalfSibling | Cousin | Parent-Child | Avuncular |
| Australian | 68 | 5 | 16 | 2 | 8 |
| Scandinavian | 193 | 1 | 4 | 24 | 5 |
| British | 307 | 6 | 6 | 8 | 3 |
| American | 262 | 2 | 28 | 23 | 31 |
Ten individuals (all with DNA derived from cell lines) showed significant loss of heterozygosity on one or, at most, two chromosomes. These individuals were not included in the analysis of the corresponding chromosomes.
Informed consent was provided by all individuals involved in this study, and all affected individuals satisfy standardized diagnostic criteria (Poser et al. 1983; Goodkin et al. 1991; McDonald et al. 2001).
Allele Frequencies
By analyzing families from four populations together as a single data set, we have tacitly assumed that these are all drawn from the same homogenous population (Northern European). To test this assumption, we calculated Wright’s F statistic (FST), which measures the extent to which observed heterozygosity falls below that which would be expected if all studied families had the same genetic background. Employing heterozygosity and allele frequency estimates derived from the full 730-family data set using the PEDSTATS and MERLIN programs (Center for Statistical Genetics) and averaging across all 4,506 markers, we found an FST value of 0.0019, which indicates a modest degree of population substructure (difference in the genetic background of the included families), equivalent to that seen in older cohorts of the homogeneous Icelandic population (Helgason et al. 2005). We then used the MERLIN program (Center for Statistical Genetics) (Abecasis et al. 2002) to estimate separately the allele frequencies among the founders in each of the four populations for each of the 4,506 independent markers. After Bonferroni correction, no marker showed statistically significant difference in allele frequency across the four populations. The average difference in the frequency of an allele between any two populations was only 3.6%, an amount that would not be expected to significantly influence the results of linkage analysis. In keeping with the four populations included in the screen having a common genetic background, the extent of difference in the estimated allele frequency was found to be highly dependent on the number of samples typed from each compared population. The smaller the sample size used to estimate an allele frequency, the greater the variance in the resultant estimate. Thus, between the American and British populations, there are only 15 markers (0.3%) that showed a difference of >10% in allele frequency, whereas, between the Australian and Scandinavian populations, 361 markers (8.0%) showed such a difference. Similarly, the frequency of DR15 carriage among the typed founders from the four populations did not show any statistically significant difference.
For each marker in the linkage panel, Illumina provided ethnic subgroup–specific allele frequencies. For whites, these frequencies were based on the genotyping of 82 unrelated CEPH individuals. When we compared these CEPH frequencies with those estimated from the founders in our total set of 730 families, we found no statistically significant differences after Bonferroni correction. Repeating the linkage analysis with use of the CEPH allele frequencies rather than those derived from the data itself did not produce any substantial change in the results (the average change in the multipoint LOD score was just 0.01, the greatest increase was just 0.21, and the greatest reduction was just 0.30), which confirms that our analysis is robust to modest misspecification of allele frequencies.
Genotyping
Each individual provided a venous blood sample. Cell lines were established from 390 individuals, and the DNA used in this screen was extracted from a growth of these lines. In the remaining individuals, DNA was extracted directly from blood by use of standard methods. In some cases, additional DNA was generated by WGA (see the “WGA” section).
Each sample included in the analysis was genotyped in four highly multiplexed assays containing up to 1,536 markers (SNPs) per tube. The assay products from these were hybridized to high-density, bead-based microarrays and were imaged on a submicron-resolution scanner. This work was performed by the Illumina BeadLab service facility in San Diego. The Illumina genotyping system is fluorescence based, with one color specifying the first allele (A) and a second color specifying the second allele (B). Homozygous samples (AA or BB) therefore generate a signal exclusively/predominantly in the corresponding single color only, whereas heterozygous samples (AB) generate a signal in both colors. Since samples vary in their performance, each marker produces three clusters of results within color-signal space, one for each genotype. The tighter and more distinct the three different clusters are, the more accurate the genotyping. As part of the Illumina genotyping process, a quality measure known as a “GenCall score” is determined for each genotype. This metric measures how close a genotype is to the centre of the cluster of other samples assigned the same genotype, as compared with the centers of the clusters for the two other genotypes. This metric varies from 0 to 1, such that the higher the value, the more reliable the genotype. A sample giving a signal close to the center of a cluster that is tight and distinct from the other two clusters will have a high GenCall score.
Genotyping data from the class II HLA gene DRB1 was generated using several different typing methods with varying resolution. To maintain consistency across data sets, we thus coded the HLA-DRB1 locus in terms of the presence or absence of the DRB1*1501 allele (the multiple sclerosis–associated allele). These data were available for 2,448 (91%) of the individuals.
WGA
For 456 individuals, the total amount of DNA available was less than the minimum recommended for the Illumina system; therefore, for each of these, we produced additional DNA by WGA with use of the Molecular Staging REPLI-g kit, in accordance with the manufacturer's recommended conditions (Dean et al. 2002). WGA was performed more than once for some individuals, so that, in total, genotyping was attempted for 508 samples of WGA-derived DNA. Since initial results with these samples were disappointing, we also attempted genotyping in residual genomic DNA, when possible. Among these 456 individuals, typing in residual genomic DNA was attempted for 401, so that just 55 individuals had genotyping attempted only in WGA-derived DNA.
Only 104 of the 508 WGA-derived DNA samples yielded usable genotypes. As expected, the likelihood that genotypes could be generated from WGA-derived DNA correlated with the starting concentration of genomic DNA used in the production reaction: the mean starting concentration was 5.9 ng/μl in the samples that failed to genotype and 17.4 ng/μl in those that worked. The 104 successfully typed WGA-derived DNA samples provided genotypes for 100 individuals, 4 of whom were successfully genotyped in duplicate.
Duplicate Genotyping
In total, 138 individuals were successfully genotyped in duplicate, and 5 individuals were successfully genotyped in triplicate, providing 148 independent comparisons (>854,000 duplicate genotypes). Among these comparisons, there were only 688 genotypes that were different, and, in each instance, only one allele differed. Since each genotype includes two alleles and was typed twice, our results indicate a crude error rate of 0.02% (i.e., 688 alleles in error of 854,000×2×2 alleles called). When we considered the GenCall scores associated with the erroneous genotypes, we observed that 67% of the 688 errors involved genotypes with a GenCall score of <0.7, whereas only 5.7% of all genotypes had GenCall scores below that threshold. Similarly, consideration of the genotyping call rate (a crude surrogate for quality) among the markers associated with erroneous genotypes revealed that 63% of the error genotypes occurred in the 46 markers with genotyping call rates <99%. To balance the amount of usable data with its accuracy, we set the following stringent but arbitrary thresholds. We chose to include only genotypes with a GenCall score >0.7, among which we would expect the crude error rate to be <0.01%. We also chose to exclude all markers with a genotyping call rate of <98% for quality genotypes (i.e., those with GenCall scores >0.7) among the quality samples and to exclude all samples with a call rate of <98% for quality genotypes (GenCall >0.7) among quality markers. These thresholds resulted in the exclusion of 474 markers and 21 samples (as described above).
Among the 148 comparisons, 52 involved only genomic DNA, 92 involved both WGA-derived and genomic DNA, and 4 involved only WGA-derived DNA. The crude error rate in the 52 comparisons that were based on only genomic DNA (0.012%) was less than half that seen among the 96 comparisons involving at least one WGA-derived sample (0.025%). In view of this evidence, for greater reliability, genotypes derived from genomic samples were included in preference to those derived from WGA-derived samples when both were available, except when the genotyping success rate in the genomic sample was substantially less than that from the WGA-derived DNA.
Across the 730 families typed for the 4,506 markers finally included in the multipoint analysis (>12 million genotypes), only 254 Mendelian inconsistencies were observed, which indicates a final approximate error rate of only 0.002%.
Statistical Analysis
The pedigree structure for each included family was confirmed by comparing the expected and observed identity-by-state (IBS) allele sharing for each pair of related individuals with use of the Graphical Relationship Representation (GRR) program (Center for Statistical Genetics) (Abecasis et al. 2001b). Pairs of individuals were also compared across families to exclude any duplication. Marker heterozygosity, genotyping success rate, and evidence of deviation from HWE were determined using the PEDSTATS program (Center for Statistical Genetics) (Wigginton et al. 2005).
LD between autosomal markers was tested using the Haploview program (which considers both consecutive and nonconsecutive pairs) (Barrett et al. 2005) and between X-linked markers, by use of the PDTPHASE program (which considers only consecutive pairs) (UNPHASED Web site) (Dudbridge 2003). The PDTPHASE program (UNPHASED Web site) was also used to test for evidence of association with disease, after removal of Mendelian inconsistencies with use of the PedCheck program (O’Connell and Weeks 1998).
Since there is no currently available software capable of combining information from the pseudoautosomal and sex-linked regions of the X chromosome to enable a single integrated linkage analysis, we treated the sex-linked and two pseudoautosomal regions as three independent chromosomes. However, the pseudoautosomal regions could not be treated as straightforward autosomes, since, at the transition point from the sex-linked to pseudoautosomal regions, the sharing of the paternally derived chromosome is determined exclusively by the sex of the pair considered, the fragment being shared for same-sex siblings but unshared for opposite-sex siblings. Since multiple sclerosis affects females more often than males, paternally derived pseudoautosomal alleles are expected to show excess sharing. Analysis of this region is further complicated by the extreme difference in the sex-specific recombination rates, which are substantially higher in the paternally derived haplotype. Therefore, in each pseudoautosomal region, linkage analysis was confined to the maternally derived chromosomes.
Apart from the pseudoautosomal regions on the X chromosome, multipoint nonparametric linkage analysis was performed using the MERLIN (Center for Statistical Genetics) and MINX (Merlin in X) programs (Abecasis et al. 2002), with default settings and allele frequencies estimated from the founders. In this linkage analysis, improbable genotypes were first removed using the program’s wipe function (Abecasis et al. 2002); again, default settings were accepted. In total, 5,692 improbable genotypes were removed from 2,888 marker family combinations. Single-point LOD scores are provided in data file 1 (online only).
MERLIN’s simulation function (Center for Statistical Genetics) was used to empirically estimate the significance of the proportion of the genome that showed excess allele sharing. Twenty whole-genome replicates of the screen (460 chromosomes) were analyzed under the null hypothesis of no linkage. The proportion of the genome with a positive LOD score was then determined in each whole-genome replicate.
In the pseudoautosomal regions, we restricted analysis to a single randomly chosen sib pair from each of the simple nuclear families (n=646) and used the MAPMAKER/SIBS program (MIT Genome Center) (Kruglyak and Lander 1995) to determine the maximum likelihood identical-by-descent allele sharing in each case. Considering the sex-concordant (n=375) and -discordant (n=271) pairs separately, we then counted the number of pairs sharing and not sharing the maternal haplotype and tested any difference using a χ2 test. On the short arm, there was no excess sharing, whereas, on the long arm, there was nominally significant excess maternal haplotype sharing (P=.001). As expected, there was no significant difference between the sharing observed in the pseudoautosomal regions and that measured in the adjacent X-linked regions; results for the pseudoautosomal regions are not included in the figure.
The potential influence of transmission-ratio distortion (TRD) was sought using the GENEHUNTER++sad program (Kruglyak et al. 1996; Lemire et al. 2004). This program incorporates the novel allele-sharing test statistic Sad, which depends not only on excess allele sharing between affected relatives but also on the lack of allele sharing between phenotypically discordant relatives. The Sad statistic is therefore substantially more resistant to the presence of TRD than the analogous Whittemore and Halpern (1994) Spairs statistic. The difference in the nonparametric linkage (NPL) scores obtained using these two allele-sharing test statistics (NPL_ Spairs and NPL_ Sad) thus provides a measure of the evidence for the presence of TRD. Since GENEHUNTER (MIT Genome Center) (Kruglyak et al. 1996) performs an exact multipoint test for linkage, memory requirements limit the number of markers that can be included in any one analysis, especially within larger families. As a result of this limit, the genome was analyzed on the basis of 98 overlapping fragments, with an average of 54 markers per fragment.
To estimate the λs attributable to individual linkage peaks, we measured the maximum likelihood zero allele-sharing probability in the affected sib pairs from the 646 simple nuclear families, using the MAPMAKER/SIBS program (MIT Genome Center) (Kruglyak and Lander 1995).
Two approaches were used to examine the data for the presence of loci interacting with the known DRB1*1501 association; each approach was applied twice, once favoring those carrying the associated DRB1 allele (1501+) and then again favoring those not carrying the associated allele (1501−). In the first approach, the data were stratified on the basis of the presence or absence of the DRB1*1501 allele in the affected individuals by appropriately recoding the affection status prior to analysis with MERLIN (with default setting as described above) (Center for Statistical Genetics). In this recoding (stratified) analysis, 409 families were informative for linkage in the 1501+ analysis (i.e., included at least two affected individuals, each carrying at least one DRB1*1501 allele), and 187 families were informative for linkage in the 1501− analysis (i.e., included at least two affected individuals who each carry no DRB1*1501 alleles). This approach ignores affected-relative pairs that are discordant for DRB1*1501 status and allows pairs from the same family to be included in each subset. In the second approach, we performed an ordered-subset analysis (OSA [Duke Center for Human Genetics]) (Hauser et al. 2004) of the data, using the chromosome 6 family-specific maximum LOD score (MLS) within the 2-cM region spanning HLA-DRB1 as the covariate. In this method, families are ranked according to the covariate, then sequentially combined so that subset-specific LOD scores can be recalculated (using GENEHUNTER plus [MIT Genome Center] [Kruglyak et al. 1996; Kong and Cox 1997]) at each point in the genome. The ordered subset of families giving the greatest total LOD score is thereby identified. This procedure was performed twice, first ranking the families from highest to lowest covariate value (the “1501+” analysis) and then from lowest to highest (the “1501−” analysis). Since each OSA (Duke Center for Human Genetics) involves a considerable degree of multiple testing, empirical methods are employed to judge the nominal significance of any evidence of linkage seen in an ordered subset. Only regions with subsets involving >10% of the total number of families and with nominal significance of <5% are reported (a more extensive list of results is available in data file 2 [online only]).
Neither of the approaches described above could be employed on chromosome 6, because of the inevitable presence of linkage with the DRB1 gene. On this chromosome, we tested only those 44 families in which no typed individuals were positive for DRB1*1501. This ultra-negative group of families is modest in number and therefore lacks power but has the virtue of being essentially independent of DRB1*1501.
Results
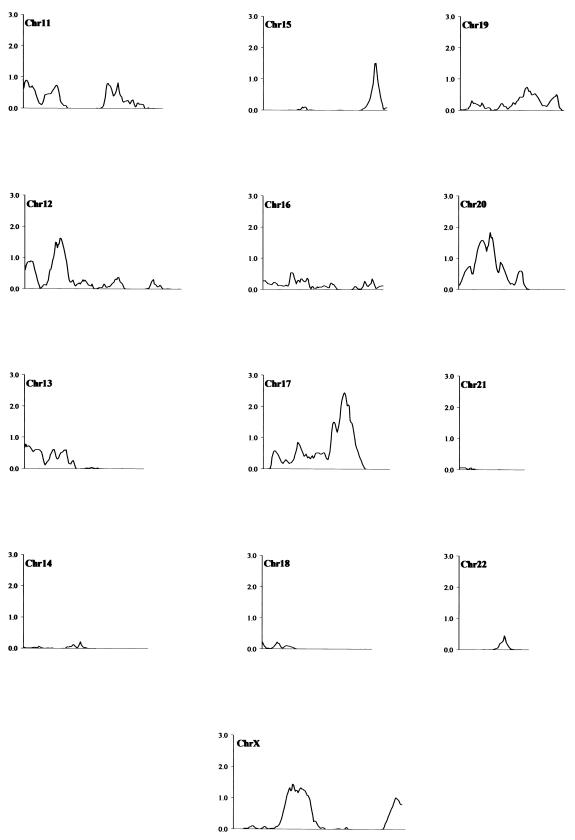
The results of multipoint nonparametric linkage analysis are shown in figure 1. Unequivocal linkage is demonstrated in the region of the MHC. Two other regions show suggestive evidence of linkage: chromosomes 17q23 and 5q33. The most negative LOD score seen anywhere in the genome is only −0.79, and overall 63.9% of the genome (outside chromosome 6) showed a positive LOD score rather than the expected 50%. Simulation studies under the null hypothesis of no linkage demonstrated that the mean proportion of the genome with a positive score is 49.7% (SD 6.7%). None of the replicate screens showed an excess of positive scores greater than that observed in the actual screen, which indicates an empirical significance of <5%. This bias in favor of positive scores confirms, again as in previous studies, more allele sharing than would be expected by chance. Table 5 summarizes the results from the six highest MLS peaks. In each case, the extent of allele sharing observed among affected siblings was used to estimate the λs attributable to individual loci (Risch 1990). This screen confirms that, outside the MHC region under the assumption of complete homogeneity, there are no loci with λs>1.2.
Figure 1.
Multipoint nonparametric linkage analysis performed using MERLIN (Center for Statistical Genetics). The figure includes one graph for each chromosome; the length of the X-axis is proportional to the genetic length of the corresponding chromosome, and the Y-axis scale is 0–3 in each case, except on chromosome 6, where the scale is 0–12.
Table 5.
Top Results from the Linkage Analysis
| Chromosome | MLS | SiblingAllele Sharing(%) | λs |
| 6p21 | 11.66 | 58.5 | 1.51 |
| 17q23 | 2.45 | 53.8 | 1.18a |
| 5q33 | 2.18 | 54.0 | 1.19a |
| 20p12 | 1.83 | 54.0 | 1.09a |
| 3p26 | 1.74 | 53.4 | 1.16a |
Since the evidence for linkage at these loci is not statistically robust, these estimates provided upper limits for the size of effects attributable to underlying susceptibility factors (Terwilliger et al. 2002).
To determine the extent to which the observed linkage peaks and background excess sharing might reflect underlying TRD, we compared the NPL scores obtained using the Spairsand Sad allele-sharing test statistics (see the “Material and Methods” section). The observed difference ranged from +0.80 to −0.82 and averaged +0.02. The overall excess of score seen with the Spairs statistic as compared with the Sad statistic suggests that at least some part of the excess background-allele sharing observed in the screen might be due to the presence of TRD. However, there does not seem to be any region in which the presence of TRD has exerted a major effect. The Spairs statistic gave higher scores than did the Sad statistic at each of the peaks listed in table 5, but these inflations were modest; increases were only 0.32, 0.25, 0.45, 0.18, and 0.51 at chromosomes 6p21, 17q23, 5q33, 20p12, and 3p26, respectively. In short, there is no evidence to support the argument that TRD is responsible for the linkage peaks observed in this study.
Stratification of the data for the associated HLA allele DRB1*1501 did not uncover any new region of statistically significant linkage. The highest LOD score seen in the 1501+ group of families is 2.18 on chromosome 5q33, which corresponds exactly to the third-highest LOD score seen in the total analysis and thereby suggests that the locus may exert its effects in concert with DRB1. The highest LOD score in the 1501− group is 1.61 on chromosome 2p25, in a region in which no linkage was seen in the combined analysis. The proportion of the genome providing positive LOD scores is inflated in both the 1501+ (62.5%) and 1501− (60.9%) analyses. No evidence of linkage was seen in the analysis of 1501− families on chromosome 6; in the HLA region, the LOD score was just 0.17.
In the OSA (Duke Center for Human Genetics), we found four additional regions of potential interest—chromosome 12q24 (nominal P=.008) emerging from the OSA that builds on families with evidence of linkage in the HLA region and chromosomes 19p13 (nominal P=.001), 1q43 (nominal P=.032), and 7q21 (nominal P=.033) emerging from the OSA that builds on families not showing evidence of linkage in the HLA region. After appropriate correction for the number of genomic regions considered, only the chromosome 19p13 result remains interesting, just reaching the corrected 5% significance threshold.
Each of the 5,282 markers passing quality standards was also tested for evidence of association with disease; after appropriate Bonferroni correction, no significant association was identifiable. The marker rs575208 from chromosome 1p13 showed the most-extreme evidence for association, with a corrected P value of 0.06. Importantly, none of the markers from the MHC region showed evidence of association with the disease, which indicates that the LOD score in this region has not been inflated by allelic association with the disease. Nominal (uncorrected) P values for each marker are provided in data file 1 (online only).
Discussion
To our knowledge, we have completed the largest and most powerful linkage screen ever performed for multiple sclerosis. The genotyping data included is highly accurate and virtually complete, making false-negative results from a lack of genome coverage extremely unlikely. Unequivocal linkage is demonstrated in the MHC region, and suggestive linkage is identified on chromosomes 17 and 5. OSA (Duke Center for Human Genetics) identifies a further locus on chromosome 19 that acts independent of HLA.
Although linkage to the MHC region is expected, given its established association with multiple sclerosis, the substantially greater LOD score attained in this screen compared with previous studies (Ebers et al. 1996; Haines et al. 1996; Sawcer et al. 1996; Kuokkanen et al. 1997; Broadley et al. 2001; Coraddu et al. 2001; Åkesson et al. 2002; Ban et al. 2002; Eraksoy et al. 2003; Hensiek et al. 2003; Kenealy et al. 2004) convincingly illustrates the increased power of this new study. In the individual 10-cM microsatellite-based screens, the MHC region has never shown linkage with genomewide significance (Ebers et al. 1996; Haines et al. 1996; Sawcer et al. 1996; Kuokkanen et al. 1997; Broadley et al. 2001; Coraddu et al. 2001; Åkesson et al. 2002; Ban et al. 2002; Eraksoy et al. 2003; Hensiek et al. 2003; Kenealy et al. 2004). Even in the meta-analysis of these microsatellite-based screens (GAMES and Transatlantic Multiple Sclerosis Genetics Cooperative 2003), the LOD score in the MHC region only just reached the genomewide significance threshold. Since the present study includes approximately the same number of families as the meta-analysis (GAMES and Transatlantic Multiple Sclerosis Genetics Cooperative 2003), the substantially greater LOD score from the MHC region confirms the additional power provided by the greater information extraction and genotyping accuracy of the new study. Furthermore, the allele sharing observed in the MHC region in our screen is consistent with what would be predicted from the known frequency and risk attributable to the DRB1*1501 haplotype, indicating that if secondary risk loci lie within the MHC region they are unlikely to exert more than a very modest additional effect on risk. The absence of linkage among those families uninfluenced by DRB1*1501 is further evidence against a substantial second locus in or near the MHC region. The possibility of additional risk loci within the MHC region is a popular hypothesis (Ligers et al. 2001; Marrosu et al. 2001; Harbo et al. 2004), but we have been unable to find any linkage evidence to support this hypothesis.
Although the non-MHC peaks of linkage observed in our study fail to reach genomewide statistical significance as defined by Lander and Kruglyak (1995), they provide invaluable information concerning the magnitude of effects likely to be attributable to multiple sclerosis–susceptibility genes. Given that the predicted effect sizes calculated from these linkage peaks are expected to be overestimates (Terwilliger et al. 2002), it is reasonable to conclude that the calculated λs values shown in table 5 provide a robust upper limit to the size of effect attributable to genuine loci. This result has substantial implications for future research into the genetic aspects of this disease.
In consideration of the results of this screen, it is important to remember that the power of linkage analysis falls rapidly as the effects attributable to individual genes decline (Risch and Merikangas 1996). One corollary of this limited resolution is that failure to detect linkage does not exclude the existence of genes with more-modest effects below the resolution provided by the screen. Therefore, it should not be concluded that the failure of this study to demonstrate statistically significant linkage outside the MHC region indicates that no such genes exist.
A number of confounding effects could have prevented the detection of linkage in this study. It seems reasonable to expect that susceptibility to multiple sclerosis is genetically heterogeneous and equally reasonable to expect that the relative importance of individual genes will differ between populations. In this setting, combining families from different backgrounds may actually dilute rather than strengthen the evidence of linkage. In our screen, however, we have been able to confirm that there is little difference in the genetic background of the four populations considered, as expected from their genealogically common ancestry. This concordance makes it unlikely that there will be much difference between the genetic factors that determine susceptibility in each of the subpopulations and suggests that much more is likely to have been gained from the increased power provided by the greater number of families included.
Following a similar line of reasoning, it seems logical to suggest that individuals developing multiple sclerosis due to risk alleles at one set of loci might show phenotypic differences from those developing their disease secondary to risk conferred by a different set of susceptibility loci. Although multiple sclerosis shows extreme variability in its phenotype, the evidence of familial clustering of particular phenotypic features is modest (Barcellos et al. 2002), and, at the moment, there is no robust manner in which to select subgroups of patients that might be genetically more homogeneous, even though these subgroups would be expected to include more penetrant alleles. On the other hand, it is inescapable that rare variants from multiple loci (extensive locus heterogeneity) could account for a significant part of the genetically determined susceptibility to multiple sclerosis, in which setting, our study would have little power to identify linkage to the individual contributory loci.
How to appropriately correct for multiple testing is an unresolved issue. Throughout this work, we have employed an ultraconservative approach and have applied crude Bonferroni correction. This procedure maximizes specificity but reduces power. Although the genetic model underlying susceptibility to multiple sclerosis is unknown, it seems reasonable to expect that it will involve a spectrum of relatively common risk alleles exerting only modest effects as well as less-common risk alleles with more pronounced effects (Wang et al. 2005). The significant excess of sharing seen in the genome outside chromosome 6 confirms that other non-MHC genes exist but have been missed by the limited power of this approach.
This screen provides the most definitive linkage map currently available for multiple sclerosis. Inspection of this map indicates that future studies attempting to identify genetic factors influencing the development of this disease will need to rely on association-based methods and must involve large patient cohorts. A review of the available genetic literature about multiple sclerosis shows that few studies met these criteria. A lack of concordance in the results from underpowered studies is entirely expected and provides little if any guidance as to which loci have been excluded and which are potentially relevant. In short, our screen indicates that all logical candidates will need to be reevaluated with more powerful studies.
Authors
Members of the International Multiple Sclerosis Genetics Consortium who made substantial contributions to this work are as follows: Stephen Sawcer, Maria Ban, Mel Maranian, Tai Wai Yeo, and Alastair Compston, University of Cambridge Department of Clinical Neurosciences, Addenbrooke’s Hospital, Cambridge, United Kingdom; Andrew Kirby, Mark J. Daly, Philip L. De Jager, Emily Walsh, Eric S. Lander, John D. Rioux, and David A. Hafler, The Broad Institute, Massachusetts Institute of Technology, Cambridge, MA; Philip L. De Jager, John D. Rioux, and David A. Hafler, Center for Neurologic Diseases, Brigham and Women’s Hospital, Philip L. De Jager, Adrian Ivinson, Eric S. Lander, John D. Rioux, and David A. Hafler, Harvard Medical School, and Adrian Ivinson, Harvard Center for Neurodegeneration and Repair, Boston; Jacqueline Rimmler, Simon G. Gregory, Silke Schmidt, and Margaret A. Pericak-Vance, Duke University Medical Center, Center for Human Genetics, Durham, NC; Eva Åkesson and Jan Hillert, Department of Neurology, Karolinska University Hospital–Huddinge, Karolinska Institute, Huddinge, Sweden; Pameli Datta and Annette Oturai, Department of Neurology, and Lars P. Ryder, Department of Clinical Immunology, Copenhagen University Hospital, Rigshospitalet, Copenhagen; Hanne F. Harbo, Department of Neurology, Ullevål University Hospital, and Anne Spurkland, Department of Anatomy, Institute of Basal Medical Sciences, University of Oslo, Oslo; Kjell-Morten Myhr, Department of Neurology, Haukeland University Hospital, Bergen, Norway; Mikko Laaksonen, Turku Immunology Centre and Department of Virology, University of Turku, Turku, Finland; David Booth, Robert Heard, and Graeme Stewart, University of Sydney, Institute for Immunology and Allergy Research, Westmead Millennium Institute, and Westmead Hospital, Sydney; Robin Lincoln, Lisa F. Barcellos, Stephen L. Hauser, and Jorge R. Oksenberg, Department of Neurology, School of Medicine, University of California at San Francisco, San Francisco; Lisa F. Barcellos, Division of Epidemiology, School of Public Health, University of California at Berkeley, Berkeley; and Shannon J. Kenealy and Jonathan L. Haines, Center for Human Genetics Research, Vanderbilt University Medical Center, Nashville.
Supplementary Material
Acknowledgments
We thank the representatives and technical staff from Illumina for their extremely efficient and helpful service—in particular, Mark Hansen, Sandy McBean, and Sarah Shaw Murray. This work was supported by the International Multiple Sclerosis Genetics Consortium, a National Multiple Sclerosis Society Collaborative MS Research Center Award, National Institutes of Health grant NS032830, the Medical Research Council (U.K.), and the Wellcome Trust. We also acknowledge the multiple sclerosis societies, from the various countries involved, for their support in the recruitment of families.
Web Resources
The URLs for data presented herein are as follows:
- Center for Statistical Genetics, http://www.sph.umich.edu/csg/abecasis/ (for MERLIN, PEDSTATS, and GRR)
- Duke Center for Human Genetics, http://www.chg.duke.edu/software/osa.html (for OSA)
- GENEHUNTER++sad, http://www.genome.mcgill.ca/~mlemire/software.html
- Haploview, http://www.broad.mit.edu/mpg/haploview/
- Illumina, http://www.illumina.com/ (for the BeadLab service facility)
- MIT Genome Center FTP Archive, http://www.broad.mit.edu/ftp/distribution/software/ (for MAPMAKER/SIBS and GENEHUNTER)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for multiple sclerosis)
- PedCheck, http://watson.hgen.pitt.edu/register/docs/pedcheck.html
- UNPHASED, http://www.rfcgr.mrc.ac.uk/~fdudbrid/software/unphased/ (for the PDTPHASE program)
References
- Abecasis GR, Cherny SS, Cardon LR (2001a) The impact of genotyping error on family-based analysis of quantitative traits. Eur J Hum Genet 9:130–134 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001b) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 [DOI] [PubMed] [Google Scholar]
- ——— (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Åkesson E, Oturai A, Berg J, Fredrikson S, Andersen O, Harbo HF, Laaksonen M, Myhr KM, Nyland HI, Ryder LP, Sandberg-Wollheim M, Sorensen PS, Spurkland A, Svejgaard A, Holmans P, Compston A, Hillert J, Sawcer S (2002) A genome-wide screen for linkage in Nordic sib-pairs with multiple sclerosis. Genes Immun 3:279–285 [DOI] [PubMed] [Google Scholar]
- Ban M, Stewart GJ, Bennetts BH, Heard R, Simmons R, Maranian M, Compston A, Sawcer SJ (2002) A genome screen for linkage in Australian sibling-pairs with multiple sclerosis. Genes Immun 3:464–469 [DOI] [PubMed] [Google Scholar]
- Barcellos LF, Oksenberg JR, Green AJ, Bucher P, Rimmler JB, Schmidt S, Garcia ME, Lincoln RR, Pericak-Vance MA, Haines JL, Hauser SL (2002) Genetic basis for clinical expression in multiple sclerosis. Brain 125:150–158 [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- Boyles AL, Scott WK, Martin ER, Schmidt S, Li Y, Ashley-Koch A, Bass MP, Schmidt M, Pericak-Vance MA, Speer MC, Hauser ER (2005) Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley S, Sawcer S, D’Alfonso S, Hensiek A, Coraddu F, Gray J, Roxburgh R, Clayton D, Buttinelli C, Quattrone A, Trojano M, Massacesi L, Compston A (2001) A genome screen for multiple sclerosis in Italian families. Genes Immun 2:205–210 [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Merette C, Xie X, Townsend L, Gilliam TC, Ott J (1993) Molecular and statistical approaches to the detection and correction of errors in genotype databases. Am J Hum Genet 53:1137–1145 [PMC free article] [PubMed] [Google Scholar]
- Carton H, Vlietinck R, Debruyne J, De Keyser J, D’Hooghe MB, Loos R, Medaer R, Truyen L, Yee IM, Sadovnick AD (1997) Risks of multiple sclerosis in relatives of patients in Flanders, Belgium. J Neurol Neurosurg Psychiatry 62:329–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compston DA, Batchelor JR, McDonald WI (1976) B-lymphocyte alloantigens associated with multiple sclerosis. Lancet 2:1261–1265 [DOI] [PubMed] [Google Scholar]
- Coraddu F, Sawcer S, D’Alfonso S, Lai M, Hensiek A, Solla E, Broadley S, Mancosu C, Pugliatti M, Marrosu MG, Compston A (2001) A genome screen for multiple sclerosis in Sardinian multiplex families. Eur J Hum Genet 9:621–626 [DOI] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA 99:5261–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G, McLoughlin H, Brady R, Adelstein AM, Tallett-Williams J (1976) Multiple sclerosis among immigrants in Greater London. Br Med J 1:861–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F (2003) Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 25:115–121 [DOI] [PubMed] [Google Scholar]
- Ebers GC, Kukay K, Bulman DE, Sadovnick AD, Rice G, Anderson C, Armstrong H, et al (1996) A full genome search in multiple sclerosis. Nat Genet 13:472–476 [DOI] [PubMed] [Google Scholar]
- Ebers GC, Sadovnick AD, Dyment DA, Yee IM, Willer CJ, Risch N (2004) Parent-of-origin effect in multiple sclerosis: observations in half-siblings. Lancet 363:1773–1774 [DOI] [PubMed] [Google Scholar]
- Ebers GC, Sadovnick AD, Risch NJ, Canadian Collaborative Study Group (1995) A genetic basis for familial aggregation in multiple sclerosis. Nature 377:150–151 [DOI] [PubMed] [Google Scholar]
- Eichorst H (1896) Uber infantile und hereditare multiple sclerose. Virch Arch 146:173–193 [Google Scholar]
- Eraksoy M, Kurtuncu M, Akman-Demir G, Kilinc M, Gedizlioglu M, Mirza M, Anlar O, Kutlu C, Demirkiran M, Idrisoglu HA, Compston A, Sawcer S (2003) A whole genome screen for linkage in Turkish multiple sclerosis. J Neuroimmunol 143:17–24 [DOI] [PubMed] [Google Scholar]
- GAMES, Transatlantic Multiple Sclerosis Genetics Cooperative (2003) A meta-analysis of whole genome linkage screens in multiple sclerosis. J Neuroimmunol 143:39–46 [DOI] [PubMed] [Google Scholar]
- Goodkin DE, Doolittle TH, Hauser SS, Ransohoff RM, Roses AD, Rudick RA (1991) Diagnostic criteria for multiple sclerosis research involving multiply affected families. Arch Neurol 48:805–807 [DOI] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA (2005) Complement factor H variant increases the risk of age-related macular degeneration. Science 308:419–421 [DOI] [PubMed] [Google Scholar]
- Haines JL, Ter-Minassian M, Bazyk A, Gusella JF, Kim DJ, Terwedow H, Pericak-Vance MA, et al (1996) A complete genomic screen for multiple sclerosis underscores a role for the major histocompatability complex. Nat Genet 13:469–471 [DOI] [PubMed] [Google Scholar]
- Haines JL, Terwedow HA, Burgess K, Pericak-Vance MA, Rimmler JB, Martin ER, Oksenberg JR, Lincoln R, Zhang DY, Banatao DR, Gatto N, Goodkin DE, Hauser SL, The Multiple Sclerosis Genetics Group (1998) Linkage of the MHC to familial multiple sclerosis suggests genetic heterogeneity. Hum Mol Genet 7:1229–1234 [DOI] [PubMed] [Google Scholar]
- Harbo HF, Lie BA, Sawcer S, Celius EG, Dai KZ, Oturai A, Hillert J, Lorentzen AR, Laaksonen M, Myhr KM, Ryder LP, Fredrikson S, Nyland H, Sorensen PS, Sandberg-Wollheim M, Andersen O, Svejgaard A, Edland A, Mellgren SI, Compston A, Vartdal F, Spurkland A (2004) Genes in the HLA class I region may contribute to the HLA class II-associated genetic susceptibility to multiple sclerosis. Tissue Antigens 63:237–247 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M, Guo SW, Risch N (1996) Affected-sib-pair interval mapping and exclusion for complex genetic traits: sampling considerations. Genet Epidemiol 13:117–137 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M (2004) Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol 27:53–63 [DOI] [PubMed] [Google Scholar]
- Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K (2005) An Icelandic example of the impact of population structure on association studies. Nat Genet 37:90–95 [DOI] [PubMed] [Google Scholar]
- Hensiek AE, Roxburgh R, Smilie B, Coraddu F, Åkesson E, Holmans P, Sawcer SJ, Compston DA (2003) Updated results of the United Kingdom linkage-based genome screen in multiple sclerosis. J Neuroimmunol 143:25–30 [DOI] [PubMed] [Google Scholar]
- Hogancamp WE, Rodriguez M, Weinshenker BG (1997) The epidemiology of multiple sclerosis. Mayo Clin Proc 72:871–878 [DOI] [PubMed] [Google Scholar]
- Hohol MJ, Orav EJ, Weiner HL (1999) Disease steps in multiple sclerosis: a longitudinal study comparing disease steps and EDSS to evaluate disease progression. Mult Scler 5:349–354 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, del Bosque-Plata L, Oda Y, Yoshiuchi I, Colilla S, Polonsky KS, Wei S, Concannon P, Iwasaki N, Schulze J, Baier LJ, Bogardus C, Groop L, Boerwinkle E, Hanis CL, Bell GI (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603 [DOI] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium, Sawcer SJ, Maranian M, Singlehurst S, Yeo T, Compston A, Daly MJ, De Jager PL, Gabriel S, Hafler DA, Ivinson AJ, Lander ES, Rioux JD, Walsh E, Gregory SG, Schmidt S, Pericak-Vance MA, Barcellos L, Hauser SL, Oksenberg JR, Kenealy SJ, Haines JL (2004) Enhancing linkage analysis of complex disorders: an evaluation of high-density genotyping. Hum Mol Genet 13:1943–1949 [DOI] [PubMed] [Google Scholar]
- Jersild C, Svejgaard A, Fog T (1972) HL-A antigens and multiple sclerosis. Lancet 1:1240–1241 [DOI] [PubMed] [Google Scholar]
- Kenealy SJ, Babron M-C, Bradford Y, Schnetz-Boutaud N, Haines JL, Rimmler JB, Schmidt S, Pericak-Vance MA, Barcellos LF, Lincoln RR, Oksenberg JR, Hauser SL, Clanet M, Brassat D, Edan G, Yaouanq J, Semana G, Cournu-Rebeix I, Lyon-Caen O, Fontaine B, for the American-French Multiple Sclerosis Genetics Group (2004) A second-generation genomic screen for multiple sclerosis. Am J Hum Genet 75:1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Kuokkanen S, Gschwend M, Rioux JD, Daly MJ, Terwilliger JD, Tienari PJ, Wikström J, Palo J, Stein LD, Hudson TJ, Lander ES, Peltonen L (1997) Genomewide scan of multiple sclerosis in Finnish multiplex families. Am J Hum Genet 61:1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Hooper AB, Huntsman JW, Ward RH (1983) Evaluating pedigree data. I. The estimation of pedigree error in the presence of marker mistyping. Am J Hum Genet 35:241–262 [PMC free article] [PubMed] [Google Scholar]
- Lemire M, Roslin NM, Laprise C, Hudson TJ, Morgan K (2004) Transmission-ratio distortion and allele sharing in affected sib pairs: a new linkage statistic with reduced bias, with application to chromosome 6q25.3. Am J Hum Genet 75:571–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligers A, Dyment DA, Willer CJ, Sadovnick AD, Ebers G, Risch N, Hillert J, the Canadian Collaborative Study Groups (2001) Evidence of linkage with HLA-DR in DRB1*15-negative families with multiple sclerosis. Am J Hum Genet 69:900–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrosu MG, Murru R, Murru MR, Costa G, Zavattari P, Whalen M, Cocco E, Mancosu C, Schirru L, Solla E, Fadda E, Melis C, Porru I, Rolesu M, Cucca F (2001) Dissection of the HLA association with multiple sclerosis in the founder isolated population of Sardinia. Hum Mol Genet 10:2907–2916 [DOI] [PubMed] [Google Scholar]
- McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, McFarland HF, Paty DW, Polman CH, Reingold SC, Sandberg-Wollheim M, Sibley W, Thompson A, van den Noort S, Weinshenker BY, Wolinsky JS (2001) Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol 50:121–127 [DOI] [PubMed] [Google Scholar]
- Mumford CJ, Wood NW, Kellar-Wood H, Thorpe JW, Miller DH, Compston DA (1994) The British Isles survey of multiple sclerosis in twins. Neurology 44:11–15 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olerup O, Hillert J (1991) HLA class II-associated genetic susceptibility in multiple sclerosis: a critical evaluation. Tissue Antigens 38:1–15 [DOI] [PubMed] [Google Scholar]
- Poser CM, Paty DW, Scheinberg L, McDonald WI, Davis FA, Ebers GC, Johnson KP, Sibley WA, Silberberg DH, Tourtellotte WW (1983) New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol 13:227–231 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet 46:222–228 [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Robertson NP, Fraser M, Deans J, Clayton D, Walker N, Compston DA (1996) Age-adjusted recurrence risks for relatives of patients with multiple sclerosis. Brain 119:449–455 [DOI] [PubMed] [Google Scholar]
- Robertson NP, O’Riordan JI, Chataway J, Kingsley DP, Miller DH, Clayton D, Compston DA (1997) Offspring recurrence rates and clinical characteristics of conjugal multiple sclerosis. Lancet 349:1587–1590 [DOI] [PubMed] [Google Scholar]
- Sadovnick AD, Baird PA, Ward RH (1988) Multiple sclerosis: updated risks for relatives. Am J Med Genet 29:533–541 [DOI] [PubMed] [Google Scholar]
- Sawcer S, Jones HB, Feakes R, Gray J, Smaldon N, Chataway J, Robertson N, Clayton D, Goodfellow PN, Compston A (1996) A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat Genet 13:464–468 [DOI] [PubMed] [Google Scholar]
- Stewart GJ, Teutsch SM, Castle M, Heard RN, Bennetts BH (1997) HLA-DR, -DQA1 and -DQB1 associations in Australian multiple sclerosis patients. Eur J Immunogenet 24:81–92 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Haghighi F, Hiekkalinna TS, Goring HH (2002) A bias-ed assessment of the use of SNPs in human complex traits. Curr Opin Genet Dev 12:726–734 [DOI] [PubMed] [Google Scholar]
- Wang WY, Barratt BJ, Clayton DG, Todd JA (2005) Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet 6:109–118 [DOI] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Wigginton JE, Cutler DJ, Abecasis GR (2005) A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 76:887–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Dyment DA, Risch NJ, Sadovnick AD, Ebers GC (2003) Twin concordance and sibling recurrence rates in multiple sclerosis. Proc Natl Acad Sci USA 100:12877–12882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Dyment DA, Sadovnick AD, Rothwell PM, Murray TJ, Ebers GC (2005) Timing of birth and risk of multiple sclerosis: population based study. BMJ 330:120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester R, Ebers G, Fu SM, Espinosa L, Zabriskie J, Kunkel HG (1975) B-cell alloantigen Ag 7a in multiple sclerosis. Lancet 2:814 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.