Abstract
An autosomal recessive syndrome of nonprogressive cerebellar ataxia and mental retardation is associated with inferior cerebellar hypoplasia and mild cerebral gyral simplification in the Hutterite population. An identity-by-descent mapping approach using eight patients from three interrelated Hutterite families localized the gene for this syndrome to chromosome region 9p24. Haplotype analysis identified familial and ancestral recombination events and refined the minimal region to a 2-Mb interval between markers D9S129 and D9S1871. A 199-kb homozygous deletion encompassing the entire very low density lipoprotein receptor (VLDLR) gene was present in all affected individuals. VLDLR is part of the reelin signaling pathway, which guides neuroblast migration in the cerebral cortex and cerebellum. To our knowledge, this syndrome represents the first human lipoprotein receptor malformation syndrome and the second human disease associated with a reelin pathway defect.
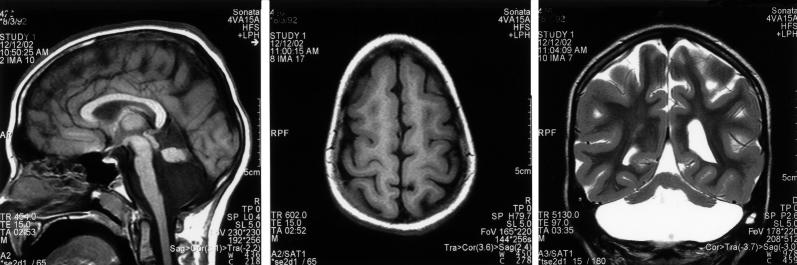
An autosomal recessive syndrome of nonprogressive cerebellar ataxia, mental retardation, and cerebellar hypoplasia was recognized and described in the Hutterite population in the 1980s (Schurig et al. 1981; Pallister and Opitz 1986). This constellation of findings was referred to as “dysequilibrium syndrome” (DES [MIM 224050]) on the basis of the clinical similarity to a group of patients with syndromic nonprogressive cerebellar ataxia initially described by Hagberg et al. in 1972 (Hagberg et al. 1972). As part of a larger project to characterize cerebellar disorders in the Hutterite population, we identified 12 Hutterites with DES (hereafter, “DES-H” refers to DES in the Hutterite population) and further described the clinical and neuroimaging features (Glass et al., in press). DES-H is a nonprogressive syndrome characterized by moderate-to-profound mental retardation, delayed ambulation, and predominantly truncal ataxia. Additional features include strabismus and pes planus in the majority of patients, seizures in 40% of patients, and short stature in 15% of patients. Magnetic resonance imaging (MRI) demonstrates inferior cerebellar hypoplasia and mild cortical gyral simplification (fig. 1).
Figure 1.
MRI demonstrating characteristic brain malformations seen in patients with DES-H. A, Sagittal T1W. B, Axial T1W. C, Coronal T2W spin echo images showing a small cerebellum in a fluid-filled but normal-sized posterior fossa. The superior half of the vermis is formed, but the inferior half is not. The pons is abnormally small. The cerebral cortex is simplified. Reprinted with permission (Glass et al., in press).
The Hutterites originated from one of several Anabaptist groups formed during the Protestant Reformation in the 16th century and have lived on the North American prairies since the late 1800s (Hostetler 1985). The population now numbers >40,000 individuals, the majority of whom are descendants of 89 founders (Nimgaonkar et al. 2000). The unique social characteristics of this population have resulted in virtual genetic isolation. In a reconstructed 1981 census of 25,223 Hutterites in North America, the average inbreeding coefficient was 0.038, with a range of 0–0.164 (T. M. Fujiwara and K. Morgan, unpublished data). Over 30 different autosomal recessive conditions have been described in the Hutterite population (Innes et al. 1999). Given the population's characteristics, it is likely that each of these rare alleles was introduced by a single ancestor.
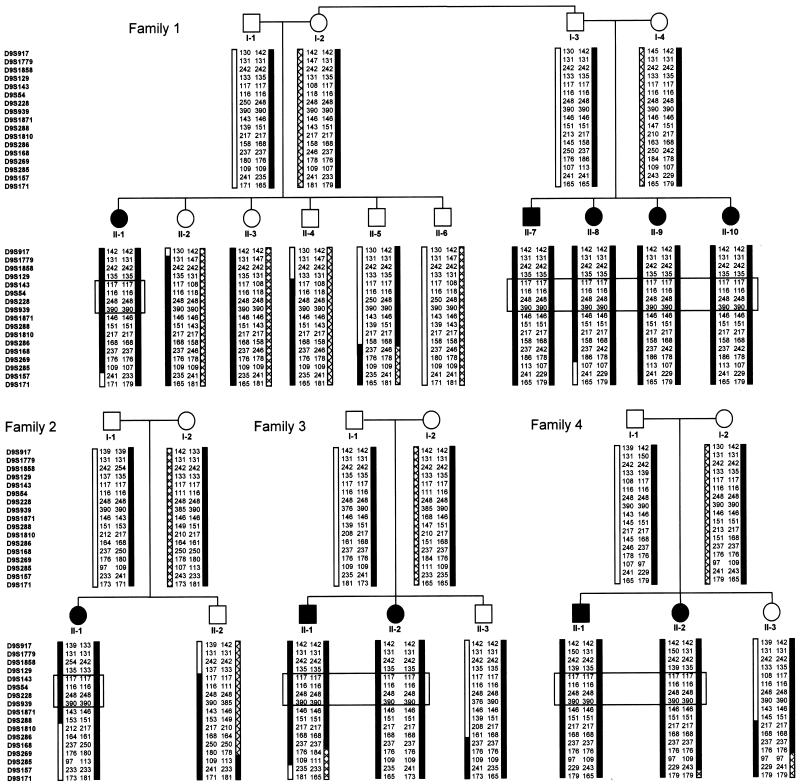
We used an identity-by-descent mapping approach to localize the gene for DES-H to chromosome region 9p24. DNA samples from the eight patients of families 1, 2, and 3 (fig. 2) were genotyped using a set of 400 polymorphic microsatellite markers (ABI PRISM Linkage Mapping Set version 2 [Applied Biosystems]) with an average spacing of <10 cM and heterozygosity of 0.79. The amplified PCR products were analyzed on an ABI 377 automated sequencer (Applied Biosystems), and the data were processed using GeneScan (version 3.7) and Genotyper (version 3.7) software (Applied Biosystems). Data from the genome scan identified homozygosity in at least five of the eight initial patients, with allele sharing in the remaining patients, for eight different loci. For these candidate regions, DNA from the eight patients and their family members (parents and unaffected siblings) was genotyped using the marker of interest as well as additional flanking microsatellite markers from the 5-cM Linkage Mapping Set (ABI PRISM Linkage Mapping Set version 2) and/or from the Généthon, deCode, or Marshfield maps (Human Genome Resources Web site). Haplotype construction by visual inspection within the nuclear families allowed seven of these regions to be excluded on the basis of lack of segregation of the haplotype with the disorder.
Figure 2.
Haplotype analysis of the Hutterite families. Affected individuals are identified by blackened symbols and unaffected family members by unblackened symbols. In family 1, individuals I-2 and I-3 are full siblings. Haplotypes were constructed by visual inspection, with the assumption of a minimal number of recombination events. The marker order was determined from the physical map of the region. For the children in each family, the allele on the left was inherited from the father and the allele on the right from the mother. A patterned bar indicates haplotypes for chromosome 9 markers, a black bar indicates the alleles associated with the DES-H disease allele, and a gray region indicates a region in which a recombinant event occurred. An observed paternal recombination event in unaffected sibling II-3 from family 4 indicates a proximal limit of D9S286 for the DES-H locus. A recombinant defining the distal boundary for the DES-H locus was not identified. The boxed markers correspond to the smallest region of shared homozygosity and the minimal region for the DES-H gene. D9S129 was predicted to be the distal limit, on the basis of inferred ancestral recombination events that resulted in different D9S129 alleles in affected individuals II-1 from family 2 and the siblings II-1 and II-2 from family 4. D9S1871 was predicted to be the proximal limit on the basis of an inferred recombinant in one of the ancestors of the father in family 2, who carried a different allele on the disease chromosome at this locus.
A three-marker haplotype generated using the genome scan marker D9S288 and two flanking markers, D9S1858 (distal) and D9S1810 (proximal), from the telomeric region of the p arm of chromosome 9 segregated with DES-H in the families, and seven of the initial eight patients were homozygous for the shared haplotype. Additional markers were genotyped in the region for these eight patients, as well as for two additional patients from family 4 and parents and unaffected siblings from all families. Multipoint linkage calculations (Kruglyak et al. 1996; University of Pittsburgh Department of Human Genetics Web site) between DES-H and the markers indicated in figure 2 revealed a LOD score >4.0 for 11 markers from D9S917 to D9S1810, with a maximum of 4.3, providing statistical evidence of linkage to the telomeric end of the short arm of chromosome 9, in region 9p24. This analysis was performed under the assumption of autosomal recessive inheritance with complete penetrance. Marker-allele frequencies were estimated from the observed alleles in all the genotyped individuals. Although the families were known to be related in multiple different ways (Glass et al., in press), the four pedigrees shown in figure 2 were used to calculate linkage, since they were expected to provide sufficient power. The frequency of the DES-H allele was set at 0.03333, on the basis of an observed population frequency of 1/900 (Glass et al., in press) and under the assumption of Hardy-Weinberg equilibrium. This is probably an overestimate of the true DES-H allele frequency, given the level of inbreeding in the Hutterites.
The extended haplotypes demonstrated both familial and ancestral recombination events (fig. 2). On the basis of familial recombination events, the minimal region for the DES-H locus was 18.1 cM (11.0 and 25.6 cM on the female and male, respectively, Marshfield genetic maps), between the p telomere and D9S286. Under the assumption of identity by descent and inferred ancestral recombination, the DES-H locus was predicted to lie in a 5.5-cM interval between D9S129 and D9S1871 (genetic distance estimated from the physical map for D9S129 and from the Marshfield genetic map and the physical map for D9S1871), in which all patients were homozygous for the same allele at D9S143, D9S54, D9S228, and D9S939. The 5.5-cM DES-H candidate region between D9S129 and D9S1871 corresponded to a 2-Mb interval and contained 10 transcripts, of which 6 encoded characterized genes.
The critical interval contained an obvious candidate, the VLDLR gene, which encodes a receptor for reelin. The reelin signaling pathway guides neuroblast migration in the cerebral cortex and cerebellum (D’Arcangelo et al. 1995; Rice and Curran 2001; Tissir and Goffinet 2003). VLDLR consists of 19 exons and spans ∼40 kb (Sakai et al. 1994). Attempts to amplify the VLDLR coding exons by PCR repeatedly failed in patients but not in controls or unaffected family members. Lack of amplification of exons 1A and 19 suggested a homozygous deletion of the entire VLDLR coding region (data not shown). Part of exon 1 of the KCNV2 gene (GenBank accession number AL354723), ∼70 kb proximal to VLDLR, was present in patients, as determined by PCR amplification. Exon 4 of the SMARCA2 gene (GenBank accession number AL138755), 430 kb distal to VLDLR, was identified in patients by PCR amplification of the trinucleotide repeat PMC303366P11 (UniSTS) (data not shown). These results were consistent with a deletion of <500 kb.
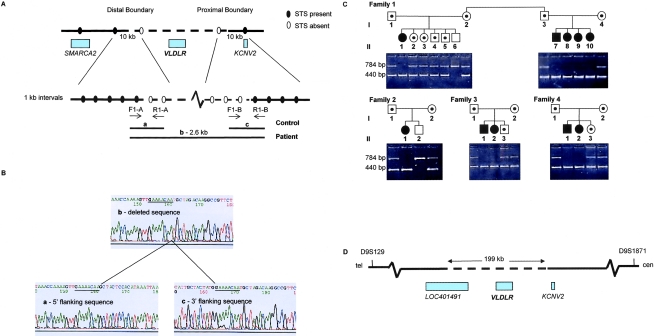
To determine the extent of the deleted sequence encompassing VLDLR, we used a sequence-tagged site (STS) walking strategy (fig. 3A). This approach generated a 2.6-kb junction fragment, which was amplified and sequenced in patients with DES-H and was compared with sequences generated by amplifying the two STSs flanking the proximal and distal boundaries in control DNA (fig. 3B). A homozygous deletion of 199,163 bp was present in each patient. A three-primer reaction was designed to allow for coamplification of both the normal and deleted alleles. Genomic DNA from all patients, parents, and siblings was amplified, and results were consistent with the linkage and haplotype data; each parent was a carrier of the deletion (fig. 3C). An 8-bp direct repeat, GAAAACAA (fig. 3B), was present at the breakpoints. Such short direct repeats have been found to mark the endpoints of many pathogenic deletions, which suggests that the repeats predispose to slippage during DNA replication that results in deletion of the intervening sequence and one of the repeat sequences (Antonarakis et al. 2001).
Figure 3.
Characterization of the VLDLR deletion. A, STS walking strategy used to identify the deletion boundaries. STSs were scored as “present” or “absent” in the genomic DNA of patients with DES-H. The dashed line represents the deletion. Blackened circles indicate that STSs are present, and unblackened circles indicate that STSs are not present in patient genomic DNA. The distal boundary was narrowed to a 69-kb region 130 kb from VLDLR by use of STSs present in UniSTS; the proximal boundary was estimated to be within the 70-kb region between VLDLR and KCNV2, on the basis of the presence of exon 1 of the KCNV2 gene in genomic DNA of patients, by use of PCR amplification. STSs were then generated using the chromosome 9 genomic contig (GenBank accession number NT_008413) at 10-kb intervals, to walk along the chromosome in both the 69-kb distal and 70-kb proximal regions. Within these 10-kb intervals, STSs were generated every 1 kb (all primer sequences and amplification conditions available on request). The forward primer of the most-proximal STS present in the distal boundary and the reverse primer of the most-distal STS present in the proximal boundary in genomic DNA of patients were used to amplify a 2.6-kb fragment spanning the deletion. Because of the large size of the deletion, these primers could not be used to amplify control DNA; instead, forward and reverse primers of the two flanking STSs for each of the boundary regions were used. B, Sequence of deletion boundaries. The products a, b, and c were sequenced using the ABI PRISM BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) and an ABI PRISM 377 automated sequencer. A direct 8-bp repeat (underlined) was present in the sequenced junction fragment of a patient with DES-H (b) and in the 5′ and 3′ flanking sequences of control DNA (a and c). C, Segregation analysis. Genomic DNA from patients and family members was amplified using a three-primer reaction mix, which generated a 784-bp fragment from a normal chromosome and a 440-bp fragment from a chromosome with the VLDLR deletion. Affected individuals are identified by blackened symbols, unaffected family members by unblackened symbols, and carriers by a small blackened circle within the symbol. The 440-bp product was identified in the patients with DES-H, the 784-bp and 440-bp products in carriers, and the 784-bp product in noncarriers. D, Map of the deleted region. The deleted segment was 199 kb in size and contained the entire VLDLR gene and part of LOC401491. KCNV2, which is proximal to VLDLR, was not contained within the deletion.
The 199-kb deletion contained the entire VLDLR gene and part of a hypothetical gene, LOC401491 (GenBank [accession number AK092343]) (fig. 3D). The latter gene is supported by the presence of a 3,167-bp transcript, which is widely expressed and is predicted to encode a 109-aa protein. The entire coding region of this gene falls within the DES-H deletion. The sequence shows 80% homology to the ALU6 human Alu subfamily. Given the extent of the deletion, it is also possible that the expression of flanking genes is also affected because of the absence of intergenic regulatory elements. Comparison of the clinical findings of the patients with DES-H with the Vldlr-deficient mouse model (see below) supports homozygous loss of VLDLR as the major contributor to the DES-H phenotype.
VLDLR is part of the reelin signaling pathway, which guides neuroblast migration in the developing cerebral cortex and cerebellum (D’Arcangelo et al. 1995; Rice and Curran 2001; Tissir and Goffinet 2003). In the cortex, Cajal-Retzius cells in the preplate secrete reelin to guide the cells of the cortical plate. In the cerebellum, the granule cells secrete reelin, which guides migration of Purkinje cells. VLDLR and apolipoprotein E receptor type 2 (ApoER2) are transmembrane receptors for reelin. With reelin binding, the cytoplasmic adaptor protein disabled-1 (Dab1) docks to the NPxY sequence in the intracellular region of VLDLR and ApoER2 and becomes phosphorylated (Howell et al. 1997; Trommsdorff et al. 1999), which triggers an intracellular signaling cascade that allows neuroblasts to complete migration and adopt their ultimate positions in laminar structures in the CNS.
Much of what is known about the reelin pathway comes from the study of a number of mouse models (reviewed by Lambert de Rouvroit and Goffinet [1998] and Tissir and Goffinet [2003]). In mice, deficiency of reelin results in the reeler phenotype (D’Arcangelo et al. 1995), which is characterized by impaired motor coordination, tremors, and ataxia. Reeler-like mice result from mutations in Dab1 (Howell et al. 1997) and double mutations in the lipoprotein receptors, Vldlr and ApoER2 (Trommsdorff et al. 1999). In reeler and reeler-like mice, the preplate forms normally, with migration of cortical neurons from the germinative zones. However, as the neurons approach their destination, they fail to recognize their proper location and orientation. Later-migrating neurons fail to split the preplate into the marginal zone and subplate and instead form layers below the preplate with a reverse “outside-in” pattern compared with normal cortex. Reelin is also involved in the radial glial scaffold in the hippocampus. In reeler mutants, the distinct “reeling” gait is caused by profound hypoplasia of the cerebellum, in which the normal cerebellar folia are missing. Vldlr knockout mice with intact ApoER2 appear neurologically normal (Frykman et al. 1995). However, the Vldlr-deficient mouse cerebellum is small, with reduced foliation and heterotopic Purkinje cells (Trommsdorff et al. 1999). Neurons in the Vldlr-deficient murine cortex fail to distribute normally after reaching their assigned layer. The clinical and pathological features of the Vldlr-deficient mouse are less severe than those seen in the reeler mouse.
Like reeler, mutations in the human orthologue reelin (RELN) cause significant disruption of neuroblast migration and architecture in the cerebellum and cerebral cortex. A syndrome of autosomal recessive lissencephaly with cerebellar hypoplasia (LCH) has been shown to be caused by mutations in RELN in two families (Hong et al. 2000). The phenotype in these patients was characterized by hypotonia, ataxia, and developmental delay, with lack of unsupported sitting and profound mental retardation with little or no language development. Seizures and congenital lymphedema were also present. In contrast, patients with DES-H have moderate motor delays, with most eventually walking unsupported, and predominantly moderate-to-severe mental retardation (Schurig et al. 1981; Glass et al., in press). Neuroimaging of patients with a RELN mutation demonstrated moderate lissencephaly, with an anterior greater than posterior gradient, a malformed hippocampus, and profound cerebellar hypoplasia with complete absence of detectable cerebellar folia (Hong et al. 2000). In DES-H, the primary abnormality found on neuroimaging was marked hypoplasia of the cerebellar hemispheres and inferior vermis (fig. 1). A simplification and thickening of the cerebral cortex was also present, but this was a much milder feature in comparison with that of patients with a RELN mutation. In DES-H, the hippocampi appeared to undergo normal developmental rotation compared with the fetal configuration of the hippocampi in the patients with a RELN mutation. Deficiency of VLDLR in both mice and humans is associated with a less-severe phenotype than that seen with deficiency of reelin.
Six subtypes of LCH have been defined (Ross et al. 2001). In this group of syndromes, the term “lissencephaly” refers to both the classic pattern of pachygyria/agyria, with marked thickening of the gray matter, and to less-severe phenotypes, ranging from mild pachygyria to simplification of the cortical gyral pattern with near-normal gray-matter thickness. Cerebellar manifestations range from relatively preserved hemispheres to marked cerebellar hypoplasia with foliation defects. The human phenotype associated with mutations in RELN has been classified as “LCHb” (Ross et al. 2001). The malformations seen in DES-H also fall within the LCH spectrum, with mild cerebral gyral simplification and more-marked cerebellar hypoplasia.
VLDLR also functions as a peripheral lipoprotein receptor for VLDL triglycerides in mice. The low density lipoprotein receptor (LDLR) can mask the effect of VLDLR on lipoprotein metabolism (Tacken et al. 2000; reviewed by Takahashi et al. [2004]). Vldlr-deficient mice are protected from obesity after exposure to a high-fat, high-calorie (HFC) diet and have increased plasma triglycerides after HFC feeding (Goudriaan et al. 2001). Of the patients with DES-H, 50% had a BMI <18.5 (underweight), which suggests that there may be some protection from obesity.
In summary, a homozygous deletion of the very low density lipoprotein receptor causes an autosomal recessive syndrome of cerebellar hypoplasia in the Hutterite population. We propose that this condition now be referred to as “VLDLR-associated cerebellar hypoplasia” (VLDLR-CH) to recognize the molecular pathogenesis. To our knowledge, this condition represents the first human lipoprotein receptor malformation syndrome and the second human disease associated with a reelin pathway defect. The clinical manifestations and MRI findings observed in this group of patients provide insight into the effects of homozygous loss of VLDLR on the development of the human CNS. It will be important to determine whether mutations in VLDLR and/or other genes in the reelin pathway cause cerebellar hypoplasia in patients from other populations.
Acknowledgments
We thank all of the Hutterite families for their active participation in this research. We appreciate the efforts and support of physicians Coleen Adams and Karen Barlow and genetic counselors Carol Farr, Jackie Morris, and Caroline Powell during the clinical characterization phase of this research. We thank Drs. Richard Hawkes, Harvey Sarnat, N. Torben Bech-Hansen, A. Micheil Innes, and Kenneth Morgan for critical review of the manuscript. This work was supported by a short-term project grant from the University of Calgary, an Alberta Children’s Hospital Foundation grant, and the Canadian Genetic Diseases Network (Networks of Centres of Excellence Program). S.F. was supported by a summer studentship from the Alberta Heritage Foundation for Medical Research. Drs. McLeod and Parboosingh contributed equally to this work, as senior investigators.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for KCNV2 [accession number AL354723], SMARCA2 [accession number AL138755], LOC401491 [accession number AK092343], and reference genomic sequence [accession number NT_008413])
- Human Genome Resources, http://www.ncbi.nlm.nih.gov/ (for Généthon, deCode, and Marshfield genetic maps)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DES) [PubMed]
- UniSTS, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search=unists (for PMC303366P11 and STSs used in walking strategy)
- University of Pittsburgh Department of Human Genetics, http://www.hgen.pitt.edu/ (for Mega2 version 3.0)
References
- Antonarakis SE, Krawczak M, Cooper DN (2001) The nature and mechanisms of human gene mutation. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 343–377 [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T (1995) A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374:719–723 10.1038/374719a0 [DOI] [PubMed] [Google Scholar]
- Frykman PK, Brown MS, Yamamoto T, Goldstein JL, Herz J (1995) Normal plasma lipoproteins and fertility in gene-targeted mice homozygous for a disruption in the gene encoding very low density lipoprotein receptor. Proc Natl Acad Sci USA 92:8453–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass HC, Boycott KM, Adams C, Barlow K, Scott JN, Chudley AE, Fujiwara T, Morgan K, Wirrell E, McLeod DR. Autosomal recessive cerebellar hypoplasia in the Hutterite population: a syndrome of nonprogressive cerebellar ataxia with mental retardation. Dev Med Child Neurol (in press) [DOI] [PubMed] [Google Scholar]
- Goudriaan JR, Tacken PJ, Dahlmans VE, Gijbels MJ, van Dijk KW, Havekes LM, Jong MC (2001) Protection from obesity in mice lacking the VLDL receptor. Arterioscler Thromb Vasc Biol 21:1488–1493 [DOI] [PubMed] [Google Scholar]
- Hagberg B, Sanner G, Steen M (1972) The dysequilibrium syndrome in cerebral palsy: clinical aspects and treatment. Acta Paediat Scand 61:1–63 [PubMed] [Google Scholar]
- Hong SE, Shugart YY, Huang DT, Shahwan SA, Grant PE, Hourihane JO, Martin ND, Walsh CA (2000) Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat Genet 26:93–96 10.1038/79246 [DOI] [PubMed] [Google Scholar]
- Hostetler J (1985) History and relevance of the Hutterite population for genetic studies. Am J Med Genet 22:453–462 10.1002/ajmg.1320220303 [DOI] [PubMed] [Google Scholar]
- Howell BW, Hawkes R, Soriano P, Cooper JA (1997) Neuronal position in the developing brain is regulated by mouse disabled-1. Nature 389:733–737 10.1038/39607 [DOI] [PubMed] [Google Scholar]
- Innes AM, Wrogemann K, Zelinski T, Coghlan G, Maendel S, Maendel M, Evans J, Greenberg CR (1999) Delivery of genetic services to the Manitoba Hutterites in the molecular era. Am J Hum Genet Suppl 65:A216 [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lambert de Rouvroit C, Goffinet AM (1998) The reeler mouse as a model of brain development. Adv Anat Embryol Cell Biol 150:1–106 [PubMed] [Google Scholar]
- Nimgaonkar VL, Fujiwara TM, Dutta M, Wood J, Gentry K, Maendel S, Morgan K, Eaton J (2000) Low prevalence of psychoses among the Hutterites, an isolated religious community. Am J Psychiatry 157:1065–1070 10.1176/appi.ajp.157.7.1065 [DOI] [PubMed] [Google Scholar]
- Pallister P, Opitz J (1986) Disequilibrium syndrome in Montana Hutterites. Am J Med Genet 22:567–569 10.1002/ajmg.1320220314 [DOI] [PubMed] [Google Scholar]
- Rice DS, Curran T (2001) Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci 24:1005–1039 10.1146/annurev.neuro.24.1.1005 [DOI] [PubMed] [Google Scholar]
- Ross ME, Swanson K, Dobyns WB (2001) Lissencephaly with cerebellar hypoplasia (LCH): a heterogeneous group of cortical malformations. Neuropediatrics 32:256–263 10.1055/s-2001-19120 [DOI] [PubMed] [Google Scholar]
- Sakai J, Hoshino A, Takahashi S, Miura Y, Ishii H, Suzuki H, Kawarabayasi Y, Yamamoto T (1994) Structure, chromosome location, and expression of the human very low density lipoprotein receptor gene. J Biol Chem 269:2173–2182 [PubMed] [Google Scholar]
- Schurig V, Van Orman A, Bowen P (1981) Nonprogressive cerebellar disorder with mental retardation and autosomal recessive inheritance in Hutterites. Am J Med Genet 9:43–53 10.1002/ajmg.1320090109 [DOI] [PubMed] [Google Scholar]
- Tacken PJ, Teusink B, Jong MC, Harats D, Havekes LM, van Dijk KW, Hofker MH (2000) LDL receptor deficiency unmasks altered VLDL triglyceride metabolism in VLDL receptor transgenic and knockout mice. J Lipid Res 41:2055–2062 [PubMed] [Google Scholar]
- Takahashi S, Sakai J, Fujino T, Hattori H, Zenimaru Y, Suzuki J, Miyamori I, Yamamoto TT (2004) The very low-density lipoprotein (VLDL) receptor: characterization and functions as a peripheral lipoprotein receptor. J Atheroscler Thromb 11:200–208 [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM (2003) Reelin and brain development. Nat Rev Neurosci 4:496–505 10.1038/nrn1113 [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J (1999) Reeler/disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97:689–701 10.1016/S0092-8674(00)80782-5 [DOI] [PubMed] [Google Scholar]