Abstract
Spondyloepiphyseal dysplasia (SED) encompasses a heterogeneous group of disorders characterized by shortening of the trunk and limbs. The autosomal dominant SED type Kimberley (SEDK) is associated with premature degenerative arthropathy and has been previously mapped in a multigenerational family to a novel locus on 15q26.1. This locus contains the gene AGC1, which encodes aggrecan, the core protein of the most abundant proteoglycan of cartilage. We screened AGC1 for mutations and identified a single–base-pair insertion, within the variable repeat region of exon 12 in affected individuals from the family with SEDK, that introduces a frameshift of 212 amino acids, including 22 cysteine residues, followed by a premature stop codon. This is the first identification of an AGC1 mutation causing a human disorder. This finding extends the spectrum of mutated genes that may cause SED and thus will aid in the molecular delineation of this complex group of conditions.
The spondyloepiphyseal dysplasia (SED) phenotype encompasses a heterogeneous group of chondrodysplasias with radiographic abnormalities of the vertebral bodies and proximal epiphyses (Horton 2002). The autosomal dominant SED congenita (MIM 183900) and the X-linked recessive SED tarda (MIM 313400) are specific subtypes that are caused by mutations in the type II collagen gene (COL2A1 [MIM 120140]) and the sedlin gene (SEDL [MIM 300202]), respectively. Autosomal dominant SED type Kimberley (SEDK [MIM 608361]) is a mild autosomal dominant form of SED that was identified in a multigenerational South African family of U.K. white descent. The phenotype of SEDK is short stature (<5th percentile for age), stocky build, and early-onset osteoarthritis of the weight-bearing joints. Radiographic features are flattened vertebral bodies, with sclerosis and end-plate irregularity, and flattened femoral epiphyses (Anderson et al. 1990). Previous linkage studies excluded linkage to COL2A1 (Anderson et al. 1990), and a genome screen identified a novel locus for this disorder on chromosome 15q26.1 (Eyre et al. 2002). The candidate gene AGC1 (MIM 155760) was initially excluded from the linked region, but revised genotyping performed using a SNP within exon 18 (NCBI dbSNP accession number rs2280467), coupled with the variable repeat region (VNTR) within exon 12 (Eyre et al. 2002) and a SNP within intron 1 (NCBI dbSNP accession number rs4932200), confirmed that the gene is within the critical region (fig. 1) (Eyre et al. 2005).
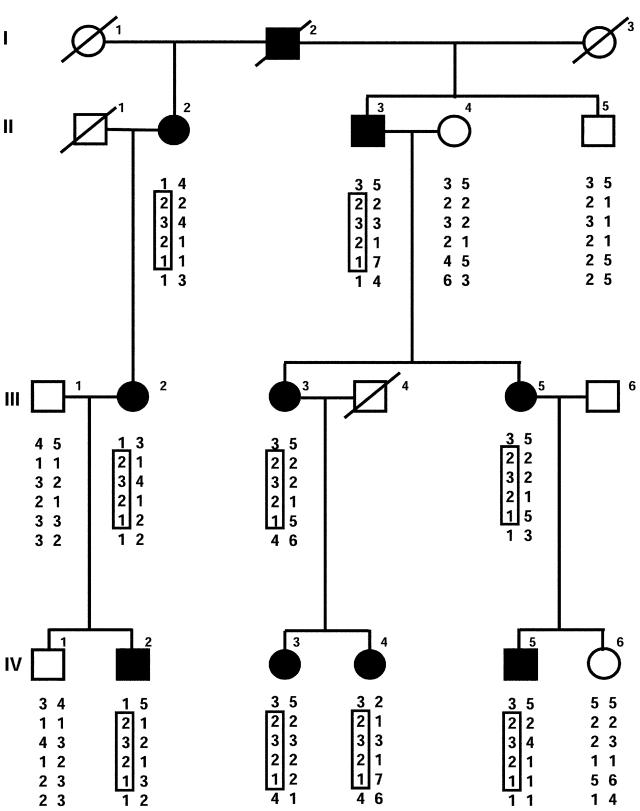
Figure 1.
Pedigree of family with SEDK. Blackened circles and squares represent affected females and males, respectively. Haplotypes for subjects who were genotyped are given under the symbols. The disease-linked haplotype is boxed. Genotypes of microsatellite markers are as published previously (Eyre et al. 2002). The marker order (top to bottom) is D15S205, AGC1 intron 1 SNP, AGC1 VNTR, AGC1 exon 18 SNP, D15S116, and D15S1004.
The AGC1 gene was considered to be a strong candidate gene for SEDK because it encodes the core protein of aggrecan, a major component of the cartilage extracellular matrix (ECM), and because AGC1 mutations have previously been found to cause forms of chondrodysplasia in the mouse and chick. The lethal mouse disorder cartilage matrix deficiency (cmd), characterized by cleft palate and short limbs, tail, and snout, is caused by homozygosity of a 7-bp deletion that results in a frameshift and a premature termination of the aggrecan core protein (Watanabe et al. 1994). Heterozygotes for the cmd mutation appear normal at birth but, within 28 d after birth, are dwarfed and have age-related spinal misalignment that leads to premature death (Watanabe et al. 1997). The phenotype of the cmd-Bc mouse is similar to that of cmd and results from the near-complete deletion of the murine aggrecan gene (Krueger et al. 1999). Homozygosity for a premature stop codon in the chick aggrecan gene causes nanomelia, a lethal disorder of cartilage development in which embryos have shortened and malformed limbs (Li et al. 1993). Heterozygosity for the nanomelia mutation results in a shortening of the long bones of the legs (Landauer 1965).
The 19-exon structure of the human AGC1 gene has been described in detail elsewhere (Doege et al. 1991; Valhmu et al. 1995). Briefly, exon 1 is noncoding and is separated from the remaining coding exons by a 13-kb intron. Exons 2–19 encode the aggrecan core protein, which comprises a central glycosaminoglycan attachment domain flanked by three globular domains. The amino terminal G1 (hyaluronan binding) and G2 globular domains plus an interglobular domain are encoded by exons 3–10. The keratan sulfate attachment domain is encoded by exon 11 and part of exon 12. The remainder of exon 12 encodes the chondroitin sulfate attachment domains (CS1 and CS2), including a VNTR of 13–33 repeats of 57 nt within the CS1 domain (Doege et al. 1997). The G3 domain, containing the C-lectin domain, has a role in intracellular trafficking and in secretion from the cell and is encoded by alternatively spliced exons from exons 13 to 18 (Domowicz et al. 2000) (fig. 2).
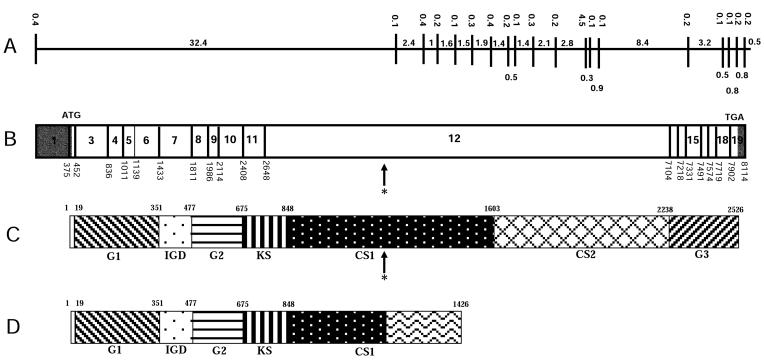
Figure 2.
Structure of the human AGC1 gene. A, Genomic organization of the AGC1 gene. Introns are represented by horizontal lines, exons are shown as vertical lines. Intron and exon sizes are shown in kb. B, cDNA structure of the AGC1 gene, showing the location of the exons. Exon sizes are shown in bp. The translation start and stop codons are labeled with ATG and TGA, respectively. The untranslated regions are shaded. The asterisk (*) marks the single-nucleotide (C) insertion at nucleotide 4015. C, Structure of the wild-type AGC1 protein. The location of each domain is marked by the amino acid numbers, with the initiation Met as amino acid 1. CS1 and CS2 = chondroitin sulfate attachment domains 1 and 2, respectively; G1, G2, and G3 = globular domains 1, 2, and 3, respectively; IGD = interglobular domain; KS = keratan sulfate attachment domain. The position of the insertion at amino acid 1214 is indicated by an asterisk (*). D, Structure of the mutant AGC1 protein identified in the SEDK-affected family. The 212-aa missense addition at the C-terminal is represented by a wave pattern.
Affected individuals of the family with SEDK were screened by PCR followed by direct sequencing (PCR primer sequences and conditions are available upon request) for mutations within the 19 exons (but excluding the VNTR region) and for splice-acceptor and/or splice-donor sequences of AGC1. The analysis of these exons did not identify any potential mutations. The linked VNTR allele (allele 3) in the family with SEDK (fig. 1) could be amplified as a 1.5-kb PCR fragment, but it was intractable to direct sequencing, because of its repeating sequence. Therefore, the VNTR region was first amplified, as described elsewhere (Doege et al. 1997), from genomic DNA of family members III-5, IV-4, and IV-5, who had genotypes of 2/3, 3/3, and 3/4, respectively (fig. 1). The PCR products were cloned into the pGEM-T Easy vector (Promega), and the clones were analyzed to determine which allele they contained. Clones containing the linked allele 3 from III-5 (9 clones) and IV-5 (3 clones) were distinguished from those with the unlinked alleles 2 and 4 on the basis of size, by separation on a 0.6% (weight/volume) agarose gel. For individual IV-4 (homozygous for allele 3), 10 clones were partially sequenced from both the 5′ and the 3′ ends, and the linked allele was distinguished from the unlinked allele on the basis of an A sequence variant (7 clones) versus a G sequence variant (3 clones) in the third repeat from the 3′ end (fig. 3).
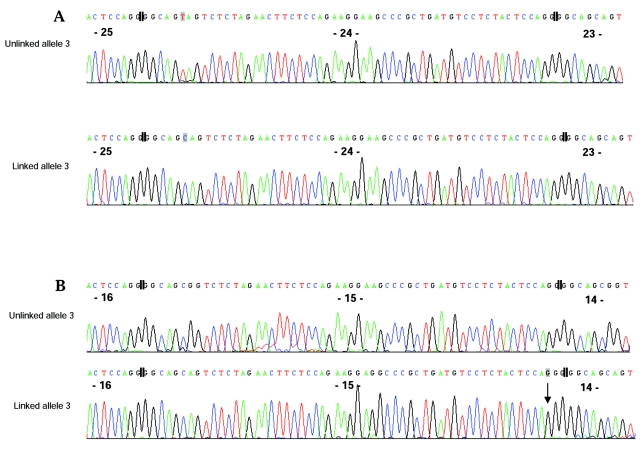
Figure 3.
Sequence of AGC1 VNTR clones from individual IV-4, shown in reverse orientation. The division between repeats is indicated by double vertical lines, and repeat numbers are shown in bold type. Sequence variations are indicated by a shaded box around the nucleotide. A, The sequence trace shows the SNP used to distinguish the linked from the unlinked VNTR allele 3 clones. B, The sequence trace shows a 1-bp insertion of a C in the 15th repeat (arrow) in the linked but not in the unlinked allele 3.
Deletion sets were generated from the 3′ ends of the two cloned alleles from each individual by use of the Erase-a-base system protocol (Promega). Briefly, cloned VNTR alleles in the sense orientation were digested with the restriction enzymes SpeI and NsiI, which have sites within the pGEM-T Easy multiple-cloning site but not within the VNTR, followed by timed exonuclease III digestion, S1 nuclease digestion, and religation. Deleted subclones representing each allele were sequenced from the 3′ end of the inserted sequence by use of the SP6 vector primer. Sequence variation between repeats allowed the deleted clones for each allele to be aligned. From this, we determined that allele 3 contained 27 repeats of the 57-nt VNTR sequence. In the 15th repeat of the linked allele 3 from III-5, IV-4, and IV-5, a unique insertion of a single C was identified (fig. 3). The predicted effect of this mutation would be a frameshift, altering the sequence of the 212 aa that follow (including the introduction of 21 cysteine residues), followed by a premature stop codon within the CS1 domain (fig. 4).
Figure 4.
Predicted translation of the normal and mutant VNTR allele 3 clones. Cysteine residues are boxed. The sequence starts at amino acid 944 of the aggrecan core protein sequence.
To confirm that the insertion was not the result of a sequencing artifact, Kozak sequences were inserted into the cloned alleles of IV-4 and IV-5, and the clones were used as templates for in vitro transcription by the RiboMax large scale RNA T7 production kit (Promega), followed by in vitro translation by the FlexiRabbit Reticulocyte Lysate System in the presence of radiolabeled 35S-cysteine or 14C–amino acid mix. The cloned linked and unlinked VNTR alleles in the sense orientation were linearized with ApaI and SacII. The two oligonucleotide primers, 5′-CAGCCACCATGGCGC-3′ and 5′-GCCATGGTGGCTGGGCC-3′, were synthesized and annealed together and were then ligated into the linearized plasmids, thereby introducing a Kozak sequence plus an inframe ATG 5′ of the AGC1 VNTR sequence. In vitro transcription and translation were performed, and the translation products were separated by SDS-PAGE (4%–12%) and were visualized by autoradiography.
The 14C–amino acid mix labeled both the normal and the mutant translation products (fig. 5A), whereas, as expected from the predicted frameshift, 35S-cysteine labeled only the mutant translation products (fig. 5B). The faster migration of the mutant, compared with that of the normal translation product, cannot be attributed to their 1.4-kD difference in estimated molecular weight but is presumably the result of the increased hydrophobicity and altered proline content of the mutant protein, which are caused by the frameshift (as discussed by Iakoucheva et al. [2001]).
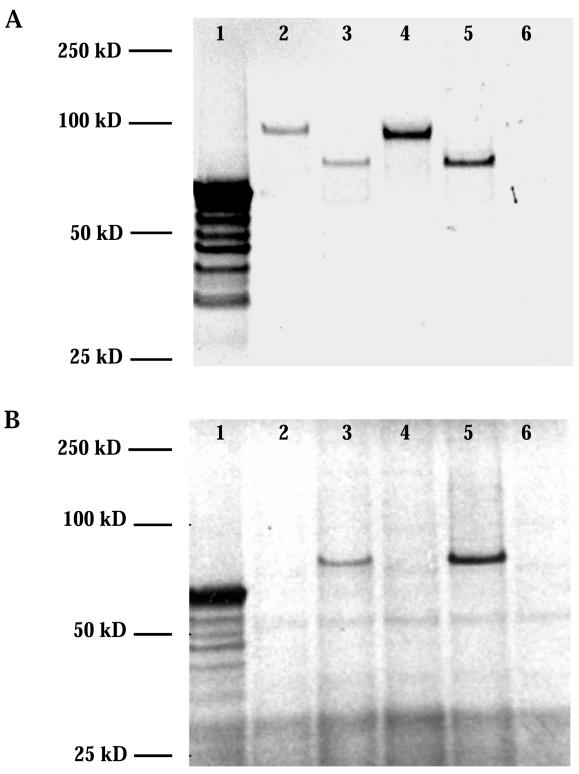
Figure 5.
Products of the in vitro transcription and translation of normal and mutant VNTR allele 3 clones after separation by SDS-PAGE under reducing conditions. A, Translation in the presence of radiolabled 14C–amino acid mix. Lane 1, luciferase control (Promega); lane 2, individual IV-4 normal allele 3; lane 3, individual IV-4 mutant allele 3; lane 4, individual IV-5 normal allele 4; lane 5, individual IV-5 mutant allele 3; lane 6, no-RNA template control. B, Translation in the presence of 35S-cysteine. Lane 1, luciferase control (Promega); lane 2, individual IV-4 normal allele 3; lane 3, individual IV-4 mutant allele 3; lane 4, individual IV-5 normal allele 4; lane 5, individual IV-5 normal allele 3; lane 6, no-RNA template control.
Amplification refractory mutation system (ARMS) was used to confirm the inheritance of the mutation by affected individuals. Allele-specific amplification was performed from genomic DNA with the use of the wild-type–specific sense primer 5′-GAAGTTCTAGAGACTGCTGCCCC-3′ and the destabilized mutant-specific sense primer 5′-GAAGTTCTAGAGACTGCTGCCACC-3′ in separate PCR reactions, with each sense primer paired with the VNTR generic antisense primer 5′-AGGTCCCCTACCGCAGAGGTAGAA-3′. A 50-μl PCR reaction included 20 pmol of either sense primer, 20 pmol of the generic antisense primer, 0.2 mM dNTPs, 2.5 U of Taq DNA polymerase (Bioline), 10 × PCR buffer, and 2 mM MgCl2. The annealing temperature was 65°C, and amplification was performed for 29 cycles on a Cetus Thermal Cycler (PerkinElmer). All affected individuals of generation IV were shown to have the mutated sequence on the linked allele 3 but not on the unlinked alleles (fig. 6). Similar screening of the alleles from 108 unrelated U.K. white control individuals did not identify this mutation.
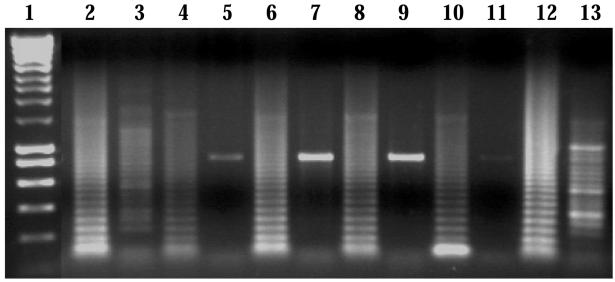
Figure 6.
ARMS products after electrophoresis on a 1% (weight/volume) agarose gel. Lane 1, 1-kb ladder (Bioline). Lanes 2, 4, 6, 8, 10, and 12, the products obtained when amplified with the normal oligonucleotide. Lanes 3, 5, 7, 9, 11, and 13, the products obtained when amplified with the destabilized mutant oligonucleotide. DNA was amplified from individual IV-1 (lanes 2 and 3), individual IV-2 (lanes 4 and 5), individual IV-3 (lanes 6 and 7), individual IV-4 (lanes 8 and 9), individual IV-5 (lanes 10 and 11), and individual IV-6 (lanes 12 and 13).
The cosegregation of the molecular defect with SEDK-affected members of the family and its absence in 216 alleles from a control population support the idea that heterozygosity for the 1-bp insertion in AGC1 is the cause of SED in this kindred. The position of the mutation predicts the synthesis of a truncated protein from the mutant allele that is ∼60% of the normal size. The truncated protein would lack half of the CS1 domain, the complete CS2 domain, and the G3 domain but would include novel sequence of 212 aa (fig. 2). In comparison, the nanomelia mutation is the transversion of G→T in a codon for Glu (GAA) that introduces a premature stop codon (but no frameshift) in the chick-specific, 20-aa repeat region of the CS2 domain; hence, the truncated protein lacks part of the CS2 domain and the G3 domain. The nanomelia mutation causes a reduction in the steady-state levels of aggrecan mRNA to ∼6% of normal levels (Stirpe et al. 1987), but the residual mRNA is translated. The truncated protein is retained and accumulates in the endoplasmic reticulum (ER) (Li et al. 1993; Vertel et al. 1993) but is not secreted into the ECM. The cmd mutation is similar to the nanomelia mutation, in that it causes a reduction in the steady-state levels of aggrecan mRNA to 41% of normal levels in the homozygote and to 81% of normal levels in the heterozygote (Watanabe et al. 1997). In contrast to the nanomelia mutation, however, the potentially truncated 32-kD cmd polypeptide has not been detected in the ER but, as expected, is absent from the ECM. The cmd mutation thus shows that the reduction or absence of aggrecan in the ECM is sufficient to cause chondrodysplasia. What is not clear, though, is whether the retention of the truncated protein by nanomelic chondrocytes impairs their function and thereby exacerbates the phenotype of the reduction (in the heterozygote) or absence (in the homozygote) of aggrecan in the ECM. The cmd and nanomelia mutations therefore predict that the SEDK AGC1 mutation may result in reduced mRNA from the mutant allele and that the truncated translation product may be retained and may accumulate within the ER. Further studies are required to establish whether the truncated protein, with its additional novel residues, is synthesized and, if so, to determine both its effect on other proteins within the ER and on chondrocyte function and its contribution to the SEDK phenotype.
This is the first report of an AGC1 mutation causing a heritable form of human chondrodysplasia. The AGC1 gene is thus a strong candidate for as-yet uncharacterized forms of SED. Genotype-phenotype correlations will inform studies aimed at understanding the molecular consequences of specific AGC1 mutations on chondrocyte function and skeletogenesis.
Acknowledgments
We thank Mike Briggs, Karl Kadler, Kieran Mellody, and Tobias Starborg, for helpful advice. This work was supported by the Biotechnology and Biological Sciences Research Council, United Kingdom; the Arthritis Research Campaign, United Kingdom; and The Wellcome Trust, United Kingdom.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/index.html (for accession numbers rs2280467 and rs4932200)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SED congenita, X-linked SED tarda, COL2A1, SEDL, SEDK, and AGC1)
References
- Anderson IJ, Tsipouras P, Scher C, Ramesar RS, Martell RW, Beighton P (1990) Spondyloepiphyseal dysplasia, mild autosomal dominant type is not due to primary defects of type II collagen. Am J Med Genet 37:272–276 10.1002/ajmg.1320370223 [DOI] [PubMed] [Google Scholar]
- Doege KJ, Coulter SN, Meek LM, Maslen K, Wood JG (1997) A human-specific polymorphism in the coding region of the aggrecan gene: variable number of tandem repeats produce a range of core protein sizes in the general population. J Biol Chem 272:13974–13979 10.1074/jbc.272.21.13974 [DOI] [PubMed] [Google Scholar]
- Doege KJ, Sasaki M, Kimura T, Yamada Y (1991) Complete coding sequence and deduced primary structure of the human cartilage large aggregating proteoglycan, aggrecan: human-specific repeats, and additional alternatively spliced forms. J Biol Chem 266:894–902 [PubMed] [Google Scholar]
- Domowicz MS, Pirok EW III, Novak TE, Schwartz NB (2000) Role of the C-terminal G3 domain in sorting and secretion of aggrecan core protein and ubiquitin-mediated degradation of accumulated mutant precursors. J Biol Chem 275:35098–35105 10.1074/jbc.275.45.35098 [DOI] [PubMed] [Google Scholar]
- Eyre S, Roby P, Wolstencroft K, Spreckley K, Aspinwall R, Bayoumi R, Al-Gazali L, Ramesar R, Beighton P, Gleghorn L, Wallis G (2005) Identification of a locus for a form of spondyloepiphyseal dysplasia on chromosome 15q26.1: exclusion of aggrecan as a candidate gene. J Med Genet 42:e34 [PMC free article] [PubMed] [Google Scholar]
- Eyre S, Roby P, Wolstencroft K, Spreckley K, Aspinwall R, Bayoumi R, Al-Gazali L, Ramesar R, Beighton P, Wallis G (2002) Identification of a locus for a form of spondyloepiphyseal dysplasia on chromosome 15q26.1: exclusion of aggrecan as a candidate gene. J Med Genet 39:634–638 10.1136/jmg.39.9.634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton WA (2002) Chondrodysplasias: disorders of cartilage matrix proteins in connective tissue and its heritable disorders. In: Royce PM, Steinmann B (eds) Connective tissue and its heritable disorders. Wiley-Liss, New York, pp 909–937 [Google Scholar]
- Iakoucheva LM, Kimzey AL, Masselon CD, Smith RD, Dunker AK, Ackerman EJ (2001) Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci 10:1353–1362 10.1110/ps.40101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger RC Jr, Kurima K, Schwartz NB (1999) Completion of the mouse aggrecan gene structure and identification of the defect in the cmd-Bc mouse as a near complete deletion of the murine aggrecan gene. Mamm Genome 10:1119–1125 10.1007/s003359901176 [DOI] [PubMed] [Google Scholar]
- Landauer W (1965) Nanomelia, a lethal mutation of the fowl. J Hered 56:131–138 [DOI] [PubMed] [Google Scholar]
- Li H, Schwartz NB, Vertel BM (1993) cDNA cloning of chick cartilage chondroitin sulfate (aggrecan) core protein and identification of a stop codon in the aggrecan gene associated with the chondrodystrophy, nanomelia. J Biol Chem 268:23504–23511 [PubMed] [Google Scholar]
- Stirpe NS, Argraves WS, Goetinck PF (1987) Chondrocytes from the cartilage proteoglycan-deficient mutant, nanomelia, synthesize greatly reduced levels of the proteoglycan core protein transcript. Dev Biol 124:77–81 10.1016/0012-1606(87)90461-1 [DOI] [PubMed] [Google Scholar]
- Valhmu WB, Palmer GD, Rivers PA, Ebara S, Cheng JF, Fischer S, Ratcliffe A (1995) Structure of the human aggrecan gene: exon-intron organization and association with the protein domains. Biochem J 309:535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertel BM, Walters LM, Grier B, Maine N, Goetinck PF (1993) Nanomelic chondrocytes synthesize, but fail to translocate, a truncated aggrecan precursor. J Cell Sci 104:939–948 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Kimata K, Line S, Strong D, Gao LY, Kozak CA, Yamada Y (1994) Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nat Genet 7:154–157 10.1038/ng0694-154 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Nakata K, Kimata K, Nakanishi I, Yamada Y (1997) Dwarfism and age-associated spinal degeneration of heterozygote cmd mice defective in aggrecan. Proc Natl Acad Sci USA 94:6943–6947 10.1073/pnas.94.13.6943 [DOI] [PMC free article] [PubMed] [Google Scholar]