Abstract
Intracellular bacteria of the genus Chlamydia cause numerous typically chronic diseases, frequently with debilitating sequelae. Genetic determinants of disease susceptibility after infection with Chlamydia bacteria are unknown. C57BL/6 mice develop severe pneumonia and poor immunity against Chlamydia after moderate respiratory infection whereas BALB/c mice are protected from disease and develop vigorous Th1 immunity. Here we show that infected C57BL/6 macrophages release more NO synthesized by NO synthase 2 (NOS2) than BALB/c macrophages and have lower mRNA concentrations of arginase II, a competitor of NOS2 for the common substrate, l-arginine. Reduction, but not elimination, of NO production by incomplete inhibition of NOS2 abolishes susceptibility of C57BL/6 mice to Chlamydia-induced disease. Thus, the quantity of NO released by infected macrophages is the effector mechanism that regulates between pathogenic and protective responses to chlamydial infection, and genes controlling NO production determine susceptibility to chlamydial disease.
Intracellular bacteria of the genus Chlamydia are among the most common human pathogens and are the leading bacterial cause of sexually transmitted disease and preventable blindness (1). Over the last decade, Chlamydia pneumoniae has been identified in a high proportion of atherosclerotic lesions, suggesting a role for chlamydial infection in coronary heart disease (2, 3), and recently has been detected in brain lesions of patients with Alzheimer's disease (4). In early trials, vaccination against Chlamydia trachomatis surprisingly increased rate and severity of the naturally acquired chlamydial eye disease, trachoma, whereas in other trials the rate of disease declined but the severity increased (5, 6). The high seroprevalence of C. pneumoniae infection is not accompanied by equally high organism isolation or disease rates (7). Collectively, these data support the notion that some individuals react with increased sensitivity to repeated exposure to chlamydial agents whereas others develop a protective response. Similarly, animal studies indicate that genetic determinants of the host response to Chlamydia spp. play a decisive role in the outcome of chlamydial infection (8, 9).
In our laboratory, we found that the susceptibility of the BALB/c and C57BL/6 inbred mouse strains to chlamydial disease differed widely in a model of lung infection with a virulent ruminant abortifacient strain of Chlamydia psittaci (10). At the peak of the early inflammatory response on days 3–5 after intranasal (i.n.) inoculation of high doses of C. psittaci [0.5–1 × 107 inclusion-forming units (IFU)], naïve C57BL/6 mice appeared healthy whereas BALB/c mice showed clinical signs of disease, including ruffled fur and labored respiration. C57BL/6 mice also had a higher LD50 than BALB/c mice for the 12-day period after infection (10). We attributed these effects to differences in the innate immune responses. In repeated lung infection, however, C57BL/6 mice primed by low-level i.n. infection developed severe interstitial pneumonia after secondary challenge whereas BALB/c mice were completely protected from clinical disease (10). The lung disease in C57BL/6 mice exhibited extensive interstitial infiltrates of macrophages, lymphocytes, and neutrophils characteristic of chlamydial disease (1), which resulted in profound lung weight increases over those of naïve controls. Resistant BALB/c mice had prominent peribronchiolar lymphocytic cuffs but minimal lung weight increases (10).
We further addressed this intriguing dichotomy of the disease response by using clinically more relevant, nonlethal chlamydial inocula and show here that differences in the NO release by infected macrophages are responsible for the differential susceptibility of C57BL/6 and BALB/c mice to primary and repeated chlamydial disease. Elevated NO synthesis in C57BL/6 mice provides protection against high-dose infection with virulent chlamydiae, but suppresses adaptive immunity at more moderate infection. This results in reduced elimination of chlamydiae at moderate infection, precipitates development of increased interstitial pneumonia, and delays resolution of the pneumonia in disease-susceptible C57BL/6 mice compared with resistant BALB/c mice. This proposed role of NO in regulating chlamydial disease reinforces the double-edged nature of this molecule with the potential for beneficial as well as deleterious effects (11), and that both too much and too little NO may have negative effects on protective immunity.
Materials and Methods
Chlamydial Lung Infection.
C. psittaci strain B577 (ATCC VR-656) and C. pneumoniae strain CDC/CWL-029 (ATCC VR-1310) were grown and purified as described (12). Six- to 8-week-old female mice [Harlan Sprague–Dawley; NO synthase 2 (NOS2)−/− (13), and NOS2+/+ C57BL/6 control mice, The Jackson Laboratory] were fed 19.9% protein, 1.33% l-arginine rodent chow (Harlan Teklad LM-485) and i.n. inoculated as described (12). At high-dose inoculation, some BALB/c mice developed lethal disease after 7 days. Moribund mice were killed before termination of the experiment on day 12 to obtain unbiased results. Drinking water (pH 2.5) with l-norvaline (Sigma) was prepared freshly every other day. Average daily consumption was 2.7 ml drinking water for BALB/c mice and 3.2 ml for C57BL/6 mice. Aminoguanidine (AG)-treated mice received 2 i.p. injections per day of 0.2 ml PBS-AG hemisulfate (Sigma).
Lungs or macrophages were suspended (10% wt/vol) in guanidinium isothiocyanate–Triton X-100-based RNA/DNA stabilization reagent (Roche Molecular Biochemicals). DNA was extracted by glass filter absorption (Roche Molecular Biochemicals), mRNA with biotin-oligo(dT), and streptavidin magnetic beads (Roche Molecular Biochemicals).
Data were analyzed by two-tailed Student's t test.
Immune Parameters.
Chlamydial elementary bodies were lysed by boiling in 0.065 M Tris⋅HCl (pH 7.0), 0.17 M DTT, 2% SDS, and 10% glycerol and washed by 5× ultrafiltration (Microcon YM-3, Millipore) in PBS-20 mM DTT. Twenty-four hours after footpad injection of 25 μl antigen solution containing 0.5 μg chlamydial protein, delayed type hypersensitivity (DTH) was determined by measuring the increase in footpad thickness with a spring-equipped dial thickness gauge (12). For antibody ELISA, chlamydial lysate (0.4 μg protein/well) was coated onto white microtiter plates, and antibodies in 1:100 diluted sera were detected with biotinylated anti-mouse IgG1 or IgG2a goat antiserum (Southern Biotechnology Associates) and streptavidin-peroxidase and luminol chemiluminescent substrate (Kirkegaard & Perry Laboratories).
Total splenic CD4+ T cells of mice on day 4 after infection with 8.1 × 105 IFU C. psittaci were determined by flow cytometry with FITC-anti mouse CD4 and phycoerythrin-anti-mouse CD3 (PharMingen). Total splenocytes were cultured in RPMI 1640 medium with 10% FBS (Life Technologies, Grand Island, NY) and 5 μg ovalbumin or chlamydial lysate protein/ml in white flat-bottom 96-well cell culture plates. Cells were pulsed with BrdUrd and permeabilized (Roche Molecular Biochemicals), and BrdUrd incorporated into DNA was detected by anti-BrdU–peroxidase and luminol.
Thioglycolate-elicited peritoneal phagocytes were incubated at 106 cells/ml/well in 24-well tissue culture plates in RPMI 1640 medium containing 50 μM l-arginine and 10% FBS. Adherent macrophages were infected with 107 IFU C. psittaci/ml and lysed in situ in RNA/DNA stabilization reagent, and mRNA was extracted.
Cumulative NO production was determined by the Griess reaction (R&D Systems) as total serum nitrite concentration.
Quantitative PCR (qPCR).
C. psittaci B577 fluorescence resonance energy transfer (FRET)–qPCR was performed as described (14). In the FRET-qPCR for C. pneumoniae, the C. psittaci B577 probe was replaced by the C. pneumoniae-specific probe 5′-CACATTAAGTTCTTCAACTTTAGGTTT-(fluorescein)-3′.
For real-time reverse transcription (RT)–qPCR oligo(dT)-primed mRNA was reverse-transcribed with Thermoscript reverse transcriptase (Life Technologies). RT reactions were diluted to 80 μl with 10 mM Tris⋅HCl (pH 8.5), 0.1 mM EDTA (T10E0.1) and 5-μl aliquots were used for qPCR (14). Thermal cycling was performed in a LightCycler (Roche Molecular Biochemicals) for 0 s at 95°C and 6 s at 70°C. SYBR green fluorescence was acquired after 10 s equilibration at 84–86°C, ≈1–2°C below the Tm of the respective amplicon. Mouse-specific primers were used at 1 μM: β-actin, sense, 5′-CTCCTCCTGAGCGCAAGTACTCTGTGT-3′; β-actin, antisense, 5′-GTGCACGATGGAGGGGCCGGACTCAT-3′; NOS2, sense, 5′-CACTTGGATCAGGAACCTGAAGCCC-3′; NOS2, antisense, 5′-CTTTGTGCTGGGAGTCATGGAGCCG-3′; arginase I, sense, 5′-AGCTGGGGATTGGCAAGGTGATGGA-3′; arginase I, antisense, 5′-AGCCCTGTCTTGTAAATTTCTTCTGTGA-3′; arginase II, sense, 5′-CTGTAGCTATAGTCGGAGCCCCTTTCT-3′; and arginase II, antisense, 5′-GTGGCATCCCAACCTGGAGAGC-3′.
Standard templates were prepared by agarose gel purification of PCR fragments amplified with deoxythymidine 5′-triphosphate (dTTP) instead of dUTP. TTP amplicons were confirmed by automated DNA sequencing, quantified by PicoGreen fluorescence (Molecular Probes, and diluted in T10E0.1 containing 5 μg/ml sheared plasmid (pGEM) DNA.
A FRET RT-qPCR for detection of polymorphisms at position 3083 of the murine NOS2 locus used NOS2 primers and reagents described above. Probes were used at 0.5 μM: NOS2 downstream 5′-(LightCycler Red 640)-CATCCTCATTGGGCCTGGTACG-(phosphate)-3′, NOS2 upstream 5′-TGAGGACCCCTTCCAGCCTT-(fluorescein)-3′. Thermal cycling parameters were 0 s at 95°C, 3 s at 59°C followed by fluorescence acquisition, and 10 s at 72°C.
Results
Differential Susceptibility of BALB/c and C57BL/6 Mice to Chlamydia-Induced Disease.
In this study, we evaluated induction of lung disease by a moderate i.n. inoculum (8.1 × 105 IFU C. psittaci) to reflect the typical chronic and nonlethal human chlamydial disease (1). We initially characterized the time course and type of differential responses in BALB/c and C57BL/6 mice to this infection. At the time of maximum lung disease on day 12, C57BL/6 mice had more than two times the lung weight increase of BALB/c mice (Fig. 1A), thus reversing the pattern of disease resistance at high inocula. Lungs of C57BL/6 mice had severe interstitial pneumonia with extensive mononuclear inflammatory cell infiltrates and large areas of lung consolidation whereas those of BALB/c mice appeared grossly and microscopically normal with only a few small foci of peribronchiolar mononuclear cell infiltrates. The more severe disease in C57BL/6 mice was accompanied by significantly elevated total chlamydial lung burden, reduced elimination of chlamydiae, and poor resolution of interstitial pneumonia (Fig. 1 A and B).
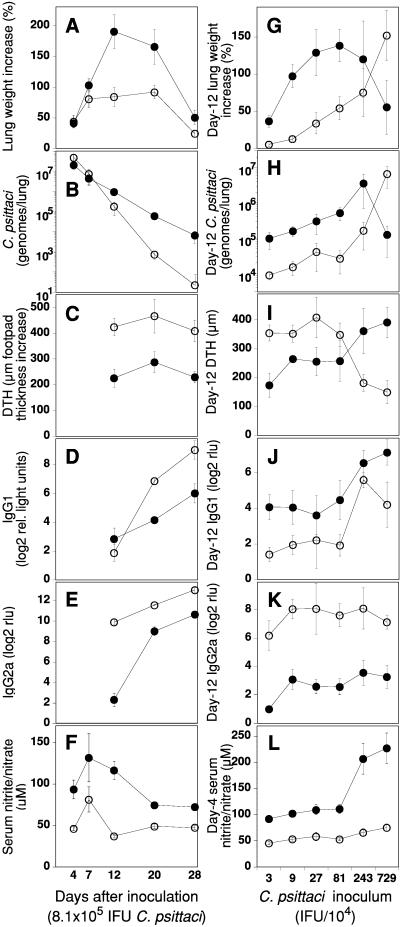
Figure 1.
Increased NO production in C57BL/6 mice after chlamydial infection is associated with enhanced disease and suppressed immunity. C57BL/6 (●) or BALB/c (○) mice were i.n. infected with 8.1 × 105 IFU C. psittaci (A–F) or a series of inocula (G–L) and killed on the indicated day after inoculation (A–F), on day 12 when interstitial pneumonia was at maximal severity (G–K), or on day 4 (L). (A and G) Time and dose dependency of chlamydial pneumonia expressed as lung weight increase over that of mock-infected mice (n = 12–18, combined data of four experiments, ± SEM). BALB/c mice have an 80% lung weight increase whereas C57BL/6 mice have a 190% increase over the average naïve lung weight of 116.3 mg. (B and H) Total chlamydial lung burden expressed as C. psittaci genomes per lung determined by FRET-qPCR. (C and I) DTH response to C. psittaci as increase in footpad thickness 24 h after antigen injection. (D, E, J, and K) IgG1 and IgG2a antibodies against C. psittaci. (F and L) Total serum nitrite/nitrate as parameter of cumulative NO production. Dependency of NO production in response to the chlamydial inoculum is shown for day 4, at the maximum of the early, innate response to the chlamydial infection.
The differential susceptibility to Chlamydia-induced interstitial pneumonia corresponded to highly significant differences in DTH and antibody responses to C. psittaci antigens (Fig. 1 C–E). C57BL/6 mice had significantly lower DTH and IgG2a responses at all time points compared with BALB/c mice, but a higher IgG1 response on day 12 (Fig. 1D). On day 12, the proliferation index of infected C57BL/6 splenocytes stimulated with C. psittaci antigen was 5.7-fold (±0.4; SEM) greater than of splenocytes stimulated with ovalbumin, whereas the index of BALB/c splenocytes was 17.0-fold (±2.1; n = 8/group; PC57BL/6:BALB/c = 0.0003) greater. Thus, BALB/c mice developed vigorous immunity against Chlamydia and rapidly eliminated organisms and disease whereas C57BL/6 mice showed suppressed immunity and exacerbated disease.
Next, we examined the patterns of day-12 disease induced by a wide range of C. psittaci dose levels. At low i.n. doses (0.3–8.1 × 105 IFU C. psittaci), C57BL/6 mice again developed substantially more severe disease and higher chlamydial lung burden than BALB/c mice (Fig. 1 G and H). At higher doses, which approached the BALB/c day-12 LD50 (4 × 106 IFU C. psittaci), C57BL/6 mice developed less severe disease than BALB/c mice, consistent with our earlier results (10). Still higher infection doses of 107 IFU or greater typically resulted in early death of many C57BL/6 mice between days 3 and 6 caused by acute pulmonic shock, whereas BALB/c mice succumbed to interstitial pneumonia between days 7 and 11 (10). Serum IgG2a concentrations in C57BL/6 mice averaged 10% of those in BALB/c mice (Fig. 1K) whereas day-12 IgG1 concentrations were marginally higher than those of BALB/c mice (Fig. 1J). The DTH response of BALB/c mice infected with the highest dose declined below that of corresponding C57BL/6 mice (Fig. 1I). These results confirmed the dichotomy of C57BL/6 disease susceptibility, with resistance at high dose and susceptibility at clinically relevant, low-dose chlamydial infection, and a reversed susceptibility pattern in BALB/c mice.
Analysis of NO Production.
To analyze the immunosuppressive mechanism, we evaluated total serum nitrite/nitrate as an indicator of cumulative NO production. Macrophage-released NO synthesized by NOS2, the high output NOS isoenzyme, acts as a regulator of lymphocyte growth and is immunosuppressive through the induction of apoptosis in activated T cells (15). C57BL/6 mice produced significantly more NO than BALB/c mice at all time points after infection (Fig. 1F). When the early innate NO response to a range of chlamydial doses was tested, the magnitude of NO output correlated with increasing chlamydial doses in C57BL/6 mice whereas BALB/c mice showed little change in NO output (Fig. 1L). These data suggested that NO could in fact be the immunosuppressive factor, and that the level of NO release possibly determined the disease outcome of chlamydial infection. We proceeded to further characterize the cells producing NO and how this production was regulated.
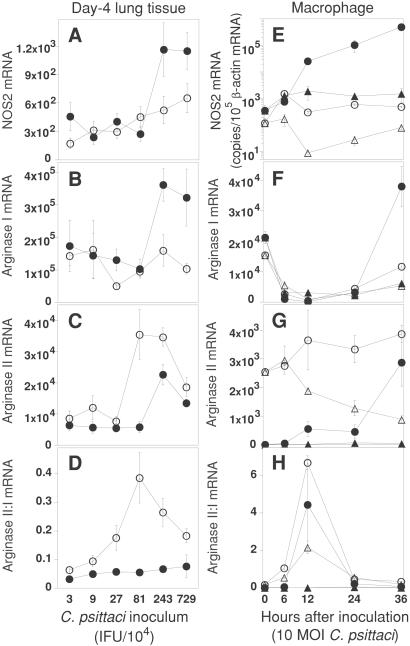
NOS2 expression in Chlamydia-infected lung tissue was examined by immunohistochemical staining. Lesions in infected lungs (Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org) were either double-stained (cryosections) or serial sections single-stained (formalin-fixed sections) with antibodies against C. psittaci, NOS2, or the macrophage maturation marker F4/80. NOS2 was virtually exclusively expressed in both mouse strains by F4/80-positive lung macrophages, which were also the main target cells for chlamydial infection (Fig. 4). This finding with C. psittaci is similar to that of C. pneumoniae, which also principally targets macrophages (16). However, infected macrophages of both mouse strains stained with similar intensity for NOS2, thus NOS2 protein levels in macrophages did not explain the differences in NO release. To obtain more precise data on NOS2 expression, we analyzed NOS2 mRNA levels in day-4 infected lung tissue by RT-qPCR. After high-dose chlamydial inoculation, C57BL/6 mice had significantly higher NOS2 transcript levels than BALB/c mice; however, at doses of 8.1 × 105 IFU C. psittaci or less NOS2 mRNA concentrations did not differ between BALB/c and C57BL/6 mice (Fig. 2A). Based on these data, we excluded different NOS2 enzyme levels as the cause of C57BL/6:BALB/c differential macrophage NO release when mice were infected with the clinically relevant chlamydial doses.
Figure 2.
BALB/c mice express arginase II in response to chlamydial infection more rapidly and at higher levels than C57BL/6 mice. BALB/c (○) or C57BL/6 (●) mice were i.n. infected with a series of C. psittaci inocula (A–D) and transcripts in lung tissue were determined on day 4 (n = 12–18, combined data of three experiments). Peritoneal macrophages (E–H) of BALB/c (○, infected; ▵, mock-infected) or C57BL/6 (●, infected; ▴, mock-infected) mice were infected with C. psittaci and specific mRNA levels were determined by RT-qPCR and expressed as transcripts per 105 β-actin transcripts (n = 6, one of two experiments is shown). (A and E) NOS2 transcripts in Chlamydia-infected lung tissue or macrophages. (B and F) Arginase I transcripts. (C and G) Arginase II transcripts. (D and H) Arginase II:arginase I transcripts.
We considered a mutant NOS2 enzyme having impaired activity as another possible explanation for the low NO production of BALB/c mice. The NOS2 genes of a number of inbred mouse lines have identical ORFs (17, 18), but the BALB/c ByJ mouse strain is an exception (GenBank accession no. AF06591920). This NOS2 gene differs by a single nonsynonymous polymorphism at position 3083, which results in the replacement of a phenylalanine residue by a serine residue at a position close to the putative NADPH ribose binding site of the reductase domain of murine NOS2 (17, 18). We amplified a DNA fragment containing the polymorphic position from genomic DNA of several BALB/c and C57BL/6 mice and probed the amplicons with FRET probes in qPCR. No deviation from the consensus murine NOS2 sequence was observed. DNA sequencing also failed to confirm a mutation. Thus, it is unlikely that a mutant NOS2 with impaired enzymatic activity is the cause of the low NO production in BALB/c mice during lung infection with C. psittaci.
Because reduced NO production in BALB/c mice was not associated with reduced expression of NOS2 or a mutant NOS2, we considered differential regulation of substrate availability as the most likely mechanism of the differential NO output (19). l-arginine is the substrate for NOS2 as well as for two arginase isoenzymes, cytosolic arginase I (GenBank accession no. U5180522) and extrahepatic mitochondrial arginase II (20, 21). These two isoenzymes hydrolyze l-arginine to urea and ornithine, and thus may reduce NO production by depleting the intracellular pool of l-arginine (22, 23). In vitro induction of either arginase isoenzyme in murine macrophages effectively down-regulates NO release and prevents NO-mediated apoptosis (22, 24). To determine whether differences in arginase expression were associated with the differential NO release in these mouse strains, we examined mRNA levels in infected lung tissue by RT-qPCR. The distinctive feature of BALB/c lung tissue, exposed to low to intermediate doses of chlamydiae, was an elevated level of arginase II transcripts, clearly visualized in the arginase II/arginase I transcript ratio, which was significantly higher than in C57BL/6 mice; in contrast, the level of arginase I mRNA in BALB/c lungs did not differ significantly from that in C57BL/6 lungs (Fig. 2 B–D). To examine whether the tissue levels of these transcripts reflected macrophage transcript levels, we tested the in vitro response of macrophages to chlamydial infection and found profound differences between the mouse strains. Of the NOS2, arginase I, and arginase II genes, BALB/c macrophages selectively and highly transcribed only the arginase II gene immediately after stimulation (Fig. 2 E–H). In contrast, C57BL/6 macrophages responded more slowly, i.e., after 24 h, but did so with sustained high levels of transcription, particularly of the NOS2 and arginase I genes. These data are consistent with the low responsiveness reported for BALB/c macrophages to a variety of stimuli when compared with C57BL/6 mice (25).
Reversal of Susceptibility Phenotype by Modulation of NOS2 Substrate Catalysis.
Collectively, these experiments suggested that profound differences in macrophage l-arginine metabolism might be the cause of the differences in NO production by BALB/c and C57BL/6 macrophages. This finding is consistent with a preferential NOS2-mediated production of NO and citrulline by lipopolysaccharide/IFN-γ-stimulated C57BL/6 macrophages, and with a preferential arginase-mediated production of ornithine and urea by BALB/c macrophages, as reported by Mills et al. (26) and proposed as a concept of M-1 and M-2 macrophages. We tested the role of l-arginine metabolism in NO production by treating infected mice with l-norvaline, a competitive inhibitor of arginase (23, 27), with the rationale to increase intracellular l-arginine levels, and consequently NO production, of infected mice. At a dose of 1.5% l-norvaline in drinking water, but not at lower or higher doses, BALB/c mice produced more NO and partially reverted to a disease phenotype (Fig. 3A).
Figure 3.
Modulation of NO production with arginase and NOS2 inhibitors reverses chlamydial disease phenotypes in an inhibitor dose-dependent manner in primary and secondary infection with C. psittaci or C. pneumoniae. (A) Mice were i.n. inoculated with 8.1 × 105 IFU C. psittaci and received the arginase inhibitor, l-norvaline, in drinking water. l-norvaline (1.5%) application significantly increases serum nitrite/nitrate and chlamydial lung disease in BALB/c (○), but not in C57BL/6 (●) mice (PDiseaseBALB/c:BALB/c+1.5%l-norvaline = 0.024, PchlamydiaeBALB/c:BALB/c+1.5%l-norvaline = 0.049; BALB/c, 62.3 μM day-4 serum nitrite/nitrate ± 5.9, SEM; BALB/c + 1.5% l-norvaline, 91.6 ± 7.9; n = 12; P = 0.007; C57BL/6, 109.72 ± 12.8; C57BL/6 + 1.5% l-norvaline, 123.22 ± 38.8; n = 12; P = 0.68). Lower or higher application does not change the disease in BALB/c mice (n = 8–20, combined data of four experiments). (B) Mice were i.n. inoculated with 8.1 × 105 IFU C. psittaci, and beginning 1 day earlier, the NOS2 inhibitor AG was administered. AG application significantly reduced serum nitrite/nitrate (BALB/c, 48.7 μM day-4 serum nitrite/nitrate ± 2.7, n = 26; C57BL/6, 89.8 ± 7.5, n = 26, PBALB/c:C57BL/6 = 0.0002; C57BL/6 + 6 mg/kg per day AG, 73.2 ± 3.8, n = 24, P0mgAG:6mgAG = 0.035; C57BL/6 + 200 mg/kg per day AG, 39.7 ± 2.9, n = 18, P0mgAG:200mgAG = 0.00006). Daily administration of 6 mg AG per kg body mass, but not of higher or lower doses, completely reverses the severe day-12 disease phenotype of C57BL/6 mice to the protected phenotype of BALB/c mice, as measured by lung weight increase or total chlamydial lung burden (PDiseaseC57BL/6:C57BL/6+6mgAG = 0.0000001, PchlamydiaeC57BL/6:C57BL/6+6mgAG = 0.0002). BALB/c mice in all dosage groups do not show significant changes in their day-12 disease (n = 6–24, combined data of five experiments). (C) Mice received a low 3 × 104 IFU C. psittaci priming i.n. infection, and 5 weeks later a challenge of 6 × 106 IFU C. psittaci. A dose of 100 mg/kg AG per day completely abolished disease of C57BL/6 mice on day 7 after secondary infection (PDiseaseC57BL/6:C57BL/6+AG = 0.003, PchlamydiaeC57BL/6:C57BL/6+AG = 0.35). BALB/c disease is marginally increased, and chlamydial burden is substantially, but not significantly, higher (n = 10, combined data of two experiments). (D) Mice primed with 3 × 105 IFU C. pneumoniae were challenged 5 weeks later with 3 × 107 IFU C. pneumoniae. With AG treatment, both mouse strains are free of gross lung disease and show reduced chlamydial lung loads (n = 10, combined data of two experiments, PDiseaseC57BL/6:C57BL/6+AG = 0.019, PchlamydiaeC57BL/6:C57BL/6+AG = 0.14; PDiseaseBALB/c:BALB/c+AG = 0.21, PchlamydiaeBALB/c:BALB/c+AG = 0.12).
l-arginine homeostasis is complex (19), and we had anticipated that it would be difficult to consistently reverse the resistant disease phenotype of BALB/c mice to the susceptible one by NOS2-substrate augmentation by using arginase inhibition. However, direct inhibition of the high-output NO-producing enzyme, NOS2, might reliably accomplish reduced NO production, and thus more completely reverse the C57BL/6 disease phenotype. If NO were a quantitative, nonredundant regulator of chlamydial disease, then optimal NO reduction by partial NOS2 inhibition should mitigate NO immunosuppression and abolish disease but not NO signaling and chlamydiastasis. We probed a reversal of the susceptible phenotype of C57BL/6 mice with AG, a selective, competitive inhibitor of NOS2 (28). Intraperitoneal administration of AG reduced NO production of infected C57BL/6 mice in a dose-dependent manner. Six milligrams AG per kg body mass daily administered to naive C. psittaci-infected mice completely protected C57BL/6 mice from disease and greatly reduced day-12 chlamydial lung burden (Fig. 3B). Other doses of AG did not change disease outcomes and numbers of chlamydiae in lung tissue.
Because chlamydial disease typically results from numerous episodes of reinfection, we tested the protective effect of NO reduction in a model of secondary lung disease 5 weeks after i.n. priming of mice with a low chlamydial inoculum. Daily administration of AG completely abolished the development of interstitial pneumonia in C57BL/6 mice on day 7 after secondary infection with C. psittaci (Fig. 3C) or C. pneumoniae (Fig. 3D). It reduced the chlamydial lung burden by more than 10-fold, although this required a higher dose of 100 mg per day (Fig. 3 C and D). These results demonstrate that macrophage NO regulates disease versus protection after primary and repeated chlamydial infection. Reduction, but not elimination, of NO production changed C57BL/6 mice from a disease-susceptible to a completely resistant phenotype whereas intrinsically low NO-producing BALB/c mice remained unaffected. Elimination of disease in C57BL/6 mice was consistent, but required different levels of NOS2 inhibition, depending on the level of chlamydial challenge and the specific immune status. We propose, therefore, that NO is a quantitative, nonredundant regulator of chlamydial disease.
To better understand the complex, nonlinear mechanisms of disease protection by reduced release of NO from macrophages, we studied the kinetics of chlamydial pneumonia under AG influence. C57BL/6 mice treated with 6 mg AG/kg per day were protected from disease and resembled BALB/c mice in many aspects (Fig. 5, which is published as supporting information on the PNAS web site). When C57BL/6 mice were untreated or were highly NOS2-inhibited (treated with 200 mg AG/kg per day), both groups of mice developed severe disease that closely paralleled each other. This finding is consistent with our earlier experiment in which NOS2+/+ and NOS2−/− C57BL/6 mice developed day-12 disease of similar severity after i.n. inoculation of 2.7 × 105 IFU C. psittaci (NOS2+/+: 113.53 ± 38% lung weight increase; NOS2−/−: 100.49 ± 31.3%). It is also consistent with results of others who did not observe differences in C. trachomatis-induced disease between NOS2+/+ and NOS2−/− mice (29–31). The most striking observation in C57BL/6 mice protected by 6 mg/kg AG was a significant increase in day-4 pulmonary concentrations of IL-12 p70 and IFN-γ transcripts (Fig. 5). These mice also showed a significant day-4 increase in splenic CD4 T cells to the level of BALB/c mice (BALB/c, 28.7% CD4+ splenocytes ± 1.33, n = 6; C57BL/6, 23.2 ± 0.05; C57BL/6 + 6 mg/kg per day AG, 28.0 ± 0.55; PC57BL/6:C57BL/6+6mgAG = 0.005).
Discussion
In this quantitative analysis of Chlamydia-induced interstitial pneumonia, C57BL/6 mice were highly prone to severe disease after i.n. inoculation with moderate doses of chlamydiae but protected from disease after high-dose inoculation. BALB/c mice exhibited a reversed phenotype of disease susceptibility. The C57BL/6 low-dose susceptibility was first evident 1 week after inoculation of naïve animals, at a time when the nascent adaptive immune response typically redirects innate defense mechanisms. DTH, antibody, and splenocyte proliferation data indicated that the adaptive immune response was strongly suppressed in C57BL/6 mice and was moderately Th2-biased at peak disease. Protection was the result in BALB/c mice, and the protection was associated with high DTH and IgG2a antichlamydial immunity, consistent with a Th1 adaptive immune response and with the absolute requirement of MHC II-restricted IFN-γ release for clearance of chlamydial infection (12, 32–35). The suppressed adaptive immunity to chlamydiae observed in C57BL/6 mice is reminiscent of patients with scarring trachoma caused by chronic C. trachomatis infection; peripheral blood mononuclear cells of trachoma patients yield significantly reduced lymphoproliferative responses to chlamydial antigens compared with asymptomatic, C. trachomatis-infected individuals (36).
Analysis of the reversal of susceptibility to Chlamydia-induced disease suggests that early in disease the IL-12–IFN-γ signaling circuitry between macrophages, natural killer (NK), and activated Th1 cells maintains high levels of both protective cytokines only within a narrowly restricted range of NO release. In the absence of NO, IL-12 signaling in NK cells is blocked, thus reducing the IFN-γ release of NK cells (37) and rendering the innate defense ineffective against chlamydial infection. Too low a level of NO production may also explain the increased severity of secondary C. psittaci disease in BALB/c mice treated with a high dose of AG (Fig. 3C). Conversely, high macrophage NO release, driven by NK and CD4 T cell-derived IFN-γ, is known to induce immunosuppression (38, 39) and down-regulate Th1 cell development by inhibition of macrophage IL-12 synthesis (40) and selective induction of apoptosis of activated Th1 cells (41). Consistent with this concept are the significantly enhanced lung levels of IL-12 and IFN-γ as well as the day-4 increase in splenic CD4 T cells of C. psittaci-infected C57BL/6 mice protected by 6 mg/kg AG to the level of BALB/c. Collectively, these data suggest that activated CD4 T cells secreting IFN-γ indirectly trigger their own apoptosis when they activate macrophages of high NO responders but that they proliferate in the presence of low NO responder macrophages (41). We propose that the decisive factor in development of chlamydial disease is a high rate of increase in macrophage NO production in response to increasing quantities of chlamydial and host inflammatory stimuli.
NOS2-synthesized NO is equally important in human immune regulation and disease (42), even if high levels of NO are not found in supernatants of cultured human macrophages (43), and intracellular l-arginine concentration is elevated during high NO production in human airway epithelium (44). Our finding of genetically determined regulation of macrophage NO production in inbred mice suggests that similar high and low NO responder genotypes with differential susceptibility to chlamydial disease may exist in the human population. This is potentially important in the context of atherosclerosis and coronary heart disease. C. pneumoniae is found in high frequency in atherosclerotic lesions (3), and NOS2 enhances atherosclerosis in mouse models (45). Whereas infectious agents are not required for initiation or progression of atherosclerosis (46), the presence of C. pneumoniae in murine atherosclerotic lesions increases lesion size and disease progression (47, 48). Thus, repeated C. pneumoniae infection of high NO responding individuals might precipitate a vicious cycle of exacerbated and prolonged pro-atherogenic inflammatory responses (49), which would have the potential to greatly accelerate an otherwise slow progression toward coronary atherosclerosis. A corollary of this antigen-independent pathogenetic mechanism would be that T cell-mediated immunoprotection from chlamydial disease by vaccination might not be safely accomplished in high NO-responding individuals genetically at risk for chlamydial disease. By integrating the environment-dependent parameters bacterial load and l-arginine supply, and host-dependent genetic determinants of NO production, immunological decision making in chlamydial infection would be probabilistic rather than deterministic. Absent of pharmacological reduction of macrophage NO release during enhanced vaccine-mediated T cell responsiveness, only genetic triage of high NO-responding individuals from a vaccinee population might ensure minimal undesirable side effects of vaccination against Chlamydia.
Supplementary Material
Acknowledgments
We thank J. Storz for the gift of C. psittaci B577 antiserum; R. Young-White for excellent technical assistance; and S. J. Ewald, S. R. Roberts, and L. G. Wolfe for valuable comments. This work was supported by the National Institutes of Health (Grant AI 38977), Bayer AG, and a Presidential Fellowship from Auburn University.
Abbreviations
- i.n.
intranasal
- IFU
inclusion-forming units
- DTH
delayed type hypersensitivity
- NOS2
NO synthase 2
- AG
aminoguanidine
- FRET
fluorescence resonance energy transfer
- qPCR
quantitative PCR
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schachter J. In: Chlamydia. Stephens R S, editor. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 139–169. [Google Scholar]
- 2.Saikku P, Leinonen M, Mattila K, Ekman M R, Nieminen M S, Makela P H, Huttunen J K, Valtonen V. Lancet. 1988;ii:983–986. doi: 10.1016/s0140-6736(88)90741-6. [DOI] [PubMed] [Google Scholar]
- 3.Jackson L A, Campbell L A, Schmidt R A, Kuo C-C, Cappucio A L, Lee M J, Grayston J T. Am J Pathol. 1997;150:1785–1790. [PMC free article] [PubMed] [Google Scholar]
- 4.Balin B J, Gérard H C, Arking E J, Appelt D M, Branigan P J, Abrams J T, Whittum-Hudson J A, Hudson A P. Med Microbiol Immunol. 1998;187:23–42. doi: 10.1007/s004300050071. [DOI] [PubMed] [Google Scholar]
- 5.Ward M E. In: Chlamydia. Stephens R S, editor. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 171–210. [Google Scholar]
- 6.Sowa S, Sowa J, Collier L H, Blyth W A. J Hyg. 1969;67:699–717. doi: 10.1017/s0022172400042157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saikku P. In: Chlamydial Infections: Proceedings of the Ninth International Symposium on Human Chlamydial Infection, Napa, CA, June 1998. Stephens R S, editor. Berkeley: Univ. of California Press; 1998. pp. 145–154. [Google Scholar]
- 8.Yang X, HayGlass K T, Brunham R C. J Immunol. 1996;156:4338–4344. [PubMed] [Google Scholar]
- 9.Nadeau J, Arbuckle L D, Skamene E. J Inflammation. 1995;45:27–48. [PubMed] [Google Scholar]
- 10.Kaltenboeck B, Huang J, Wang M-D, Lenz S D, Gao D. In: Chlamydial Infections: Proceedings of the Ninth International Symposium on Human Chlamydial Infection, Napa, CA, June 1998. Stephens R S, editor. Berkeley: Univ. of California Press; 1998. pp. 403–406. [Google Scholar]
- 11.MacMicking J, Qiao-wen W, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Wang M-D, Lenz S D, Gao D, Kaltenboeck B. J Immunol. 1999;162:2217–2226. [PubMed] [Google Scholar]
- 13.Laubach V E, Shesely E G, Smithies O, Sherman P A. Proc Natl Acad Sci USA. 1995;92:10688–10692. doi: 10.1073/pnas.92.23.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, DeGraves F J, Gao D, Feng P, Schlapp T, Kaltenboeck B. BioTechniques. 2001;30:150–157. doi: 10.2144/01301rr03. [DOI] [PubMed] [Google Scholar]
- 15.Eisenstein T K, Huang D, Meissler J J, Jr, Al-Ramadi B. Immunobiology. 1994;191:493–502. doi: 10.1016/S0171-2985(11)80455-9. [DOI] [PubMed] [Google Scholar]
- 16.Moazed T C, Kuo C-C, Grayston J T, Campbell L A. J Infect Dis. 1998;177:1322–1325. doi: 10.1086/515280. [DOI] [PubMed] [Google Scholar]
- 17.Lyons C R, Orloff G J, Cunningham J M. J Biol Chem. 1992;267:6370–6374. [PubMed] [Google Scholar]
- 18.Xie Q-W, Cho H J, Calaycay J, Mumford R A, Swiderek K M, Lee T D, Ding A, Troso T, Nathan C. Science. 1992;256:225–228. doi: 10.1126/science.1373522. [DOI] [PubMed] [Google Scholar]
- 19.Mori M, Gotoh T. In: in Nitric Oxide. Ignarro L J, editor. New York: Academic; 2000. pp. 199–208. [Google Scholar]
- 20.Morris S M, Jr, Kepka-Lenhart D, Chen L C. Am J Physiol. 1998;275:E740–E747. doi: 10.1152/ajpendo.1998.275.5.E740. [DOI] [PubMed] [Google Scholar]
- 21.Iyer R K, Bando J M, Jenkinson C P, Vockley J G, Kim P S, Kern R M, Cederbaum S D, Grody W W. Mol Genet Metab. 1998;63:168–175. doi: 10.1006/mgme.1997.2669. [DOI] [PubMed] [Google Scholar]
- 22.Gotoh T, Mori M. J Cell Biol. 1999;144:427–434. doi: 10.1083/jcb.144.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C-I, Liao J C, Kuo L. Am J Physiol. 1998;274:H342–H348. doi: 10.1152/ajpheart.1998.274.1.H342. [DOI] [PubMed] [Google Scholar]
- 24.Munder M, Eichmann K, Morán J M, Centeno F, Soler G, Modolell M. J Immunol. 1999;163:3771–3777. [PubMed] [Google Scholar]
- 25.Oswald I P, Afroun S, Bray D, Petit J-F, Lemaire G. J Leukocyte Biol. 1992;52:315–322. doi: 10.1002/jlb.52.3.315. [DOI] [PubMed] [Google Scholar]
- 26.Mills C D, Kincaid K, Alt J M, Heilman M J, Hill A M. J Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.1701141. [DOI] [PubMed] [Google Scholar]
- 27.Tews J K, Harper A E. J Nutr. 1986;116:1464–1472. doi: 10.1093/jn/116.8.1464. [DOI] [PubMed] [Google Scholar]
- 28.Corbett J A, Tilton R G, Chang K, Hasan K S, Ido Y, Wang J L, Sweetland M A, Lancaster J R, Jr, Williamson J R, McDaniel M L. Diabetes. 1992;41:552–556. doi: 10.2337/diab.41.4.552. [DOI] [PubMed] [Google Scholar]
- 29.Perry L L, Feilzer K, Caldwell H D. Infect Immun. 1998;66:1265–1269. doi: 10.1128/iai.66.3.1265-1269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Igietseme J U, Perry L L, Ananaba G A, Uriri I M, Ojior O O, Kumar S N, Caldwell H D. Infect Immun. 1998;66:1282–1286. doi: 10.1128/iai.66.4.1282-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsey K H, Miranpuri G S, Sigar I M, Quellette S, Byrne G I. Infect Immun. 2001;69:5131–5137. doi: 10.1128/IAI.69.8.5131-5137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igietseme J U, Ramsey K H, Magee D M, Williams D M, Kincy T J, Rank R G. Reg Immunol. 1993;5:317–324. [PubMed] [Google Scholar]
- 33.Morrison R P, Feilzer K, Tumas D B. Infect Immun. 1995;63:4661–4668. doi: 10.1128/iai.63.12.4661-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rank R G. In: Chlamydia. Stephens R S, editor. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 239–295. [Google Scholar]
- 35.Rottenberg M E, Gigliotti Rothfuchs A C, Gigliotti D, Svanholm C, Bandholtz L, Wigzell H. J Immunol. 1999;162:2829–2836. [PubMed] [Google Scholar]
- 36.Holland M J, Bailey R L, Hayes L J, Whittle H C, Mabey D C. J Infect Dis. 1993;168:1528–1531. doi: 10.1093/infdis/168.6.1528. [DOI] [PubMed] [Google Scholar]
- 37.Diefenbach A, Schindler H, Röllinghoff M, Yokoyama W M, Bogdan C. Science. 1999;284:951–955. doi: 10.1126/science.284.5416.951. [DOI] [PubMed] [Google Scholar]
- 38.Schwacha M G, Eisenstein T K. Infect Immun. 1997;65:4897–4903. doi: 10.1128/iai.65.12.4897-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwacha M G, Meissler J J, Jr, Eisenstein T K. Infect Immun. 1998;66:5862–5866. doi: 10.1128/iai.66.12.5862-5866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang F-P, Niedbala W, Wei X-Q, Xu D, Feng G-J, Robinson J H, Lam C, Liew F Y. Eur J Immunol. 1998;28:4062–4070. doi: 10.1002/(SICI)1521-4141(199812)28:12<4062::AID-IMMU4062>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 41.Dalton D D, Haynes L, Chu C-Q, Swain S L. J Exp Med. 2000;192:117–122. doi: 10.1084/jem.192.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allione A, Bernabei P, Bosticardo M, Ariotto S, Forni G, Novelli F. J Immunol. 1999;163:4182–4191. [PubMed] [Google Scholar]
- 43.Gantt K R, Goldman T L, McCormick M L, Miller M A, Jeronimo S M B, Nascimento E T, Britigan B E, Wilson M E. J Immunol. 2001;167:893–901. doi: 10.4049/jimmunol.167.2.893. [DOI] [PubMed] [Google Scholar]
- 44.Guo F H, Comhair S A A, Zheng S, Dweik R A, Eissa N T, Thomassen M J, Calhoun W, Erzurum S C. J Immunol. 2000;164:5970–5980. doi: 10.4049/jimmunol.164.11.5970. [DOI] [PubMed] [Google Scholar]
- 45.Detmers P A, Hernandez M, Mudgett J, Hassing H, Burton C, Mundt S, Chun S, Fletcher D, Card D J, Lisnock J M, et al. J Immunol. 2000;165:3430–3435. doi: 10.4049/jimmunol.165.6.3430. [DOI] [PubMed] [Google Scholar]
- 46.Wright S D, Burton C, Hernandez M, Hassing H, Montenegro J, Mundt S, Patel S, Card D J, Hermanowski-Vosatka A, Bergstrom J D, et al. J Exp Med. 2000;191:1437–1441. doi: 10.1084/jem.191.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moazed T C, Campbell L A, Rosenfeld M E, Grayston J T, Kuo C-C. J Infect Dis. 1999;180:238–241. doi: 10.1086/314855. [DOI] [PubMed] [Google Scholar]
- 48.Hu H, Pierce G N, Zhong G. J Clin Invest. 1999;103:747–753. doi: 10.1172/JCI4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross R. N Eng J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.