Abstract
Activation of the transcription factor NF-κB is critical for the tumor necrosis factor-α (TNF-α)-induced inflammatory response. Here we report the complete gene expression profile from activated microvascular endothelial cells emphasizing the direct contribution of the NF-κB pathway. Human microvascular endothelial cell line-1 (HMEC-1) cells were modified to express dominant interfering mutants of the IKK/NF-κB signaling module and expression profiles were determined. Our results provide compelling evidence for the virtually absolute dependence of TNF-α-regulated genes on NF-κB. A constitutively active IKK2 was sufficient for maximal induction of most target genes, whereas a transdominant IκBα suppressed gene expression. Several genes with a critical role in atherogenesis were identified. The endothelial lipase (EL) gene, a key enzyme involved in lipoprotein metabolism, was investigated more in detail. Binding sites interacting with NF-κB in vitro and in vivo were identified and co-transfection experiments demonstrated the direct regulation of the EL promoter by NF-κB. We conclude that targeting the IKK/NF-κB pathway or specific genes downstream may be effective for the control or prevention of chronic inflammatory diseases such as atherosclerosis.
INTRODUCTION
The endothelium is a dynamic organ that provides a structural and functional barrier between the circulation and the surrounding tissue. The endothelial cell (EC), that forms the non-thrombogenic lining of the vessels in every organ, represents a selective semi-permeable barrier that reacts with physical and chemical stimuli regulating the hemostasis, vasomotor tone and immune response (1). Endothelial cells play a pivotal role in inflammation, which constitutes the survey strategy of the innate immune system to thwart most pathogenic threats. To date much evidence reveals that the magnitude of the inflammatory response is crucial to keep the organism homeostasis and dysregulation of it can promote disease. Highly regulated go/stop signals are required to establish multiple checkpoints [reviewed in (2)] and the EC is indeed a critical one. Its activation initiates the inflammatory response by recruiting leukocytes into the damaged tissue (1), therefore EC dysfunction contributes to the development of a chronic inflammatory response or vascular disease. Tumor necrosis factor α (TNF-α) is a potent pro-inflammatory cytokine that triggers a strong endothelial activation which results in an increased vascular permeability, the hallmark of the inflammatory response. The biological effects of TNF-α are achieved by activation of signaling cascades that elicit a specific gene expression program. One major signaling pathway involves nuclear factor-κB (NF-κB).
Activation of NF-κB/Rel transcription factors plays a central role in the regulation of diverse cellular processes such as inflammation, immune response, differentiation, proliferation, apoptosis and cancer. The mammalian Rel family consists of five members p65/RelA, RelB, c-Rel, p50 and p52 which can form homo- and/or heterodimers. They are tightly controlled by a family of inhibitory molecules (IκBs) comprising IκBα, IκBβ and IκBε, and the precursor molecules for p50 and p52, p105 and p100, respectively. In resting cells, NF-κB is inactive because of its association with IκB proteins. Thereby NF-κB is retained in the cytoplasm and DNA binding is prevented. Upon cytokine signaling, innate or adaptive immune responses, or environmental stress NF-κB activation is initiated (3). Signaling pathways converge at a multisubunit IκB kinase complex that consists of two catalytic subunits IKK1/α and IKK2/β, and the regulatory components NEMO/IKKγ and a newly identified protein ELKS (4,5). Mice deficient of IKK2 or NEMO lack cytokine-induced NF-κB activation (6). The ‘canonical’ IKKβ- and IKKγ-dependent signaling pathway involves phosphorylation of the IκB proteins at conserved serine residues in their N-terminal domain. Subsequently, they are polyubiquitinated and degraded by the proteasome. Released NF-κB translocates to the nucleus and binds to cognate DNA motifs in target genes, regulating their transcription.
The biological effects of IKK/NF-κB signaling depend on transcriptional regulation of a network of genes that contain NF-κB binding sites in their promoter or enhancer regions. To date a large list of target genes has been identified (7) and in endothelial cells we have previously shown that genetic inhibition of NF-κB by IκBα mutants or dominant negative IKKβ blocks endothelial activation by suppressing the expression of NF-κB dependent genes(8).
In this report, we have investigated the requirement of the IKK complex and NF-κB for the gene expression by performing gene profiling in a human microvascular endothelial cell line after prolonged TNF-α stimulation. Our approach used retroviral gene transfer to effectively transduce a constitutive active version of IKKβ or a dominant negative IκBα, by which the ‘canonical’ NF-κB pathway was modulated allowing a fine-tuned investigation of differential gene induction by TNF-α. Our data indicates that the IKK/NF-κB cascade governs the long-term gene response to TNF-α of microvascular endothelial cells and suggest that secondary transcriptional responses involving other regulators are submitted to NF-κB control in agreement with recent previous reports (9,10). In addition we provide compelling evidence for a pro-atherogenic program elicited by IKK/NF-κB signalosome in ECs. The transcriptional regulation of a potentially key pro-atherogenic gene, endothelial lipase (EL) by NF-κB further supports the master role of this signaling pathway in atherosclerosis.
MATERIALS AND METHODS
Cell culture
Human microvascular endothelial cell line-1 (HMEC-1) was provided by V. Gehrke (Muenster, Germany). Primary human umbilical vein endothelial cells (HUVEC) were obtained from Clonetics (Cell Systems, Germany). Primary human saphena vein endothelial cells (HSVEC) were provided by N. Marx (Ulm, Germany). HMEC-1 were cultured in endothelial basal medium (Clonetics) supplemented with 2% fetal bovine serum (FBS), 1.0 µg/ml hydrocortisone, 10 ng/ml human epidermal growth factor, 12 µg/ml bovine brain extract, 50 µg/ml gentamicin and 50 ng/ml amphotericin B. For culture of HUVEC and HSVEC medium was supplemented with 10% FBS. Experiments were performed with primary cells in passage 4. Cells were stimulated with human recombinant TNF-α (40 ng/ml, a gift from Boeheringer-Ingelheim). Protein synthesis inhibition was achieved by treatment of the cells with cycloheximide (5 µg/ml, Sigma). The ΦNX amphotropic retrovirus producers (G. Nolan, Stanford, CA) and NIH-3T3 cells were cultured in DMEM (Life Technologies) containing 10% FBS, 100 U/ml penicillin-streptomycin (PAN Systems, Germany), 1% l-Glutamine (PAN Systems, Germany), 1% non-essential aminoacids (PAN Systems, Germany) and 25 µM β-mercaptoethanol (Sigma).
Retroviral infection of endothelial cells
The pCFG5-IEGZ retroviral vectors encoding transdominant IκBα (TD IκBα) or constitutively active IKK2 (CA IKK2), have been described (8). Infected cells were selected with 200 µg/ml of Zeocin (Invitrogen) until all cells were GFP positive. As HMEC-1 expressing the CA-IKK2 altered growth characteristics over time, GFP positive cells were sorted 48 h after infection by flow cytometry using a FACSAria (Becton Dickinson) and prepared immediately for the experiments.
Flow cytometry and protein analysis
Analysis of ICAM-1 expression was performed as described (8). Flow cytometry experiments were repeated at least twice. For western blot analysis, whole cell extracts were prepared and analyzed as described (8). For analysis of periplakin expression, 70 µg of protein extracts were separated on 5% polyacrylamide gels and probed using antibodies against periplakin (Santa Cruz). For EMSA, whole cell extracts (2–5 µg protein) were incubated with radiolabeled double-stranded oligonucleotides containing the Ig-κ enhancer consensus κB site (11) or putative κB sites from the endothelial lipase gene (sequences available upon request). For supershift experiments, 5 µg of protein extract were preincubated for 30 min at room temperature with specific antibodies. And for competition assays, protein extracts were pre-incubated with non-labeled double-stranded oligonucleotides for 10 min at room temperature. Protein synthesis inhibition by cycloheximide treatment was controlled by metabolic cell labeling with [35S] Methionine (100 µCi/ml; Amersham Pharmacia Biotech).
Microarray analysis
A total of 4–5 × 106 cells were stimulated with TNF-α for 16 h or left untreated, harvested and frozen at −80°C. Cell pellets were then processed at the IZKF Microarray Facility Tuebingen (Tuebingen, Germany) where, the RNA preparation and hybridization assay were performed according to the manufacturer's instruction. Hybridization was done onto Affymetrix Humane Genome Array Chip U133A. Data were processed with Affymetrix Microarray Suite (MAS) Software 5.0 and NetAFFX Analysis Center Tools (Affymetrix Inc. CA). Further analyzes including Venn diagrams were performed using GeneSpring (Silicon Genetics/Agilent) and VisualX was used for graphical representations (available from hans.kestler@medizin.uni-ulm.de upon request).
RT–PCR analysis
Total RNA was isolated using the High Pure RNA Isolation Kit (Roche, Germany). cDNA was synthesized with M-MLV reverse transcriptase (Promega, USA) and PCR was performed using Taq DNA polymerase (Amersham Pharmacia Biotech, USA). Primers sequences are available upon request.
Cloning and transient co-transfection assays
The genomic region spanning from −1381 to +240 of the endothelial lipase gene [GenBank™/EBI Data Bank Accession no. NM_006033.1, Gene ID 9388, LIPG] was amplified with Pfx polymerase (Invitrogen) and inserted upstream of the luciferase gene (5′hLIPG) (primer sequences available upon request). The generation of reporter vectors for the putative NF-κB binding sites in LIPG promoter was done by cloning a synthetic oligonucleotide containing three copies of either the distal site (3× -L1) or the proximal site (3× -L2) upstream of a TATA-only promoter luciferase construct. All reporter vectors were confirmed by sequencing. Five µg of these constructs and as a control 100 ng of β-actin renilla expression vector were transiently co-transfected together with 15 µg of either RelA/p65 expression vector or empty vector into NIH-3T3 cells. Luciferase activities were measured after 36 h using the Dual-Luciferase Reporter Assay System (Promega, USA).
Site directed mutagenesis
For mutagenesis of the distal κB site in the 5′hLIPG vector, a mutagenic primer (5′-gagggagaagaggaggacaaagctcacgctcaggtggctctcccccgacgg-3′) was annealed to denaturate the parental plasmid and extended by PCR using Pfx polymerase (Invitrogene). Non-mutated DNA was removed by treatment with DpnI endonuclease. Positive clones were sequenced to confirm the mutation and the integrity of the remaining construct.
Chromatin immunoprecipitation assay (ChIP)
Chromatin immunoprecipitation (ChIP) was performed as described previously (12). One µg of rabbit anti-RelA/p65 (sc-372, Santa Cruz) or rabbit anti-PLCγ1 (sc-81, Santa Cruz) were used. Samples were analyzed by PCR using specific primers for the appropriate regions (primer sequences available upon request).
RESULTS
Genetic manipulation of NF-κB activity in HMEC-1 cells
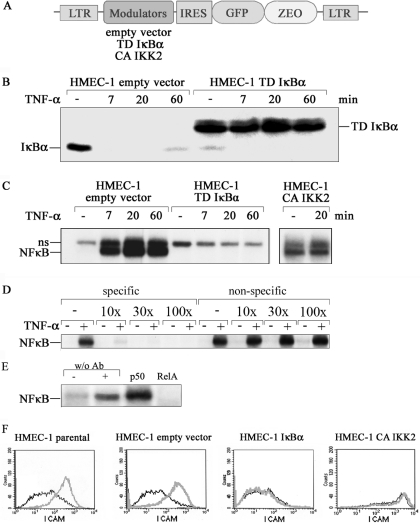
We wanted to investigate the role of the IKK/NF-κB signaling pathway in the gene expression program of microvascular endothelial cells under TNF-α stimulation. Retroviral gene transfer was used to stably express the dominant interfering mutants of the IKK/NF-κB pathway in the HMEC-1 cell line, an established microvascular endothelial cell line derived from human dermal microvasculature (13). Suppression of NF-κB signaling was achieved by using a trans-dominant IκBα protein (TD-IκBα) with serine to alanine mutations in the N-terminal domain. For selective activation of this pathway, a constitutively active version of IKK2 (CA-IKK2) bearing phosphomimetic serine to glutamate mutations in the activation loop was used (Figure 1A) (8,14,15). Consequences of the expression of these NF-κB modulating proteins were then analyzed in transduced cells. Whereas TNF-α induced rapid degradation of endogenous IκBα in cells infected with empty vector, the transdominant IκBα protein was stable under these conditions (Figure 1B). Expression of the endogenous IκBα protein was almost completely repressed in TD-IκBα infected cells consistent with earlier findings (Figure 1B) (8,15). Furthermore, stimulation of TD-IκBα-expressing cells failed to induce NF-κB as documented by electrophoretic mobility shift assay (EMSA) experiments (Figure 1C). Importantly, expression of CA-IKK2 induced NF-κB DNA binding activity already in the absence of TNF-α, which could be slightly enhanced by treatment with TNF-α (Figure 1C). Competition experiments using a mutated κB-probe confirms the specificity for NF-κB binding (Figure 1D). While supershift analyzes with specific antibodies show that the DNA binding complex is composed of p50/p65 proteins (Figure 1E).
Figure 1.
Genetic manipulation of NF-κB transduced HMEC-1 cells. (A) Schematic representation of the retrovirus used for the expression of the IκBα and IKKβ mutants. IRES, internal ribosome entry site; LTR long terminal repeat: Zeo, zeocin resistance gene; TD, transdominant; CA, constitutively active. (B) Endothelial cells stably expressing parental vector or transdominant IκBα were stimulated with TNF-α for the time intervals indicated. IκBα degradation was visualized by Western blot analysis. (C) TNF-α-induced NF-κB DNA-binding activity was studied by EMSA. NF-κB complexes are indicated; ns, non-specific band. (D) Competition EMSA. Labeled consensus κB probe was incubated with 20 min TNF-α-stimulated or control HMEC-1 whole cell extracts in the presence of increasing amounts of cold probes for consensus kB-probe (specific) or mutated kB-probe (non-specific). (E) Supershift assay. Whole cell extracts of 20 min TNF-α-stimulated or control HMEC-1 were pre-incubated with specific antibodies before NF-κB DNA-binding activity was studied by EMSA. (F) Parental HMEC-1 or cells expressing TD IκBα, CA IKK2 or empty vector, respectively, were stimulated for 16 h with TNF-α. Expression of ICAM was determined by FACS before (black line) or after TNF-α stimulation (grey line).
We had previously seen that a similar modulation of NF-κB activity in HUVEC cells affected the expression of various chemokines and cell surface proteins (8). Therefore, we analyzed infected HMEC-1 cells before and after stimulation with TNF-α for expression of ICAM-1 by FACS. Both parental HMEC-1 and empty vector transduced HMEC-1 showed an up-regulation of ICAM-1 (Figure 1F). The CA-IKK2 expressing cells showed strong ICAM-1 expression even without TNF-α stimulation, whereas no ICAM-1 expression could be induced in TD-IκBα cells (Figure 1F). Similar results were obtained for interleukin-8 and MCP-1 (data not shown).
Identification of TNF-α regulated genes in HMEC-1 cells and dependence on the IKK/NF-κB pathway
These results encouraged us to perform a global analysis of gene expression in response to TNF-α in these genetically modified cells. Gene expression profiles were obtained from three independent experiments using the Affymetrix Human Genome U133A platform. Our definition of a classical TNF-α-induced NF-κB target gene was the following: NF-κB target genes should be up-regulated in unstimulated HMEC-1 cells expressing CA-IKK2 compared to empty vector control cells. TNF-α stimulation might result in a further induction of these genes in CA-IKK2 expressing cells. These genes should be induced by TNF-α in empty vector infected cells, but not or less induced in TNF-α stimulated HMEC-1 cells expressing TD-IκBα. Corresponding criteria should hold for genes repressed by NF-κB. In the subsequent analyzes we only considered genes whose expression was altered at least 2-fold (SLR ≥ 1) in a minimum of seven out of nine possible combinations between the triplicates of each comparison.
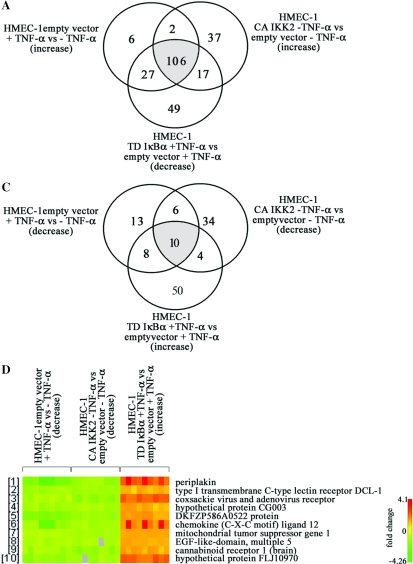
We identified 141 genes up-regulated by TNF-α in control HMEC-1 (Figure 2A). CA-IKK2 induced expression of 162 genes whereas 199 entries corresponded to genes down-regulated in TNF-α stimulated TD-IκBα cells when compared to TNF-α-stimulated control cells. The above described criteria were fully met by 106 genes which represent targets with high probability to contain NF-κB regulatory elements within their promoter/enhancer regions. It became clear that out of the 141 genes induced by TNF-α in control cells, only 6 genes (4% of the total) did not reveal evidence for an involvement of classical NF-κB for transcriptional regulation. Expression of 27 genes was induced by TNF-α in control cells and was decreased in TD-IκBα cells, yet they were not significantly increased in CA-IKK2 infected cells suggesting that NF-κB is not sufficient and other pathways are required. Surprisingly, 49 genes were decreased in TNF-α stimulated HMEC-1 transducing TD-IκBα compared to stimulated HMEC-1 transduced with the empty vector. This result might suggest a role of IκBα in gene regulation in addition to NF-κB inhibition. In this respect, recent evidence shows that IκBα recruitment to the hes1 promoter is associated with transcriptional repression (16). At the same time, 37 genes showed up-regulation in the presence of CA-IKK2 but they were not strongly induced by TNF-α stimulation in control cells. This hints at functions of IKK in addition to regulating NF-κB. Indeed, recent evidence suggests that IKK2 is involved in control of the Foxo3a transcription factor (17).
Figure 2.
Identification of TNF-α regulated genes and dependence on the IKK/NF-κB pathway. HMEC-1 lines expressing TD IκBα, CA IKK2 or empty vector were cultured to ∼80% confluence, stimulated with of TNF-α for 16 h and RNA expression profiles were determined by oligonucleotide array hybridization in three independent experiments. Venn diagrams are depicted for (A) up-regulated genes and (C) down-regulated genes. In (B and D) the graphic representations of the respective genes up- or down-regulated by TNF-α that fully meet the inclusion criteria are shown. For each gene the variation of the expression levels between the three experiments is shown.
Expression profiling results were highly consistent as is evident from the representation shown in Figure 2B. This Figure depicts the genes for which all predictions described above were met. The five genes with the highest TNF-α induced expression were ubiquitin D (average 89-fold), natural killer cell transcript 4 (NK4, average 62-fold induction), Epstein–Barr virus-induced gene 3 (EBI3, average 54-fold), chemokine (C-C motif) ligand 5 (RANTES, average 36-fold) and chromosome 1 open reading frame (ORF) 29 corresponding to hypothetical protein GS3686 (average 23-fold). These five genes are also highly regulated in HMEC-1 expressing the active IKK2 mutant protein. In addition, CA-IKK2 strongly induced expression of the bone marrow stromal cell antigen 2, viperin, chemokine (C-X-C motif) ligand 5 (epithelial-derived neutrophil-activating peptide 78), chemokine (C-X-C motif) ligand 11, baculoviral IAP repeat-containing 3 and the myxovirus resistance 2 gene. All these genes are also induced by TNF-α and repressed in TNF-α-stimulated HMEC-1 transduced with TD-IκBα. Interestingly, the gene profile obtained reflects a pro-atherogenic program induced by long-term TNF-α stimulation in ECs which is strictly regulated by IKK/NF-κB signaling pathway. (The complete and detailed list of identified genes is provided as Supplementary Data Set and will be submitted to MIAME).
TNF-α and CA-IKK2 not only induced gene expression but also a number of genes were identified which are repressed by these pathways. The corresponding Venn diagram represents 37 entries of genes down-regulated in control cells exposed to TNF-α (Figure 2C). Furthermore, expression of 54 genes decreased in CA-IKK2 cells, whereas 72 transcripts were increased in TD-IκBα HMEC-1 when stimulated with TNF-α compared to stimulated control cells. In contrast to the extremely high overlap between TNFα-targets and NF-κB-regulated genes observed for the induced genes, only 27% of the genes down-regulated by TNF-α were affected by the IKK/NF-κB interfering mutants. This indicates a wider spectrum of signaling pathways involved in gene repression by TNF-α.
TNF-α induced gene expression by RT–PCR
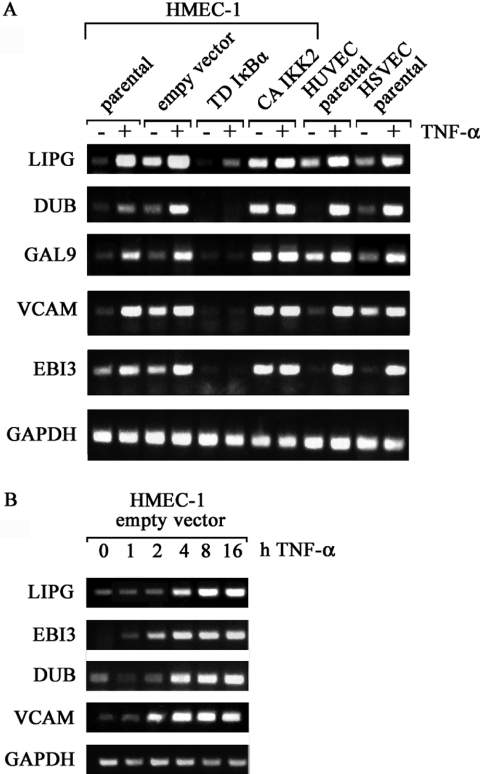
We wanted to confirm the differential expression pattern for a subset of genes by semi-quantitative RT–PCR. In addition to the analysis in the various HMEC lines, we also investigated these targets in TNF-α-stimulated HUVEC and HSVEC. This comparison allowed us to determine whether these selected genes were regulated similarly in microvasculature- and macrovasculature-derived endothelial cells and also allowed us to estimate the impact of our experimental model in gene expression. Five genes showing an induction between 5 fold and more than 20 fold in the microarray experiments were chosen: ubiquitin D (DUB), Epstein–Barr virus-induced gene-3 (EBI3), vascular cell adhesion molecule-1 (VCAM), endothelial lipase (LIPG) and galectin 9 (LGALS9) (Figure 3A).
Figure 3.
Analysis of TNF-α-induced gene expression in two additional endothelial models by RT–PCR. (A) Total RNA from HMEC-1 lines and primary HUVEC and HSVEC stimulated with TNF-α for 16 h was used for cDNA preparation and RT–PCR analysis. Specific primers for the indicated genes were used. (B), Kinetic of TNF-α-induction of different mRNAs in empty vector infected HMEC-1 is shown.
In agreement with the microarray data, TNF-α stimulation increases mRNA levels of all genes investigated in parental and empty vector transduced HMEC-1. The house-keeping gene GAPDH was used as control. The same behavior is observed for HUVEC and HSVEC when exposed to TNF-α, despite some differences in basal expression levels in unstimulated cells. As expected, TD-IκBα largely abolished TNF-α-induced expression of the investigated genes in HMEC-1 cells. In contrast, CA-IKK2 increased mRNA levels of these genes and additional TNF-α-stimulation resulted in only a minimal further increase. Therefore, IKK2 is sufficient to up-regulate expression of all genes investigated. Kinetic analyzes revealed that VCAM and EBI3 show increased expression between 1 and 2 h of TNF-α stimulation, whereas expression-levels of LIPG and DUB started to increase after 4 h (Figure 3B).
TNF-α/NF-κB suppresses periplakin expression
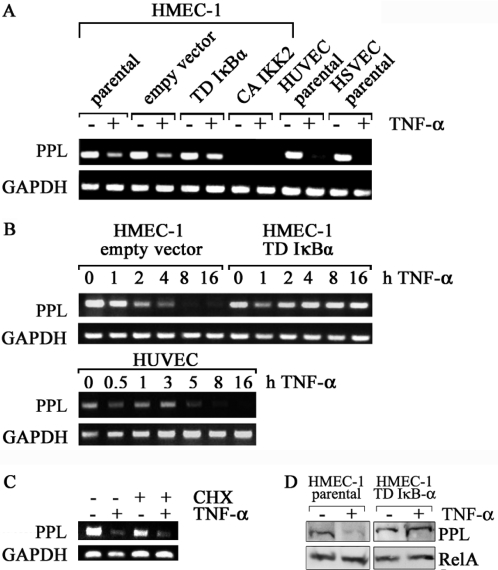
Our screen had revealed a number of genes repressed by both TNF-α and IKK2. We chose the best regulated of these genes, periplakin, for a more detailed analysis. Expression profiling had predicted a roughly 6- to 7-fold regulation and this was confirmed by RT–PCR (Figure 4A). Repression was even more pronounced in HUVEC and HSVEC. Repression was strongly attenuated when NF-κB induction was blocked by TD-IκBα. CA-IKK2 virtually eliminated baseline expression. Kinetic analyzes revealed that most of periplakin mRNA was reduced between 5 and 8 h after TNF-α induction in HUVEC and HMEC1, respectively (Figure 4B). No loss of periplakin mRNA was detected when TD-IκBα was expressed. The effect of NF-κB on TNF-α-induced repression of periplakin is most likely direct as it is independent of de novo protein synthesis as shown by the analysis of periplakin expression after pre-treatment of the cells with cycloheximide (Figure 4C). The efficiency of protein synthesis inhibition by cycloheximide was larger than 90% confirmed by in vivo [35S]-methionine cell-labeling (data not shown). Finally, western blot analysis showed periplakin down-regulation in parental HMEC-1 after TNF-α-stimulation, whereas cells expressing TD-IκBα showed no decrease at the protein level (Figure 4D).
Figure 4.
TNF-α down-regulates periplakin expression in HMEC-1, HUVEC and HSVEC. (A) Total RNA from HMEC-1 lines and primary HUVEC and HSVEC stimulated with TNF-α for 16 h was used for cDNA preparation and RT–PCR analysis. (B) Time course of periplakin mRNA down-regulation in response to TNF-α. HMEC-1 stably expressing the empty vector or transdominant IκBα, or HUVEC were stimulated with TNF-α for the indicated intervals, total RNA was analyzed by RT–PCR. (C) Expression level of periplakin was analyzed after TNF-α stimulation in the presence of cycloheximide. Confluent cells were pre-incubated with vehicle or cycloheximide (CHX) for 1 h and then treated with TNF-α or medium for 8 h. (D) Western blot analysis for periplakin (195 kDa). Parental or TD-IκBα transduced HMEC-1 were stimulated with TNF-α for 16 h and whole protein extracts were probed with a periplakin-specific . RelA/p65 was used as loading control.
The endothelial lipase gene contains functional NF-κB sites
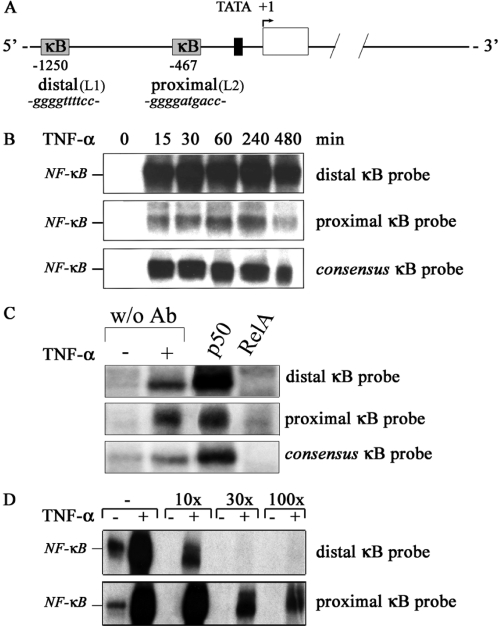
A known predisposing factor in the development of atherosclerosis is a change in high density lipoprotein (HDL) versus low density lipoprotein (LDL) levels. The endothelial lipase (gene nomenclature LIPG) identified in our gene screening is of specific interest in this respect due to its role in HDL metabolism. This recently cloned new member of the lipase family of proteins catabolizes HDL and it is the only lipase member secreted by endothelial cells (18,19). Endothelial lipase is induced by inflammatory cytokines but clear NF-κB dependence has not been demonstrated (20). Due to the potential role of this gene as predisposing factor for lipid metabolism-related diseases a more detailed promoter analysis was performed. The gene encoding endothelial lipase is located on chromosome 18. It consists of 10 exons and 9 introns. A TATA box was predicted between −30 and −22. Two κB sites were predicted in the 5′ region at −467 (proximal) and at −1250 (distal) (Figure 5A). NF-κB-binding to these sites was analyzed by EMSA. The distal site showed strong NF-κB-binding (Figure 5B), whereas the proximal site showed only weak NF-κB-binding. Supershift analyzes confirmed the binding of RelA/p65 to both putative κB sites (Figure 5C). Moreover, when a labeled consensus κB probe was incubated with stimulated HMEC-1 protein extracts in the presence of increasing amounts of oligonucleotides corresponding to the putative κB sites in endothelial lipase gene, strong competition for NF-κB binding was seen for the distal site, while the proximal site competed to a lesser extent (Figure 5D).
Figure 5.
The endothelial lipase gene contains functional NF-κB sites. (A) Schematic representation of putative NF-κB binding sites in the endothelial lipase (LIPG) gene. (B) TNF-α-induced NF-κB DNA-binding to putative κB sites in the LIPG gene was studied by EMSA. (C) Supershift assay. Whole cell extracts of 1 h TNF-α-stimulated or control HMEC-1 were pre-incubated with specific antibodies before NF-κB DNA-binding activity to putative kB-sites was studied. (D) Competition EMSA. Labeled consensus κB probe was incubated with 1 h TNF-α-stimulated or control HMEC-1 extracts in the presence of increasing amounts of cold probes for the putative κB sites.
TNF-α induces recruitment of RelA/p65 to the endothelial lipase promoter
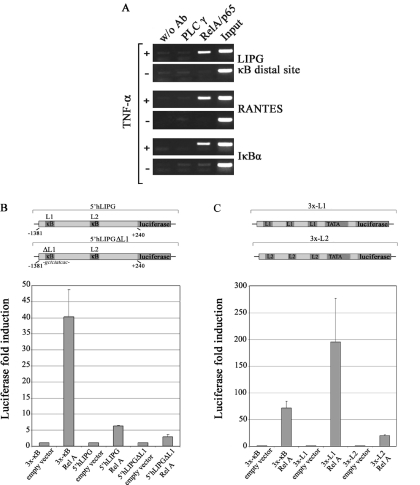
To determine whether inducible NF-κB-binding to the LIPG promoter could be observed in vivo, we performed ChIP. HMEC-1 cells were stimulated with TNF-α for 4 h and formaldehyde-cross linked chromatin was precipitated with an anti-RelA/p65 antibody. As a negative control an irrelevant antibody (anti-PLCγ1) was used (Figure 6A). The binding of RelA/p65 to the IκBα and RANTES promoters served as positive control. ChIP-analysis of these control genes revealed TNF-α-inducible RelA/p65 recruitment to the respective promoters as expected. Interestingly, inducible recruitment of RelA/p65 was detectable at the distal site of the LIPG promoter (Figure 6A) while only a weak induction was observed at the proximal site (data not shown).
Figure 6.
TNF-α-induced expression of endothelial lipase is associated with recruitment of RelA/p65 to the distal κB site. (A) Binding of RelA/p65 to regulatory regions of LIPG, RANTES and IκBα in HMEC-1 after stimulation with TNF-α for 4 h was detected by ChIP assay. Negative controls were chromatin samples to which no antibody was added (w/o Ab) or immunoprecipitated with anti-PLC-γ1 antibody. DNA purified from the lysates incubated without antibody was used as input control (Input). All experiments were performed at least in duplicates. (B) LIPG promoter fragment encompassing the region from −1381 to +240 was cloned upstream of a luciferase reporter gene. The resultant vector named 5′hLIPG was transiently co-transfected into NIH-3T3 cells together with a RelA/p65 expression vector or an empty vector as control. Site directed mutagenesis to the distal kB-site of 5′hLIPG resulted in the modified reporter vector named 5′hLIPGΔL1, which was transiently co-transfected into NIH-3T3 cells together with a RelA/p65 expression vector or an empty vector as control. (C) Luciferase reported vectors containing three copies of LIPG-kB distal (3x-L1) and proximal (3x-L2) sites were transiently co-transfected into NIH-3T3 cells with RelA/p65 expression vector or empty vector as control. A luciferase vector containing three copies of the κB-motif was used as positive control. Luciferase activities were measured after 24 h. All experiments were performed at least in duplicates.
Finally we wanted to demonstrate the direct regulation of the LIPG promoter by NF-κB in reporter co-transfection experiments. To achieve these experiments we used NIH-3T3 cells, since HMEC-1 cells were completely resistant to transient transfection. A promoter fragment encompassing the region from −1381 to +240 was cloned upstream of a luciferase reporter gene (Figure 6B). This promoter fragment, named 5′hLIPG, contained both the distal and the proximal κB sites. When this reporter was co-transfected with RelA/p65 expression vector in NIH-3T3 cells we indeed observed an induction of 6 fold in the luciferase activity. The mutation of the perfect distal κB-motif results in a reduction of RelA/p65-induction in 50% (Figure 6B). In addition, we have generated luciferase reporter constructs that contain three copies of either the distal (3× -L1) or the proximal (3× -L2) site up-stream of a TATA-only promoter. These synthetic reporters are both responsive to RelA/p65 co-transfection in NIH-3T3 cells. However, a stronger induction of luciferase is achieved by the distal motif in comparison to the proximal κB-site (Figure 6C).
DISCUSSION
The induction of gene expression by TNF-α is known to critically depend on activation of MAPK and IKK signaling. The first pathway involves phosphorylation of ERKs, JNKs and p38 members and the subsequent activation of transcription factors like Elk-1, AP-1, SRF or CREB, whereas the IKK pathway induces activation of NF-κB. Thus, it is expected that endothelial gene profile after long-term TNF-α stimulation should reflect the activation of the given range of signaling cascades. Yet, our gene expression profile suggests that the IKK/NF-κB signalosome plays the dominant role in TNF-α-induced activation of HMEC-1 cells. Almost all TNF-α-induced genes in control HMEC-1 were repressed in TD-IκBα transducing cells (94%), while the 76% of them were already up-regulated in CA-IKK2 infected cells in the absence of stimulation. A total of 106 genes equivalent to the 75% of the TNF-α-induced profile were transcriptionally controlled by CA-IKK2 and TD-IκBα. This group of genes represents the vast majority of TNF-α-response genes and the classical IKK/NF-κB signaling module is sufficient to regulate their expression probably by direct NF-κB regulatory elements in their promoter/enhancer regions.
In our study TNF-α induction of only few genes was partially or completely independent of classical IKK/NF-κB activation. The group of genes that were induced by TNF-α, repressed by TD-IκBα but not up-regulated by CA-IKK2 is of particular interest. Their transcriptional regulation requires NF-κB but NF-κB is not sufficient, suggesting that other regulators are necessary. TNF-α-induced IKK/NF-κB-independent mechanisms could involve the MAPK/AP-1 pathway. A previous report analyzed the contribution of p38-MAPK in the gene expression profile of HUVEC after 5 h TNF-α stimulation and determined that a minor number of genes underlie additional control by this pathway (10). Using the p38-MAPK inhibitor SB202190 they observed that the TNF-α-dependent up-regulation of the gene C1orf4 was totally inhibited. In our screen, TNF-α induction of this gene was comparable to the extent observed by the previous authors, but at the same time TD-IκBα was able to completely suppress this induction. In the same study, the IKK2-dependent induction of other genes such as IL8, VCAM, inhibin beta or GBP-1 was partially repressed (∼50%) by SB202190. According to our genechip data, these genes fully met the inclusion criteria and definition of a NF-κB target, showing complete repression by TD-IκBα. It has been proposed that p38-MAPK activity is required to enhance the accessibility of RelA/p65 for recruitment to a subset of genes activated in cells exposed to inflammatory stimuli in a promoter-specific manner (21). This mechanism could explain the enhanced expression observed in these NF-κB targets by p38-MAPK. Another interesting observation is the regulation of ubiquitin D. Viemman et al. (10) showed that the strong induction of this gene by TNF-α was completely inhibited in the presence of SB202190. In our genechip analysis this gene showed the strongest up-regulation by TNF-α and repression by TD-IκBα whereas CA-IKK2 induction did not reach the extent of TNF-α. In addition, the semi-quantitative RT–PCR revealed the complete absence of this mRNA in HMEC-1 transduced with TD-IκBα and stimulated by TNF-α at different time intervals (data not shown), suggesting an absolute dependence on the IKK/NF-κB module. Our results together with those presented by Viemman et al. (10) indicates that ubiquitin D expression requires synergistic activation of IKK/NF-κB and p38-MAPK, and that both pathways are essential.
Recently, Krappman et al. investigated to a deeper extent the cross-talk between NF-κB and AP-1 transcription factors in the LPS-induced gene expression in precursor B cells (9). Their results indicated that NF-κB mounts the secondary transcriptional response to LPS involving the control of AP-1 activation. In our experimental model, long-term TNF-α stimulation, was used to mimic a pathophysiological condition. This prolonged activation of the EC, might enable NF-κB to evoke a transcriptional response in cooperation with AP-1 signaling for e.g. or with pathways such as JAK/STAT activated by autocrine sources like IL-6. However, further analyzes are needed to define the integration role that NF-κB might play to orchestrate the inflammatory program elicited by TNF-α in ECs.
TNF-α stimulation also represses transcription. The analysis of periplakin, the gene that showed the strongest down-regulation by TNF-α and fully met our inclusion criteria, suggests a direct transcriptional repression mechanism by NF-κB. The mechanisms by which NF-κB down-regulates transcription appear to be diverse and are not fully understood yet (22). In a recent report, it was proposed that RelA/p65 directly represses transcription of antiapoptotic genes by a mechanism that involves a differential phosphorylation of RelA/p65, which regulates its ability to interact with corepressors such as histone deacetylase (23). In addition to direct repression, it was also proposed that NF-κB affects Sox9 and MyoD expression by an indirect mechanism involving mRNA destabilization (24).
A second goal of our study was to investigate the NF-κB dependence of genes with a link to atherogenesis. Given the current appreciation that inflammatory mechanisms are coupled to atherogenesis (25) we were eager to define the role of TNF-α-induced endothelial gene program in this process.
NF-κB target genes could be classified in different functional groups. Those with a primarily inflammatory function were predominant. Genes for monocyte, macrophage, neutrophil and T-cell chemoattractant chemokines and cytokines (RANTES, CCL20, CXCL3, CXCL2, CXCL11, CXCL5, MCP-1, IL8 and GM-CSF) were up-regulated consistent with the critical role of the endothelium in promoting leukocyte infiltration. Interestingly, the EBI-3 gene showed the strongest up-regulation among the regulated cytokines. In particular, the product of this gene is a 34 kDa secreted glycoprotein with important immunomodulatory functions which has been implicated in chronic inflammatory diseases (26–28). Up-regulation of these chemokines indeed highlights the active role of the endothelium in atheroma formation.
Genes involved in extracellular matrix (ECM) remodeling play a critical role in atherogenesis. Several genes belonging to this group were up-regulated, e.g. Cathepsin S (CTSS). CTSS is one of the most potent mammalian elastases. It is overexpressed in atherosclerotic lesions (29) and CTSS-deficiency attenuates atherosclerosis in LDLR −/− mice (30). Up-regulation of matrix metalloproteinases MMP-10 and MMP-12 further suggest an active role of the endothelium in ECM-remodeling and potentially plaque instability. MMP-12 has been associated with atherosclerosis (31). We found expression of these MMPs affected by TD-IκBα, but not by CA-IKK2, indicating that NF-κB is essential but not sufficient for their transcription. Taken together, the up-regulation of these proteases modulates vascular permeability and remodeling, leukocyte transmigration and ECM integrity. These features are critical for initiation, maintenance and resolution of atherosclerosis.
To analyze the NF-κB-dependence of genes with a functional role in atherogenesis we investigated more in detail the promoter of endothelial lipase (LIPG), a gene that fully met our inclusion criteria and that is implicated in HDL metabolism. LIPG is a new member of the triglyceride lipase family expressed in endothelial cells (18,19). Both acute and chronic inflammatory states are associated with decreased levels of HDL-cholesterol and LIPG could play an important role in this process. Recent findings revealed that inhibition of the endothelial lipase gene resulted in a 50% increase in HDL-cholesterol levels in mice (32). LIPG directly mediates cellular lipoprotein uptake (33) and apoE knockout animals also targeted at the LIPG locus show lower levels of atherosclerosis (34). In our gene profile, TNF-α stimulation resulted in a 10-fold induction of LIPG mRNA levels while CA-IKK2 induced its expression in 6-fold. RT–PCR analysis validated these results and confirmed the induction of gene expression in two primary endothelial cell models. Two putative NF-κB binding sites were predicted in LIPG 5′-flanking region. Results from EMSA, ChIP assays and co-transfection experiments suggest regulation of LIPG by NF-κB. Moreover, mutation analysis of the putative distal κB-site in the LIPG pomoter and experiments with synthetic luciferase reporter vectors containing either the distal or the proximal site give further evidence of the direct contribution of these sites in the transcriptional regulation of endothelial lipase. However, although the proximal site shows a weaker response to RelA, it might still contribute to the overall NF-κB response of the LIPG promoter. This could explain the incomplete inhibition of the 5′hLIPGΔL1 construct. Further studies are needed to determine whether the induced expression of LIPG in endothelial cells directly contributes to the development of atherosclerotic plaques.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr A. Ushmorov for important technical advice, F. Díaz for helpful collaboration during the preparation of this manuscript and Dr R. Marienfeld for critical reading of the manuscript. This work was supported by grants from the German Science Foundation (DFG SFB 451/TP A9) and the Fonds der Chemischen Industrie to T.Wirth. Funding to pay the Open Access publication charges for this article was provided by German Science Foundation (DFG SFB 451/TP A9).
Conflict of interest statement. None declared.
REFERENCES
- 1.Cines D.B., Pollak E.S., Buck C.A., Loscalzo J., Zimmerman G.A., McEver R.P., Pober J.S., Wick T.M., Konkle B.A., Schwartz B.S., et al. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91:3527–3561. [PubMed] [Google Scholar]
- 2.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh S., Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109:S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 4.Karin M., Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 5.Ducut Sigala J.L., Bottero V., Young D.B., Shevchenko A., Mercurio F., Verma I.M. Activation of transcription factor NF-kappaB requires ELKS, an IkappaB kinase regulatory subunit. Science. 2004;304:1963–1967. doi: 10.1126/science.1098387. [DOI] [PubMed] [Google Scholar]
- 6.Rudolph D., Yeh W.C., Wakeham A., Rudolph B., Nallainathan D., Potter J., Elia A.J., Mak T.W. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- 7.Pahl H.L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 8.Denk A., Goebeler M., Schmid S., Berberich I., Ritz O., Lindemann D., Ludwig S., Wirth T. Activation of NF-kappa B via the Ikappa B kinase complex is both essential and sufficient for proinflammatory gene expression in primary endothelial cells. J. Biol. Chem. 2001;276:28451–28458. doi: 10.1074/jbc.M102698200. [DOI] [PubMed] [Google Scholar]
- 9.Krappmann D., Wegener E., Sunami Y., Esen M., Thiel A., Mordmuller B., Scheidereit C. The IkappaB kinase complex and NF-kappaB act as master regulators of lipopolysaccharide-induced gene expression and control subordinate activation of AP-1. Mol. Cell. Biol. 2004;24:6488–6500. doi: 10.1128/MCB.24.14.6488-6500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viemann D., Goebeler M., Schmid S., Klimmek K., Sorg C., Ludwig S., Roth J. Transcriptional profiling of IKK2/NF-kappa B- and p38 MAP kinase-dependent gene expression in TNF-alpha-stimulated primary human endothelial cells. Blood. 2004;103:3365–3373. doi: 10.1182/blood-2003-09-3296. [DOI] [PubMed] [Google Scholar]
- 11.Lernbecher T., Muller U., Wirth T. Distinct NF-kappa B/Rel transcription factors are responsible for tissue-specific and inducible gene activation. Nature. 1993;365:767–770. doi: 10.1038/365767a0. [DOI] [PubMed] [Google Scholar]
- 12.Ushmorov A., Ritz O., Hummel M., Leithauser F., Moller P., Stein H., Wirth T. Epigenetic silencing of the immunoglobulin heavy-chain gene in classical Hodgkin lymphoma-derived cell lines contributes to the loss of immunoglobulin expression. Blood. 2004;104:3326–3334. doi: 10.1182/blood-2003-04-1197. [DOI] [PubMed] [Google Scholar]
- 13.Ades E.W., Candal F.J., Swerlick R.A., George V.G., Summers S., Bosse D.C., Lawley T.J. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J. Invest. Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 14.Huber M.A., Denk A., Peter R.U., Weber L., Kraut N., Wirth T. The IKK-2/Ikappa Balpha/NF-kappa B pathway plays a key role in the regulation of CCR3 and eotaxin-1 in fibroblasts. A critical link to dermatitis in Ikappa Balpha -deficient mice. J. Biol. Chem. 2002;277:1268–1275. doi: 10.1074/jbc.M109358200. [DOI] [PubMed] [Google Scholar]
- 15.Huber M.A., Azoitei N., Baumann B., Grunert S., Sommer A., Pehamberger H., Kraut N., Beug H., Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguilera C., Hoya-Arias R., Haegeman G., Espinosa L., Bigas A. Recruitment of IkappaBalpha to the hes1 promoter is associated with transcriptional repression. Proc. Natl Acad. Sci. USA. 2004;101:16537–16542. doi: 10.1073/pnas.0404429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu M.C., Lee D.F., Xia W., Golfman L.S., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 18.Hirata K., Dichek H.L., Cioffi J.A., Choi S.Y., Leeper N.J., Quintana L., Kronmal G.S., Cooper A.D., Quertermous T. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J. Biol. Chem. 1999;274:14170–14175. doi: 10.1074/jbc.274.20.14170. [DOI] [PubMed] [Google Scholar]
- 19.Jaye M., Lynch K.J., Krawiec J., Marchadier D., Maugeais C., Doan K., South V., Amin D., Perrone M., Rader D.J. A novel endothelial-derived lipase that modulates HDL metabolism. Nature Genet. 1999;21:424–428. doi: 10.1038/7766. [DOI] [PubMed] [Google Scholar]
- 20.Jin W., Sun G.S., Marchadier D., Octtaviani E., Glick J.M., Rader D.J. Endothelial cells secrete triglyceride lipase and phospholipase activities in response to cytokines as a result of endothelial lipase. Circ. Res. 2003;92:644–650. doi: 10.1161/01.RES.0000064502.47539.6D. [DOI] [PubMed] [Google Scholar]
- 21.Saccani S., Pantano S., Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nature Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 22.Zhong H., May M.J., Jimi E., Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 23.Campbell K.J., Rocha S., Perkins N.D. Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol. Cell. 2004;13:853–865. doi: 10.1016/s1097-2765(04)00131-5. [DOI] [PubMed] [Google Scholar]
- 24.Sitcheran R., Cogswell P.C., Baldwin A.S., Jr NF-kappaB mediates inhibition of mesenchymal cell differentiation through a posttranscriptional gene silencing mechanism. Genes Dev. 2003;17:2368–2373. doi: 10.1101/gad.1114503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 26.Devergne O., Hummel M., Koeppen H., Le Beau M.M., Nathanson E.C., Kieff E., Birkenbach M. A novel interleukin-12 p40-related protein induced by latent Epstein–Barr virus infection in B lymphocytes. J. Virol. 1996;70:1143–1153. doi: 10.1128/jvi.70.2.1143-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieuwenhuis E.E., Neurath M.F., Corazza N., Iijima H., Trgovcich J., Wirtz S., Glickman J., Bailey D., Yoshida M., Galle P.R., et al. Disruption of T helper 2-immune responses in Epstein–Barr virus-induced gene 3-deficient mice. Proc. Natl Acad. Sci. USA. 2002;99:16951–16956. doi: 10.1073/pnas.252648899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pflanz S., Timans J.C., Cheung J., Rosales R., Kanzler H., Gilbert J., Hibbert L., Churakova T., Travis M., Vaisberg E., et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4(+) T cells. Immunity. 2002;16:779–790. doi: 10.1016/s1074-7613(02)00324-2. [DOI] [PubMed] [Google Scholar]
- 29.Sukhova G.K., Shi G.P., Simon D.I., Chapman H.A., Libby P. Expression of the elastolytic cathepsins S and K in human atheroma and regulation of their production in smooth muscle cells. J. Clin. Invest. 1998;102:576–583. doi: 10.1172/JCI181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sukhova G.K., Zhang Y., Pan J.H., Wada Y., Yamamoto T., Naito M., Kodama T., Tsimikas S., Witztum J.L., Lu M.L., et al. Deficiency of cathepsin S reduces atherosclerosis in LDL receptor-deficient mice. J. Clin. Invest. 2003;111:897–906. doi: 10.1172/JCI14915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luttun A., Lutgens E., Manderveld A., Maris K., Collen D., Carmeliet P., Moons L. Loss of matrix metalloproteinase-9 or matrix metalloproteinase-12 protects apolipoprotein E-deficient mice against atherosclerotic media destruction but differentially affects plaque growth. Circulation. 2004;109:1408–1414. doi: 10.1161/01.CIR.0000121728.14930.DE. [DOI] [PubMed] [Google Scholar]
- 32.Jin W., Millar J.S., Broedl U., Glick J.M., Rader D.J. Inhibition of endothelial lipase causes increased HDL cholesterol levels in vivo. J. Clin. Invest. 2003;111:357–362. doi: 10.1172/JCI16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuki I.V., Blanchard N., Jin W., Marchadier D.H., Millar J.S., Glick J.M., Rader D.J. Endogenously produced endothelial lipase enhances binding and cellular processing of plasma lipoproteins via heparan sulfate proteoglycan-mediated pathway. J. Biol. Chem. 2003;278:34331–34338. doi: 10.1074/jbc.M302181200. [DOI] [PubMed] [Google Scholar]
- 34.Ishida T., Choi S.Y., Kundu R.K., Spin J., Yamashita T., Hirata K., Kojima Y., Yokoyama M., Cooper A.D., Quertermous T. Endothelial lipase modulates susceptibility to atherosclerosis in apolipoprotein-E-deficient mice. J. Biol. Chem. 2004;279:45085–45092. doi: 10.1074/jbc.M406360200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.