Abstract
Gfi-1B (growth factor independence-1B) gene is an erythroid-specific transcription factor, whose expression plays an essential role in erythropoiesis. Our laboratory has previously defined the human Gfi-1B promoter region and shown that GATA-1 mediates erythroid-specific Gfi-1B transcription. By further investigating the regulation of the Gfi-1B promoter, here we report that (i) Gfi-1B transcription is negatively regulated by its own gene product, (ii) GATA-1, instead of Gfi-1B, binds directly to the Gfi-1-like sites in the Gfi-1B promoter and (iii) Gfi-1B suppresses GATA-1-mediated stimulation of Gfi-1B promoter through their protein interaction. These results not only demonstrate that interaction of GATA-1 and Gfi-1B participates in a feedback regulatory pathway in controlling the expression of the Gfi-1B gene, but also provide the first evidence that Gfi-1B can exert its repression function by acting on GATA-1-mediated transcription without direct binding to the Gfi-1 site of the target genes. Based on these data, we propose that this negative auto-regulatory feedback loop is important in restricting the expression level of Gfi-1B, thus optimizing its function in erythroid cells.
INTRODUCTION
Gfi-1B (growth factor independence-1B) is an erythroid-specific Gfi-family transcriptional factor, which was identified by low stringency hybridization screening with a partial Gfi-1 cDNA probe (1). Both Gfi-1 and Gfi-1B contain a SNAG domain that mediates transcriptional repression and a zinc finger domain at its C-terminus for their DNA binding to the TAAATCAC(A/T)GCA recognition sequence (1–3). Expression of Gfi-1B is confined in erythroid lineage cells and megakaryocytes in human (4,5), whereas Gfi-1 is more abundant in the lymphopoietic thymus (6–8). So far, p21 (cip1/waf1), Socs1 and Socs3 are known as the target genes of Gfi-1B-mediated transcriptional repression (1,9). Since p21 is a cell cycle inhibitor and SOCS family members are known to suppress cytokine signaling, the functional role of Gfi-1B is considered to be important in controlling proliferation of erythrocyte/megakaryocyte-lineage cells. Its importance in erythropoiesis has been further highlighted by gene targeting experiment showing that Gfi-1B gene disruption results in embryonic lethality due to loss of red blood cell formation (10).
Enforced expression experiment in early erythroid progenitor cells has shown that Gfi-1B induces a drastic expansion of erythroblast independent of its SNAG repression domain with a parallel increase of GATA-2 expression, which is required for proliferation of erythroblasts (5). On the other hand, a recent study has shown that Gfi-1B plays a critical role in terminal differentiation through its transcription repression function (11). Likely, the function of Gfi-1B in erythropoiesis is highly dependent on cell stage and the sequence context of its targeted gene promoter. Despite the differential roles of Gfi-1B in different stages of differentiation, results of both studies indicate that elevation of Gfi-1B level alters the program of normal erythropoiesis (5,11). However, it remains unclear how Gfi-1B expression is regulated in erythroid cells and whether there is a direct relationship between Gfi-1B and other transcription factor that is involved in erythropoiesis.
The expression of many eukaryotic transcription factors has been shown to be auto-regulated positively and negatively (12–16). In most auto-regulatory cases, a given factor binds to its own promoter and either activates or represses transcription. In this study, we observed negative auto-regulation of Gfi-1B in K562 cells. By analyzing the sequence of human Gfi-1B gene promoter region (17), we found the presence of two tandem repeats of Gfi-1-like sites located at −59/−56 and −47/−44 relative to its transcription start site. Very recently, a report has demonstrated that mouse Gfi-1B directly binds to the Gfi-1 binding sites near the mRNA transcription start site of the mouse Gfi-1B promoter and is able to auto-repress its own expression (18). However, here we showed that mutations in these two Gfi-1-like sites reduced the promoter activity of the human Gfi-1B promoter in K562 cells, indicating that these sites mediate transcriptional activation rather than silencing. By detailed DNA-binding analyses, we proved that GATA-1, instead of Gfi-1B, is the main transcription factor preferentially binding to these non-typical GATA sites. Furthermore, we found that the Gfi-1B can form a complex with GATA-1, by which GATA-1-mediated transcription is repressed by Gfi-1B. Coincidentally, one recent report also showed that Gfi-1B forms a complex with GATA-1 and associates with the myc and myb promoters in mouse erythroleukemic (MEL) cells. Given the facts that overexpression of Gfi-1B in erythroid progenitors induces growth arrest and that expression of myc and myb is often associated with cell proliferation, they hypothesized that GATA-1/Gfi-1B is a repressive complex that suppresses transcription of myc and myb genes (19). Our results, on the other hand, present the first direct evidence that transcriptional repression function of Gfi-1B can work through its interaction with GATA-1 independent of its direct DNA binding to the gene promoter. Since our previous study has shown that GATA-1 is a necessary transcription factor for Gfi-1B expression, the auto-regulatory mechanism observed in this study reflects that the expression of Gfi-1B and the function of GATA-1 are mutually regulated in K562 cells.
MATERIALS AND METHODS
Cell culture
K562 cells were maintained in RPMI 1640 (Invitrogen Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone), 2 mM l-glutamine, 100 U of penicillin G per ml and 100 U of streptomycin per ml. 293T cells were grown in DMEM containing 10% FBS.
Preparation of nuclear extracts and gel-shift analysis
Nuclear extracts prepared from K562 and 293T cells and gel-shift reaction were as described previously (20). Competitive DNA oligomer, GATA-1 (C-20, Santa Cruz, CA), Gfi-1B (H-150, Santa Cruz) or FLAG (M2, Sigma) antibody was added to the nuclear extracts for 30 min on ice prior to the DNA-binding reaction. After DNA-binding reaction at room temperature for 25 min, samples were analyzed by electrophoresis at 150 V for 2.5 h through the nondenaturing 4% polyacrylamide gels. Gels were then dried for autoradiography.
Plasmid constructs and site-directed mutagenesis
Two AATC sites at −59/−56 and −47/−44 within the promoter region were simultaneously mutated to GGTC in the pGL2-hG (−325/+19) using Quick-Change Site-directed Mutagenesis Kit (Stratagene). A 3XGATA-T81-Luc reporter plasmid was constructed by inserting oligonucleotide containing three tandem copies of GATA-1 binding site (5′-GATCAGAAGCTT CACTTGATAACAGAAAGTGATAACAGAAAGTGATAATCTGTCGACCGTCGT-3′) upstream of a minimal herpes simplex thymidine kinase promoter in the pT81 luciferase vector (21), which was provided by A. Iwama (5) (Laboratory of Stem Cell Therapy, Center for Experimental Medicine, The Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo, 108-8639, Japan). Expression vector of Gfi-1B-FLAG, in which FLAG is tagged at the C-terminus of Gfi-1B, was constructed by ligating the PCR DNA fragment spanning the open reading frame region with the EcoRI/SalI digested pRK5-FLAG plasmid. To generate the expression vectors of the wild-type and deleted mutants of Myc-GATA-1, the regions covering 1–414, 1–199, 1–255 or 1–287 amino acids of mGATA-1 cDNA were amplified by PCR and inserted to the XhoI site of plasmid pCS2-MT (provided by Dave Turner, University of Michigan). The wild-type and deleted mutants of FLAG-Gfi-1B covering 1–330, 164–330 and 1–137 amino acids of hGfi-1B cDNA were each subcloned into the KpnI and XbaI sites of plasmid pCMV2 to generate the pCMV2-Gfi-1B (wild type), pCMV2-Gfi-1B (N-del) and pCMV2-Gfi-1B (C-del) constructs. Another deleted mutant construct covering 45–330 amino acids of hGfi-1B cDNA was subcloned into the HindIII and EcoRI sites of plasmid pCMV2 to generate the pCMV2-Gfi-1B (ΔSNAG) constructs. The VP16-Gfi-1B construct was generated by inserting the fragment covering 161–330 amino acids of hGfi-1B cDNA to the EcoRI site of pVP16 vector. The retroviral vector for expressing Gfi-1B with C-terminal FLAG-tag was constructed by ligating the coding region of Gfi-1B-FLAG with the HpaI-digested pS2 plasmid. All constructs were verified by dideoxy termination sequencing.
Retrovirus production and selection of stable cell line
S2 retroviral vectors expressing C-terminal FLAG-tagged wild-type Gfi-1B was transfected to the packaging cell line PT67. Stable clones were selected in the medium supplemented with G418 antibiotic (400 µg/ml). Following propagation of each clone, viral supernatant from the individual culture was collected and stored at −80°C. K562 cells were infected with viral supernatant for 48 h. Subsequently, K562 cells were kept in the medium containing G418 for further selection. Stable clone was propagated and confirmed by the detection of FLAG-tagged Gfi-1B in the cell lysates by western blotting using anti-FLAG antibodies (M2).
Transient-transfection and luciferase assays
We used electroporation for K562 cells transfection as described previously (17). 293T cells plated on 35 mm dishes were transiently transfected with a mixture containing 6 µg of lipofectamine (Invitrogen Life Technologies) and 1 µg of plasmid DNA. After transfection for 24 h, cells were washed and lysed in a reporter lysis buffer (0.5 M HEPES, pH 7.8, 0.2% Triton X-100, 1 mM CaCl2 and 1 mM MgCl2), and 50 µl of the cell lysates was mixed with 50 µl of luciferase assay buffer (Packard). The luciferase activity was measured with a luminescence counter (Packard).
Immunoprecipitation
Cells were seeded and transfected with a mixture of lipofectamine (Invitrogen Life Technologies) and plasmid DNA. After transfection for 24 h, cells were washed and lysed in 50 mM Tris–HCl buffer (pH 7.5) containing 1% Triton X-100, 150 mM NaCl, 0.1% SDS, 5 mM NaF and proteinase inhibitors (Sigma). The lysates were centrifuged at 10 000 g for 5 min and pre-cleared by incubating with normal mouse IgG and protein A beads for 30 min at 4°C. The supernatants were immunoprecipitated using M2-Ab beads at 4°C for 4 h. The beads were then washed four times by the lysis buffer and the proteins on the beads were separated by 10% SDS–PAGE.
Western blot analysis
Whole cell extracts (50 µg of protein) in RIPA buffer [50 mM Tris–HCl, pH 7.6, 150 mM NaCl, 0.1% sodium deoxycholate and SDS, 1% Triton X-100, 5 mM NaF, 5 mM Na4VO3 and proteinase inhibitors (Sigma)] were separated on a 10% SDS–polyacrylamide gel and transferred to a PVDF membrane (Millipore). The anti-GATA-1 (C-20, Santa Cruz) was diluted at 1:1000, anti-Gfi-1B (H-150, Santa Cruz), anti-β-tubulin (Sigma) and anti-Myc (9E10, Santa Cruz) at 1:2000, and anti-FLAG (M2, Sigma) at 1:5000, respectively. Enhanced chemiluminescence detection of the horseradish peroxidase reaction was performed according to the vendor's instructions (PerkinElmer Life Sciences).
RT–PCR assay
Total RNA was prepared for the synthesis of oligo(dT)-primed cDNA using the superscript II (Invitrogen, Carlsbad, CA). One micro liter of cDNA was then amplified using 30 cycles of 94°C for 30 s, 60°C for 45 s and 72°C for 1 min with specific primers using Fast-Run™ RT–PCR kit (Protech Technology Enterprise). PCR products were separated on 10% polyacrylamide gel and visualized by ethidium bromide staining. The primer sequences were endogenous Gfi-1B sense primer 5′-GGAGAAGTATCTATTTGTGC-3′, antisense primer 5′-AGTCAAGCTTCAGCACAATGGGGCCCTC-3′; Gfi-1B-FLAG sense primer 5′-CGGAATTCGCGTACCACTGTGTGAAG-3′, antisense primer 5′-GTCGTCATCGTCTTTGTAGTCCAT-3′; human hypoxanthine phosphoribosyltransferase (HPRT) sense primer 5′-AAGGACCCCACGAAGTGTTGG-3′, antisense primer 5′-AGGGAACTGATAGTCTATAG-3′; human hTK1 sense primer 5′-AGCTGCATTAACCTGCCCACTC-3′, antisense primer 5′-GCACTTGTACTGAGCAATATG-3′.
RNA interference
The pS2-Gfi-1B-FLAG retrovirus integrated K562 cells (3 × 106) were electroporated with or without the 100 nM GATA-1 siRNA (custom SMART pool, hGATA-1, NM_002049, Dharmacon Research Inc., Lafayette, CO). After transfection for 24 h, cells were harvested for the western blot and chromatin immunoprecipitation.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described previously (17). Antibodies used for immunoprecipitation were anti-FLAG (Sigma) and anti-NF-YB (C-20, Santa Cruz) antibodies. The immunoprecipitated DNA was applied to PCR using the following thermal cycling program: 95°C for 5 min, 32 cycles of 30 s at 95°C, 30 s at 52°C and 45 s at 72°C, followed by a 5 min extension time at 72°C. Thirty percent of the products were separated by 10% PAGE and visualized by ethidium bromide staining. The specific primers for the hGfi-1B promoter were sense primer: 5′-GAGTTTTATAAGTTAGAGCT-3′ and antisense primer: 5′-AAATAGATACTTCTCCTTTTTGC-3′. The size of PCR product of hGfi-1B promoter was 345 bp.
RESULTS
Gfi-1B transcription is down-regulated by ectopic expression of Gfi-1B protein
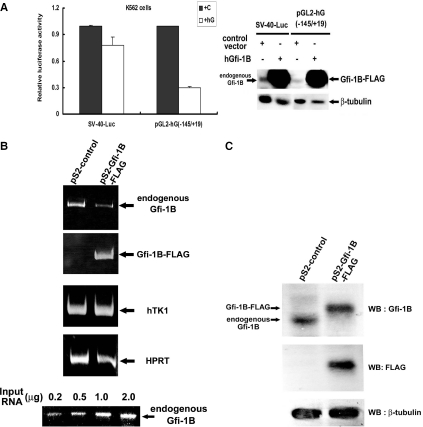
To test the effect of Gfi-1B on its own promoter, a plasmid expressing the C-terminus FLAG-tagged Gfi-1B under the control of the cytomegalovirus promoter was co-transfected with the human Gfi-1B promoter-luciferase construct into K562 cells. As shown in Figure 1A, overexpression of Gfi-1B-FLAG strongly reduced the Gfi-1B promoter activity to 30% of the control set, while a slight reduction of SV-40 promoter activity was seen under the same transfection condition, indicating promoter-specific repression by Gfi-1B. To know whether this auto-inhibition can occur under a condition where Gfi-1B expression level is elevated moderately, we constructed a retroviral expression vector of Gfi-1B-FLAG under the control of LTR promoter. K562 cells were infected by retrovirus generated from the empty and Gfi-1B-FLAG retroviral vectors containing neomycin resistance gene. Followed by clonal selection, G418 resistant clones were pooled and subjected to RT–PCR and western blot analyses. Since endogenous Gfi-1B RNA transcript contains a 5′-untranslated region (5′-UTR) covering from exon1 to exon2 of the human Gfi-1B gene (17), we amplified this 5′-UTR sequence of endogenous Gfi-1B RNA from total cellular RNA by RT–PCR to distinguish from Gfi-1B-FLAG transcript, which does not have the 5′-UTR sequence and was specifically amplified by the FLAG primer for detection. In accordance with the results from the reporter assay, cells stably expressing moderate amount of Gfi-1B-FLAG exhibited a marked reduction of the expression level of endogenous Gfi-1B transcript as compared with the cells selected from the control virus infection (Figure 1B). Similarly, the western blot analysis also showed that endogenous Gfi-1B protein was no longer detectable in cells expressing Gfi-1B-FLAG (Figure 1C), confirming that the endogenous Gfi-1B gene in K562 cells is suppressed by enforced expressed Gfi-1B-FLAG.
Figure 1.
Endogenous Gfi-1B transcription is repressed by ectopic expression of Gfi-1B in K562 cells. (A) K562 cells were transfected with the pGL2-SV40 or pGL2-hG (−145/+19) and pCMV-β-Gal together with control vector or Gfi-1B-FLAG expression vector as indicated. After transfection, each cell sample was divided into two parts. One for luciferase activity assay (left panel), and the other were subjected to western blot analysis (right panel) using antibodies against Gfi-1B (H-150) or β-tubulin. The luciferase activity was normalized by β-galactosidase activity in each sample, and luciferase activity of the Gfi-1B promoter from cells co-transfected with the control vector after normalization was set arbitrarily to 1. The data are means ± SD (n = 3) from three independent experiments. (B) K562 cells infected by control (pS2) and Gfi-1B-FLAG expression retrovirus were G418 selected and harvested for RNA preparation. RT–PCR assay were performed with 0.5 µg of each RNA sample and the specific primers for detecting endogenous Gfi-1B, TK1, HPRT and Gfi-1B-FLAG genes as described in Materials and Methods. Bottom panel shows the RT–PCR results for detecting endogenous Gfi-1B RNA in reactions containing different amounts of total input RNA from pS2 control cells. (C) Total lysates from the selected K562 cell lines as described above were subjected to western blot analysis using antibodies against Gfi-1B (H-150), FLAG (M2) and β-tubulin.
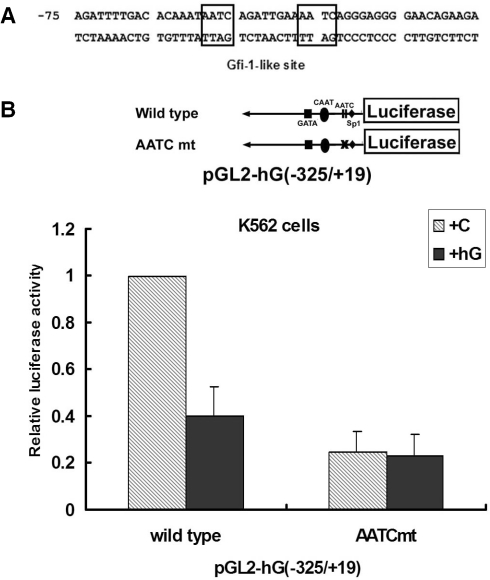
The contribution of Gfi-1-like sequences to the Gfi-1B promoter activity
Since Gfi-1B is known as a transcriptional repressor with a DNA-binding activity (1,9), we then examined whether the human Gfi-1B promoter contains typical Gfi-1 binding site. Sequence analysis of the Gfi-1B promoter has revealed two potential tandem Gfi-1-like sites (AATC) at −59/−56 and −47/−44 of the promoter region (Figure 2A). The in vivo footprint analysis also clearly showed that this region was well protected in genomic DNA of K562 cells as a contrast to no protection in that of myeloid U937 cells (data not shown). It is known that both Gfi-1 and Gfi-1B exhibit DNA-binding activity to the same consensus sequence (1,3). If this region were directly recognized by Gfi-1B, it would be also protected in U937 cells which contain Gfi-1 but not Gfi-1B (22). We then tested whether these two tandem AATC sequences play a role in negative regulation of Gfi-1B promoter. The Gfi-1B promoter reporter plasmid containing mutations of these two sites was constructed. The wild-type and mutated-type reporters were transfected into K562 cells for the promoter activity analysis (Figure 2B). Interestingly, mutation of these two AATC sequences reduced the promoter activity to 34% relative to the wild-type promoter, indicating that these two AATC sites act as critical positive, rather than negative, cis-elements in controlling the Gfi-1B promoter activity in K562 cells. Overexpression of Gfi-1B significantly decreased the luciferase activity expressed from the wild-type promoter, but not the AATC mutated-type reporter, indicating that these two AATC sites still contribute to the responsiveness to Gfi-1B-mediated repression. Accordingly, we speculated that a positive regulator binding to these two AATC sites may mediate repression effect of Gfi-1B on its own promoter.
Figure 2.
The Gfi-1-like sequences in the human Gfi-1B promoter act as a positive cis-element in transcription. (A) Schematic representation of the Gfi-1-like sites in the Gfi-1B promoter. (B) K562 cells were co-transfected with wild-type or AATC mt (−70/−41 mutation) of pGL2-hG (−325/+19) reporter construct together with pCMV-β-Gal in combination with either control vector or Gfi-1B-FLAG expression vector as indicated. The luciferase activities after normalization by β-galactosidase were calculated relative to that in the cells transfected with wild type of pGL2-hG (−325/+19) and control vector, which was set arbitrarily to 1. The data are means ± SD (n = 3) from three independent experiments.
GATA-1 binds to the AATC sites
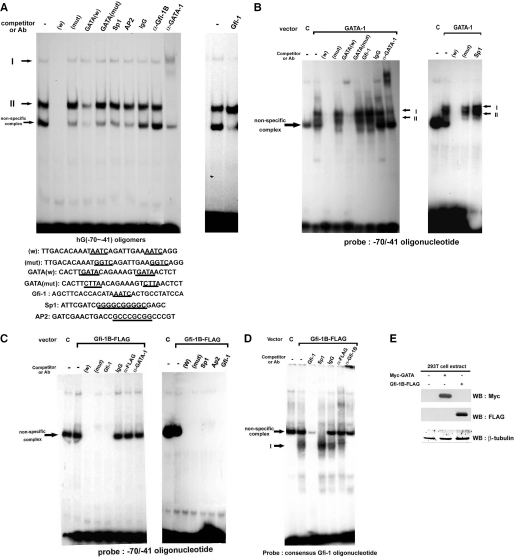
To examine whether the sequence containing two tandem repeats of AATC can be recognized by Gfi-1B, we performed the in vitro DNA gel-shift assay using radio-labeled DNA fragment covering −70/−41 region of the Gfi-1B promoter sequence. Incubation of this probe with nuclear extract of K562 cells gave rise to DNA–protein complexes I and II. These two complexes were abolished by excess amounts of the unlabeled oligomer (Figure 3A), but not by the oligomer carrying mutations in two AATC sites, indicating their specificity to the sequence containing AATC. One retarded complex seen in the assay was indicated to be non-specific, because various oligomers, including −70/−41 of Gfi-1B promoter, GATA, Gfi-1, AP2 and Sp1 consensus sequences, could also compete for its formation. Excess amount of unlabeled Gfi-1 oligomer, which contains typical AATC core sequence, was unable to compete for complex II formation, but with some capacity of competing for complex I. However, complex I formation was unaffected by pre-incubation of K562 extract with specific Gfi-1B antibody. Interestingly, complex II formation could be supershifted by the specific GATA-1 antibody and competed by excess amount of unlabeled consensus GATA oligomer, while the mutated GATA oligomer with a deviation of GATA to CTTA was unable to compete for complex II formation. Unlike the denoted non-specific complex, complex II formation was unaffected by either AP2, Sp1 or Gfi-1 oligomers. Altogether, these results strongly suggest that GATA-1 binds to the tandem AATC sites within the −70/−41 region. Since complex I could not be competed by the GATA consensus oligomer, binding of GATA-1 is unlikely to be involved in this complex formation. Although there is a third AATC site located on the reverse strand within the −70/−41 region, we found that GATA-1 was unable to bind to the AATC mutated probe, which retains the intact third AATC sequence on the reverse strand (data not shown). Nor did mutation of this third AATC site decrease the Gfi-1B promoter activity expressed in K562 cells (data not shown). Here, we excluded the possibility that GATA-1 can bind to the Gfi-1B promoter via the third AATC on the reverse strand.
Figure 3.
GATA-1 can bind to AATC sequences in the Gfi-1B promoter. (A) Gel-shift assays of the −70/−41 region of the human Gfi-1B promoter. The 32P-labeled double-stranded oligonucleotide containing −70/−41 sequence of the Gfi-1B promoter was used for the assays in the absence or presence of unlabeled competitor (80×) with 10 µg of nuclear protein extracted from K562 cells as indicated. The sequence of each competitor is shown below. Control IgG, Gfi-1B (H-150) and GATA-1 (C-20) antibodies were added to the reaction mixtures where indicated. (B) Gel-shift assays of the −70/−41 region of the human Gfi-1B promoter using 2.5 µg of nuclear extracts from 293T cells overexpressing Myc-GATA-1. For the competition experiments, various unlabeled competitor as indicated in 80-fold molar excess was included in the reaction mixture. GATA-1 (C-20) antibody was added to the reaction mixtures. (C) Gel-shift assays of the −70/−41 region of the human Gfi-1B promoter using 2.5 µg of nuclear extracts from 293T cell overexpressing Gfi-1B-FLAG. For the competition experiments, various unlabeled competitor in 80-fold molar excess was included in the reaction mixture. GATA-1 (C-20) or FLAG (M2) antibody was added to the reaction mixtures where indicated. (D) Consensus Gfi-1 DNA-binding oligomer was 32P-labeled for gel-shift assay using 2.5 µg of nuclear extracts from 293T cell overexpressing Gfi-1B-FLAG protein. Each assay contained 2.5 µg of nuclear protein extracted from 293T cells expressing Gfi-1B-FLAG protein. For the competition experiments, unlabeled competitor in 80-fold molar excess was included in the reaction mixture. Gfi-1B (H-150) or FLAG (M2) antibody was added to the reaction mixtures where indicated. (E) 293T cells were transfected the control vector, pMyc-GATA-1 and Gfi-1B-FLAG expression vector as indicated. After transfection, 30 µg of total lysate protein was subjected to SDS–PAGE and western blot analysis using antibodies against Myc (9E10), Gfi-1B-FLAG (M2), and β-tubulin.
Since 293T cells do not express Gfi-1B and GATA-1, we then ectopically expressed GATA-1 or Gfi-1B-FLAG in this cell line for nuclear extract preparation and gel-shift assays. Radio-labeled −70/−41 probe formed specific complexes I and II with nuclear extracts of cells expressing GATA-1. These two complexes were abolished by excess amount of unlabeled GATA consensus oligonucleotide but not by Gfi-1 oligomer (Figure 3B). In contrast, nuclear extract of cells expressing Gfi-1B was unable to form any specific 7complex with the −70/−41 probe (Figure 3C). One complex formed in these gel-shift assays shown in Figure 3B and C was denoted non-specific, since this complex could be competed by various unrelated oligomers when using 293T cell extracts expressing either Gfi-1B-FLAG or GATA-1 for the assays. To test whether Gfi-1B protein in this nuclear extract is functional in binding to the consensus Gfi-1 oligonucleotide, we then prepared the radio-labeled Gfi-1 consensus oligomer for the gel-shift assay. As shown in Figure 3D, nuclear extracts of cells expressing Gfi-1B-FLAG did form specific complex with consensus Gfi-1 oligomer probe, which could be super-shifted by the specific Gfi-1B or FLAG antibody and abolished by excessive amount of unlabeled consensus oligomer. Therefore, lack of complex formation between the Gfi-1B promoter probe with extracts containing Gfi-1B protein is not due to that the Gfi-1B-FLAG protein expressed in 293T cells is defective in specific DNA binding. Figure 3E showed the ectopic expression of Myc-GATA-1 and Gfi-1B-FLAG in nuclear extracts of 293T cells. These gel-shift data led us to conclude that GATA-1, instead of Gfi-1B, binds to the AATC sites within the −70/−41 region of the human Gfi-1B promoter.
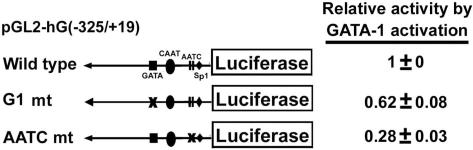
Both non-typical and typical GATA sites contribute to GATA-1-mediated transcriptional activation of Gfi-1B gene
Our previous study has shown that in K562 cells GATA-1 binds to the typical GATA-site located at −132/−129 of the Gfi-1B promoter to mediate transcriptional activation (17). Here, we further compared the contribution of the typical GATA (G1) site at −132/−129 and non-typical GATA sites (two AATC at −59/−56 and −47/−44 positions) in GATA-1-mediated transcription of the Gfi-1B promoter. Co-transfection with GATA-1 expression vector for the reporter assay indicated that either mutation of GATA site at the G1 or two AATC sites reduced the GATA-1-activating effect significantly (Figure 4). Thus, both GATA and AATC sites are recognized by GATA-1 for transcriptional activation.
Figure 4.
Both non-typical and typical GATA-1 sites contribute to GATA-1-mediated transcriptional activation of Gfi-1B genes. The pGL2-hG (−325/+19) constructs of wild type, G1 mt (−132/−129 mutation) or AATC mt (−70/−41 mutation) were each co-transfected with the GATA-1 expression vector in K562 cells, and luciferase activities were determined. The luciferase activities were standardized relative to β-galactosidase expression and expressed relative to wild type of pGL2-hG (−325/+19) that was set arbitrarily to 1. The data are means ± SD from three independent experiments.
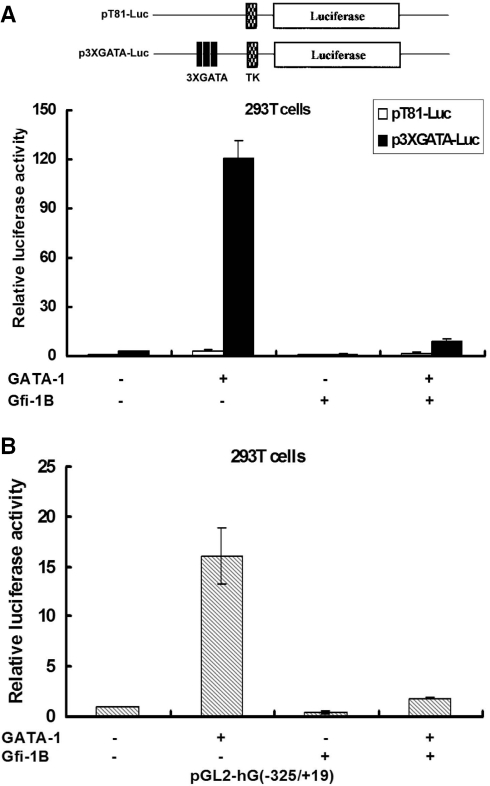
Gfi-1B inhibits GATA-1-mediated transcription
Given that GATA-1 is essential for activating the Gfi-1B promoter, we then raised the question whether GATA-1-mediated transcription is inhibited by co-expression of Gfi-1B protein. To address this question, we transfected 293T cells with 3XGATA-reporter plasmid and the control plasmid pT81-Luc with expression vector of GATA-1 in the presence or absence of Gfi-1B-FLAG co-expression (Figure 5A). It turned out that co-expression of Gfi-1B specifically repressed GATA-1-dependent activation of luciferase activity derived from 3XGATA-reporter. Similar results were also seen in the cells that were transfected with Gfi-1B promoter luciferase construct (Figure 5B). Clearly, Gfi-1B negatively auto-regulates its own promoter through repressing GATA-1-mediated transcription. Since Gfi-1B is unable to bind to GATA sequence, these data also suggest that Gfi-1B-mediated repression need not work through its binding to the recognition sequence.
Figure 5.
Gfi-1B inhibits GATA-1-mediated trans-activation in 293T cells. (A) 293T cells were transfected with a reporter plasmid containing 3XGATA-specific binding sequences or a basal thymidine kinase promoter (pT81-Luc) in the presence of the control, GATA-1, Gfi-1B or GATA-1 plus Gfi-1B expression vector as indicated. The luciferase activities after normalization by were calculated relative to that in the cells co-transfected with the control vector. The data represent averages from three independent experiments. (B) 293T cells were transfected with the pGL2-hG (−325/+19) and pCMV-β-Gal together with control vectors, GATA-1, Gfi-1B alone or GATA-1 and Gfi-1B expression vector as indicated. The luciferase activities after normalization by β-galactosidase were calculated relative to that in the cells transfected with wild type of pGL2-hG (−325/+19) and control vector, which was set arbitrarily to 1. The data represent averages from three independent experiments.
Gfi-1B interacts with GATA-1
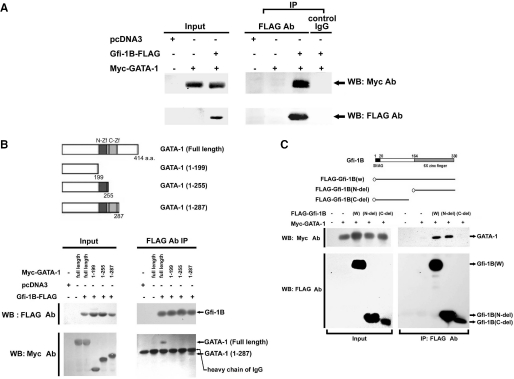
To assess the mechanism responsible for the repression of GATA-1-mediated transcription by Gfi-1B, we then tested whether Gfi-1B interacts with GATA-1 by transfecting 293T cells with expression vectors of Gfi-1B-FLAG and Myc-GATA-1 for the co-immunoprecipitation experiment. As shown in Figure 6A, Myc-GATA-1 was co-immunoprecipiated with Gfi-1B-FLAG by anti-FLAG M2 antibody in the cell lysate co-expressing Gfi-1B-FLAG and Myc-GATA-1, while no myc-GATA-1 was precipitated in the cell lysate expressing Myc-GATA-1 alone. This result suggests that co-expression of both proteins can form a specific complex in the cells. We then further defined the domain necessary for their interaction. GATA-1 contains two zinc fingers. One is the carboxyl finger spanning 257–287 amino acids which is essential for DNA binding and the other N-terminal finger at 199–255 amino acids which stabilizes its binding to the GATA sequence (23–25). We then constructed different deleted forms of pMyc-GATA-1 for the co-immunoprecipitation experiment. After co-expression with Gfi-1B-FLAG in 293T cells, we found that both the wild-type and 1–287, but not 1–199 or 1–255, forms of Myc-GATA-1, formed a complex with Gfi-1B-FLAG (Figure 6B), indicating that carboxyl finger of GATA-1 is necessary for its interaction with Gfi-1B protein.
Figure 6.
Gfi-1B interacts with GATA-1. (A) Expression vector of Myc-GATA-1 together with either the control (C) or Gfi-1B-FLAG expression vector was transfected into 293T cells. After transfection for 24 h, whole cell lysates were prepared and immunoprecipitated with anti-FLAG (M2) beads or control IgG. The immunoprecipitates were subjected to immunoblot analysis with anti-Myc antibody. The blot was reprobed with anti-FLAG (M2) antibody to confirm that Gfi-1B was successfully immunoprecipitated. The input indicated 10% of the whole cell lysate used for the co-immunoprecipitation. (B) Schematic representation of wild-type and deletion constructs of GATA-1 (upper panel). N-ZF indicates N-terminal zinc finger; C-ZF, C-terminal zinc-finger. Various deleted constructs of Myc-GATA-1 were co-transfected with Gfi-1B-FLAG into 293T cells. Cells were harvested for immunoprecipitation, and the immunoprecipitates were subjected to immunoblot analysis with anti-Myc (9E10) and anti-FLAG (M2) antibody as described above. (C) Schematic representation of deletion constructs of Gfi-1B (upper panel). Control vector, wild type, N-del (164–330) or C-del (1–137) deletion construct of FLAG-Gfi-1B was costransfected with Myc-GATA-1 into 293T cells. Cells were harvested for immunoprecipitation and the immunoprecipitates were subjected to immunoblot analysis with anti-Myc (9E10) and anti-FLAG (M2) antibody.
Gfi-1B contains a SNAG domain in the N-terminus mediating its transcriptional repression and six zinc-finger domains at its C-terminus for DNA binding (1–3). To examine the domain of Gfi-1B that is involved in complex formation with GATA-1 protein, we expressed various deleted forms of FLAG-Gfi-1B with Myc-GATA-1 in 293T cells for co-immunoprecipitation experiment. As shown in Figure 6C, deletion of carboxyl zinc-finger (C-del) abrogated the interaction of FLAG-Gfi-1B with myc-GATA-1, indicating the requirement of the carboxyl zinc-finger of Gfi-1B for its interaction with GATA-1.
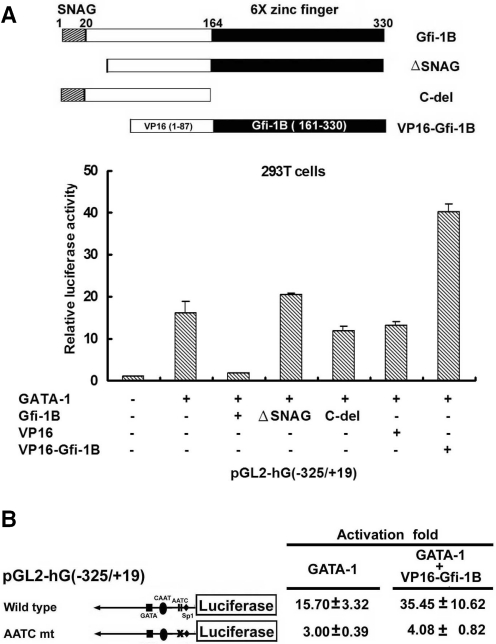
The SNAG domain of Gfi-1B is required for inhibiting GATA-1-mediated trans-activation
To know whether overexpression of Gfi-1B affects the DNA-binding activity of GATA-1, we performed the gel-shift assay using consensus GATA binding oligonucleotides as the probe with the 293T nuclear extracts expressing GATA-1. Inclusion of increasing amounts of Gfi-1B-FLAG in nuclear extract did not perturb the specific GATA-1/DNA complex formation in the binding reaction (data not shown), indicating that Gfi-1B does not interfere with DNA-binding activity of GATA-1. We then turned to determine whether the repression domain of Gfi-1B is responsible for suppressing GATA-1-mediated trans-activation. Toward this end, Gfi-1B promoter luciferase construct was co-transfected with various deleted forms of FLAG-Gfi-1B constructs and GATA-1 expression vector into 293T cells. As shown in Figure 7A, removal of SNAG repression domain relieved the repressive effect of Gfi-1B on GATA-1-mediated transcription, suggesting the SNAG domain is required for suppressing GATA-1-mediated transcription of the Gfi-1B promoter. Because the C-del Gfi-1B loses the ability to interact with GATA-1, as expected, its expression did not affect GATA-1-mediated trans-activation. Furthermore, replacement of SNAG domain by a herpes virus trans-activating protein, VP16, converted Gfi-1B to an activator for GATA-1-mediated transcriptional activation of Gfi-1B promoter. Thus, the interaction between GATA-1 and Gfi-1B does not affect the DNA-binding activity of GATA-1, and the SNAG domain of Gfi-1B contributes to its repression function on GATA-1-mediated transcription. To ascertain that the functional effect of Gfi-1B on its own promoter is indeed through GATA-1 binding to the AATC sites, we compared the effect of VP16-Gfi-1B on GATA-1-mediated activation of wild-type and AATC-mut Gfi-1B promoter reporter. As shown in Figure 7B, stimulation of GATA-1-mediated activation of Gfi-1B promoter by VP16-Gfi-1B was diminished by disrupting the AATC sites. In summary, Gfi-1B can specifically interact with GATA-1, by which GATA-1-dependent transcription can be suppressed by Gfi-1B-mediated repression via its SNAG domain.
Figure 7.
The SNAG domain of Gfi-1B is required for repressing GATA-1-mediated trans-activation. (A) 293T cells were transfected with pGL2-hG (−325/+19) together with with pCMV-β-Gal in combination with either control vector, GATA-1 alone or GATA-1 plus wild-type, ΔSNAG (45–330) or C-del (1–137) truncated FLAG-Gfi-1B expression vector as indicated. All the luciferase activities were calculated and expressed as described in the legend to Figure 5B. The data represent averages from three independent experiments. (B) 293T cells were transfected with the wild-type or AATC mt of pGL2-hG (−325/+19) constructs together with the expression vector of GATA-1 and pCMV-β-Gal in the presence of control vector or VP16-Gfi-1B plasmid. Luciferase activities were determined and expressed as described in the legend to Figure 5B. The data are means ± SD from three independent experiments.
GATA-1-dependent binding of Gfi-1B to its own promoter
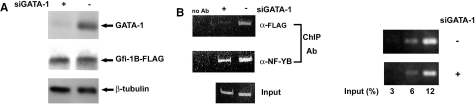
Finally, we tested whether GATA-1 is required for Gfi-1B-FLAG binding to the Gfi-1B promoter in K562 cells. Toward this end, we depleted the expression of GATA-1 in K562 cells stably expressing Gfi-1B-FLAG by transfecting these cells with siRNA of GATA-1 (Figure 8A). We then performed the chromatin immunoprecipitation experiments in Gfi-1B-FLAG expressing cells with FLAG antibody to determine the effect of GATA-1 depletion on Gfi-1B-FLAG binding to the Gfi-1B promoter region. As shown in Figure 8B, FLAG antibody was able to immunoprecipitate formaldehyde-fixed chromatin containing endogenous Gfi-1B promoter DNA from cells without GATA-1 depletion. For cells depleted of GATA-1, FLAG antibody no longer immunoprecipitated endogenous Gfi-1B promoter DNA, indicating the necessary role of GATA-1 in mediating Gfi-1B association with its own promoter in vivo. Our previous study has shown that NF-Y binds to the Gfi-1B promoter for transcriptional activation. Here, we showed that GATA-1 depletion did not significantly alter NF-Y binding with the endogenous Gfi-1B promoter region, indicating the specific effect of GATA-1 on Gfi-1B-FLAG binding to the human Gfi-1B promoter. These ChIP results further supported the role of GATA-1 as a mediator in Gfi-1B auto-regulation.
Figure 8.
GATA-1 is required for Gfi-1B-FLAG occupancy at the Gfi-1B promoter. (A) The pS2-Gfi-1B-FLAG retrovirus integrated K562 cells (3 × 106) were electroporated with or without GATA-1 siRNA. Total cell lysates from the electroporated K562 cells were subjected to western blot analysis using antibodies against GATA-1 (C-20), Gfi-1B (H-150) and β-tubulin. (B) ChIP assays were performed with equal amount of the electroporated pS2-Gfi-1B-FLAG cells (3 × 106) as described in Materials and Methods. Samples of sonicated and purified chromatin were immunoprecipitated with anti-FLAG M2 beads or anti-NF-YB (C-20) antibody as indicated, and DNA isolated from immunoprecipitated material was amplified by PCR with primers specific for the Gfi-1B promoter. Different amounts of chromatin samples prior to immunoprecipitation were also amplified to indicate the linearity of PCR for the Gfi-1B promoter and relative amount of total input chromatin.
DISCUSSION
It is well known that the auto-regulatory loop provides a mechanism by which the concentration of the transcriptional regulator is highly controlled. Gfi-1B is a transcriptional repressor, whose function has been shown to directly repress transcription of Socs1 and Socs3 in hematopoietic cells (9). Recent studies using retroviral vector-mediated expression of Gfi-1B further showed that elevated expression of Gfi-1B alters behavior of proliferation and differentiation of erythroid cells at different developmental stage (5,11), suggesting that its expression level needs to be regulated within the erythroid differentiation program. In this work, our data suggest that Gfi-1B is auto-regulated in a negative manner. However, unlike other auto-regulatory loop, here we show that the expression of Gfi-1B regulates its own promoter through its interaction with GATA-1. These results also suggest a new mode of Gfi-1B action, in which Gfi-1B negatively modulates GATA-1-mediated transcription through protein interaction without direct binding to its recognition sequence.
Our previous study has shown that GATA-1 binds to the Gfi-1B promoter at −132/−129 GATA site (17) and plays a necessary role in trans-activating the Gfi-1B promoter in K562 cells. However, GATA mutation at the −132/−129 site did not abolish GATA-1-mediated activation of the Gfi-1B transcription, indicating that additional GATA-1 recognition site is present in the promoter. Using nuclear extracts from cells overexpressing GATA-1, our gel-shift data revealed that GATA-1 also binds to the region containing two tandem AATC (GATT in the reverse strand) sites at −59/−56 and −47/−44. Moreover, GATA-1-mediated activation of Gfi-1B promoter was diminished by mutations introduced to these two AATC sites, suggesting GATA-1 works on these two non-typical GATA sites for trans-activation.
The gene targeting experiments have demonstrated that both GATA-1 and Gfi-1B genes are essential for erythropoiesis (5,10,11,26–28). The expression patterns of these two genes have been shown to be well correlated throughout successive differentiation stages from erythroid-blast-forming units to mature erythroblasts (5). Data from our studies using the promoter analysis indicate that GATA-1 works on GATA and AATC sites to activate Gfi-1B transcription, which accounts for their co-expression pattern. Interestingly, here we show that the mechanism of GATA-1-mediated transcriptional activation of Gfi-1B expression is intriguingly restricted by negative auto-regulation of Gfi-1B through their interaction, by which GATA-1-mediated transcription is repressed via the SNAG domain of Gfi-1B. Conceivably, this auto-regulatory mechanism provides a means to limit the expression level of Gfi-1B in erythroid cells. Since GATA-1 is a central regulator in terminal differentiation of erythroid cells (27,29,30), this auto-regulatory loop is particularly important in controlling the amount of Gfi-1B expression in a range that would not perturb the transcription function of GATA-1 during erythropoiesis.
Up to now, LMO-2 (31,32), FOG-1 (33), Pu.1 (34–36), CBP (37), Fli-1 (38), Ski (39), EKLF/Sp1 (40) and Herp2 (41) have been shown to interact with GATA-1. Among them, Pu.1 and Ski repress GATA-1-mediated trans-activation by inhibiting its DNA-binding activity through their physical interaction (36,39). Unlike these two negative regulators but similar to Herp2 (41), Gfi-1B did not affect DNA-binding activity of GATA-1. Rather, the inhibitory effect is dependent on its SNAG repression domain, since replacement of SNAG domain by VP16 converts Gfi-1B from a co-repressor to a co-activator in GATA-1-dependent transcription. Our finding adds Gfi-1B to the list of transcriptional regulators that interact with GATA-1. It should be mentioned that we did not find the direct interaction between these two proteins when yeast two-hybrid system was used for the interaction assay (data not shown). Therefore, it is possible that Gfi-1B and GATA-1 may require the involvement of other cellular factor or certain post-translational modification for their complex formation. If this is true, it would suggest that the inhibitory effect of Gfi-1B on GATA-1 is modulated in a cell context-dependent manner. Alternatively, it is also possible that the effect of Gfi-1B on GATA-1 function depends on other interacting factors and the gene promoter context as well. More experiments are needed to define the factors involved in their functional interaction for further understanding how the interplay between GATA-1 and Gfi-1B is regulated during terminal maturation and survival of erythroid cells.
While this manuscript was in its revision preparation, Rodriguez et al. (19) reported that Gfi-1B is present in the pull-down complexes containing GATA-1 protein in MEL cells and that Gfi-1B and GATA-1 are bound to the myc and myb promoters in vivo. Accordingly, they proposed that GATA-1 forms a repressive complex with Gfi-1B to repress expression of myc and myb, thus prohibiting proliferation of erythroid cells that express elevated level of Gfi-1B. While their hypothesis remains to be elucidated, data from our ChIP and reporter analyses demonstrated that Gfi-1B represses and binds to its own promoter in vivo dependent on the presence of GATA-1, providing the first direct evidence of Gfi-1B/GATA-1 repression mechanism. Apparently, the Gfi-1B promoter is one typical target gene for GATA-1/Gfi-1B complex-mediated repression in erythroid cells. Taken together, we propose that the expression of Gfi-1B and the function of GATA-1 in erythroid lineage cells are mutually regulated to finely tune their functions.
Although AATC is the core sequence of a consensus Gfi-1 binding site (1,3), data from different approaches in this study suggest that Gfi-1B cannot bind to this DNA region of its own promoter. First, we found that the DNA–protein complexes formed in the gel-shift assay using −70/−41 probe with nuclear extracts of K562 cells could not be competed by Gfi-1 binding consensus oligomer. Although this probe could also form another prominent complex with K562 extract, we found that all unrelated oligomers tested in this study competed for this complex formation, indicating the non-specific nature of this complex in our gel-shift assay (Figure 3A). Second, Gfi-1B expressed in 293T cells did not form any specific complex when Gfi-1B promoter sequence was used as the probe for the gel-shift assay, while the same extract could form specific complex with the Gfi-1 consensus oligomer probe. All our results suggest that these two AATC sites are preferentially recognized by GATA-1 in K562 cells for transcriptional activation of Gfi-1B gene.
Contradictory to the results shown here, the report by Vassen et al. (18) has described that in vitro translated Gfi-1B formed specific complexes with the mouse Gfi-1B promoter sequence probe covering the two AATC sites in the gel-shift assays. Their results suggest that Gfi-1B can repress its own promoter by recognizing these AATC sites directly. The discrepancy in Gfi-1B binding to these AATC sites in these two studies could be due to the difference in preparation of Gfi-1B protein for the gel-shift assays. In the present study, we used Gfi-1B expressed in nuclear extract for the binding assay, while in vitro translated Gfi-1B in reticulocyte lysate was used in their assay condition. Perhaps, co-factor present in reticulocyte lysate can assist or promote the binding of Gfi-1B to this sequence. It is worth of noting that in their report, inclusion of excess amount of unlabeled Gfi-1B promoter sequence did not abolish the DNA/protein complex formation between the consensus Gfi-1 oligomer and in vitro translated Gfi-1B, indicating a higher binding affinity of Gfi-1B to the consensus Gfi-1 oligomer than to the Gfi-1B promoter sequence. Their study has shown that enforced expression of Gfi-1B under the control of the vav promoter in transgenic mice inhibits endogenous Gfi-1B transcription in spleen but not in bone marrow, suggesting that negative auto-regulation of Gfi-1B is highly dependent on the cellular context. According to our data in K562 cell, the preferential binding of GATA-1 to the AATC sites and the interaction capability between GATA-1 and Gfi-1B make a cellular context for the negative auto-regulation of Gfi-1B through GATA-1. Nevertheless, here we do not exclude the possibility that Gfi-1B when expressed in abundant amount may still be able to recognize the Gfi-1B promoter through direct binding to the AATC sites in NIH3T3 cells that are deficient in GATA-1 expression. Our findings in K562 cells may represent a situation that an increase of Gfi-1B expression leads to a repression complex formation through interacting with GATA-1 that has occupied at the promoter. Thus, GATA-1 acts as an activator or a repression mediator by sensing the amount of Gfi-1B present in K562 cells.
Acknowledgments
The authors are grateful to Dr Atsushi Iwama for the gift of reporter construct pT81-Luc and Dr Dave Turner for providing the expression vector pCS2-MT. This research is supported by grant NSC 93-2320-B-002-110, NSC93-3112-B-002-012 and NSC93-2752-B-002-006-PAE from the National Science Council, Taiwan, R.O.C. Funding to pay the Open Access publication charges for this article was provided by the National Science Council, Taiwan, ROC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tong B., Grimes H.L., Yang T.Y., Bear S.E., Qin Z., Du K., El-Deiry W.S., Tsichlis P.N. The Gfi-1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol. Cell. Biol. 1998;18:2462–2473. doi: 10.1128/mcb.18.5.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grimes H.L., Chan T.O., Zweidler-McKay P.A., Tong B., Tsichlis P.N. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zweidler-Mckay P.A., Grimes H.L., Flubacher M.M., Tsichlis P.N. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodel B., Wagner T., Zornig M., Niessing J., Moroy T. The human homologue (GFI1B) of the chicken GFI gene maps to chromosome 9q34.13-A locus frequently altered in hematopoietic diseases. Genomics. 1998;54:580–582. doi: 10.1006/geno.1998.5601. [DOI] [PubMed] [Google Scholar]
- 5.Osawa M., Yamaguchi T., Nakamura Y., Kaneko S., Onodera M., Sawada K., Jegalian A., Wu H., Nakauchi H., Iwama A. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood. 2002;100:2769–2777. doi: 10.1182/blood-2002-01-0182. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J., Guo L., Min B., Watson C.J., Hu-Li J., Young H.A., Tsichlis P.N., Paul W.E. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity. 2002;16:733–744. doi: 10.1016/s1074-7613(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 7.Karsunky H., Mende I., Schmidt T., Moroy T. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene. 2002;21:1571–1579. doi: 10.1038/sj.onc.1205216. [DOI] [PubMed] [Google Scholar]
- 8.Gilks C.B., Bear S.E., Grimes H.L., Tsichlis P.N. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol. Cell. Biol. 1993;13:1759–1768. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jegalian A.G., Wu H. Regulation of Socs gene expression by the proto-oncoprotein GFI-1B: two routes for STAT5 target gene induction by erythropoietin. J. Biol. Chem. 2002;277:2345–2352. doi: 10.1074/jbc.M105575200. [DOI] [PubMed] [Google Scholar]
- 10.Saleque S., Cameron S., Orkin S.H. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 2002;16:301–306. doi: 10.1101/gad.959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcon L., Lacout C., Svinartchouk F., Le Couedic J.P., Villeval J.L., Vainchenker W., Dumenil D. Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood. 2005;105:1448–1455. doi: 10.1182/blood-2003-11-4068. [DOI] [PubMed] [Google Scholar]
- 12.Angel P., Hattori K., Smeal T., Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 13.Chen H., Ray-Gallet D., Zhang P., Hetherington C.J., Gonzalez D.A., Zhang D.E., Moreau-Gachelin F., Tenen D.G. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene. 1995;11:1549–1560. [PubMed] [Google Scholar]
- 14.Grass J.A., Boyer M.E., Pal S., Wu J., Weiss M.J., Bresnick E.H. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl Acad. Sci. USA. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timchenko N., Wilson D.R., Taylor L.R., Abdelsayed S., Wilde M., Sawadogo M., Darlington G.J. Autoregulation of the human C/EBP alpha gene by stimulation of upstream stimulatory factor binding. Mol. Cell. Biol. 1995;15:1192–1202. doi: 10.1128/mcb.15.3.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai S.F., Strauss E., Orkin S.H. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 17.Huang D.Y., Kuo Y.Y., Lai J.S., Suzuki Y., Sugano S., Chang Z.F. GATA-1 and NF-Y cooperate to mediate erythroid-specific transcription of Gfi-1B gene. Nucleic Acids Res. 2004;32:3935–3946. doi: 10.1093/nar/gkh719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassen L., Fiolka K., Mahlmann S., Moroy T. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 2005;33:987–998. doi: 10.1093/nar/gki243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez P., Bonte E., Krijgsveld J., Kolodziej K.E., Guyot B., Heck A.J., Vyas P., de Boer E., Grosveld F., Strouboulis J. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordeen S.K. Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques. 1988;6:454–458. [PubMed] [Google Scholar]
- 22.Duan Z., Horwitz M. Targets of the transcriptional repressor oncoprotein Gfi-1. Proc. Natl Acad. Sci. USA. 2003;100:12905–12910. doi: 10.1073/pnas.1031694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans T., Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 24.Martin D.I., Zon L.I., Mutter G., Orkin S.H. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344:444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- 25.Watamoto K., Towatari M., Ozawa Y., Miyata Y., Okamoto M., Abe A., Naoe T., Saito H. Altered interaction of HDAC5 with GATA-1 during MEL cell differentiation. Oncogene. 2003;22:9176–9184. doi: 10.1038/sj.onc.1206902. [DOI] [PubMed] [Google Scholar]
- 26.Pevny L., Simon M.C., Robertson E., Klein W.H., Tsai S.F., D'Agati V., Orkin S.H., Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 27.Shivdasani R.A., Fujiwara Y., McDevitt M.A., Orkin S.H. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujiwara Y., Browne C.P., Cunniff K., Goff S.C., Orkin S.H. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl Acad. Sci. USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantor A.B., Orkin S.H. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 30.Orkin S.H. Diversification of haematopoietic stem cells to specific lineages. Nature Rev. Genet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- 31.Wadman I.A., Osada H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osada H., Grutz G., Axelson H., Forster A., Rabbitts T.H. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proc. Natl Acad. Sci. USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsang A.P., Visvader J.E., Turner C.A., Fujiwara Y., Yu C., Weiss M.J., Crossley M., Orkin S.H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 34.Zhang P., Behre G., Pan J., Iwama A., Wara-Aswapati N., Radomska H.S., Auron P.E., Tenen D.G., Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl Acad. Sci. USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rekhtman N., Radparvar F., Evans T., Skoultchi A.I. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nerlov C., Querfurth E., Kulessa H., Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- 37.Blobel G.A., Nakajima T., Eckner R., Montminy M., Orkin S.H. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl Acad. Sci. USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisbacher M., Holmes M.L., Newton A., Hogg P.J., Khachigian L.M., Crossley M., Chong B.H. Protein–protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol. Cell. Biol. 2003;23:3427–3441. doi: 10.1128/MCB.23.10.3427-3441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueki N., Zhang L., Hayman M.J. Ski negatively regulates erythroid differentiation through its interaction with GATA1. Mol. Cell. Biol. 2004;24:10118–10125. doi: 10.1128/MCB.24.23.10118-10125.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merika M., Orkin S.H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elagib K.E., Xiao M., Hussaini I.M., Delehanty L.L., Palmer L.A., Racke F.K., Birrer M.J., Shanmugasundaram G., McDevitt M.A., Goldfarb A.N. Jun blockade of erythropoiesis: role for repression of GATA-1 by HERP2. Mol. Cell. Biol. 2004;24:7779–7794. doi: 10.1128/MCB.24.17.7779-7794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]