Abstract
Myocyte enhancer factor-2 (MEF2) transcription factors are activated by p38 mitogen-activated protein kinase during neuronal and myogenic differentiation. Recent work has shown that stimulation of this pathway is antiapoptotic during development but proapoptotic in mature neurons exposed to excitotoxic or other stress. We now report that excitotoxic (N-methyl-D-aspartate) insults to mature cerebrocortical neurons activate caspase-3, -7, in turn cleaving MEF2A, C, and D isoforms. MEF2 cleavage fragments containing a truncated transactivation domain but preserved DNA-binding domain block MEF2 transcriptional activity via dominant interference. Transfection of constitutively active MEF2 (MEF2C-CA) rescues MEF2 transcriptional activity after N-methyl-D-aspartate insult and prevents neuronal apoptosis. Conversely, dominant-interfering MEF2 abrogates neuroprotection by MEF2C-CA. These results define a pathway to excitotoxic neuronal stress/apoptosis via caspase-catalyzed cleavage of MEF2. Additionally, we show that similar MEF2 cleavage fragments are generated in vivo during focal stroke damage. Hence, this pathway appears to have pathophysiological relevance in vivo.
Paradoxically, we and others have shown that activation of the p38 mitogen-activated protein kinase (MAPK) pathway can lead to either antiapoptotic or proapoptotic effects on neurons (1–4). An important transcription factor that is activated by the p38 stress kinase pathway and thought to mediate these life and death events, at least in part, is myocyte enhancer factor-2 (MEF2) (5, 6). There are four MEF2 proteins (MEF2A, -B, -C, and -D). They represent transcription factors in the MADS (MCM1-agamous-deficiens-serum response factor) family, signifying that they contain a MADS box and act in concert with other MADS domain factors as well as other classes of transcription factors (7–10). Previously, we had cloned and characterized MEF2C and shown that it was the predominant MEF2 family member in cerebrocortical neurons (7), with lesser contributions of MEF2A and -D in this neuronal population (11, 12). Olson and colleagues have shown that MEF2 proteins are also highly expressed in cardiac myocytes and induce myogenesis in conjunction with basic helix-loop-helix transcription factors, as judged from both in vitro and in vivo models; other groups have reported concordant findings (13–15). Moreover, recent work from our group and others has shown that in vitro transfection of stem-like P19 cells or murine embryonic stem cells with a constitutively active form of MEF2C (MEF2C-CA) was antiapoptotic and induced a phenotype with both muscle and neuronal characteristics (4, 16). Forced expression of a dominant-interfering form of MEF2C (MEF2C-DN) prevented retinoic-acid-induced neurogenesis of these cells. Additionally, when constitutively active MEF2A was transfected into postmitotic cerebellar granule cell neurons in the face of an insult, such as extracellular K+ withdrawal, the MEF2 activity promoted survival (i.e., was antiapoptotic) (17).
On the other hand, inhibition of the p38-MEF2 pathway has been shown to prevent neuronal apoptosis induced by N-methyl-d-aspartate (NMDA) or related insults in a variety of in vitro and in vivo paradigms, suggesting that the pathway can be proapoptotic under some conditions (1–3). Heretofore, the basis for this dualistic nature of the p38-MEF2 pathway as either a pro- or antiapoptotic trigger has remained elusive.
Caspases are proteases required for the process of apoptosis, cleaving sensitive proteins specifically at consensus sites encompassing an aspartate residue (18–22). Caspases are synthesized as inactive proenzymes and activated in a proteolytic cascade after exposure to apoptotic signals. A direct interaction between caspases and MEF2 proteins has not been previously demonstrated. While the current study was under review, Li et al. (23) reported cleavage of two MEF2 isoforms (MEF2A and -D) in cerebellar granule cells after K+ withdrawal. Polycaspase inhibitors prevented this cleavage. However, that report did not demonstrate a direct interaction of caspases with MEF2 and was unable to find the exact cleavage sites of MEF2. Therefore, many questions still remain, including that of direct MEF2 cleavage by caspases, which specific caspases may be involved, the mechanism of action of cleaved MEF2 participating in neuronal cell death pathways, and the functional implications of MEF2 cleavage in vivo. Here we show that exposure of cerebrocortical neurons to an excitotoxic concentration of NMDA, a stimulus known to activate the p38 pathway as well as caspase enzymes (1–3, 24–30), leads to direct cleavage of MEF2A, -C, and -D by caspases. We observed similar cleavage of MEF2 after focal ischemia/reperfusion in vivo. Caspase target sites on MEF2 proteins are all in the transactivation domain, with cleavage leading to the formation of endogenous dominant-interfering forms of MEF2. We show that such dominant-negative MEF2s are able to block transcriptional activation by intact MEF2, thereby contributing to neuronal apoptosis. We conclude that the p38-MEF2 pathway signals to produce survival-promoting gene regulation, but when interrupted by excessive caspase activation, this pathway becomes proapoptotic.
Materials and Methods
Plasmids and Transfections.
MEF2-DN (pcDNA1-MEF2C-1–105flag) and MEF2C-CA (pcDNA1-MEF2C-1–117/VP16) have been described previously (4). The construct pcDNA1-MEF2C-1–105flag retains the DNA-binding domain of MEF2 and acts as a dominant-interfering form because of truncation in the transactivation domain at amino acid residue 105 with replacement of the remainder of the sequence with a flag tag for labeling purposes. Conversely, despite the fact that it contains a truncated version of the MEF2C transactivation domain, the construct pcDNA1-MEF2C-1–117/VP16 is a constitutively active form of MEF2 because it encodes a fully active VP16 transactivation domain. The MEF2-dependent luciferase reporter gene was constructed by insertion of two tandem copies of the MEF2 site from the brain creatine kinase enhancer upstream of the basal promoter of the embryonic myosin heavy-chain gene. Point mutations (Asp to Ala) were introduced into pcDNA3-MEF2A, -MEF2C, and -MEF2D expression vectors by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Truncated MEF2 cDNAs were amplified with cloned Pfu polymerase and inserted into pcDNA3. Cerebrocortical neurons from embryonic day 16 rat embryos were cultured as described (24). After 17 days in vitro, cells were transfected with the various constructs (1.5–2 μg) by using LipofectAMINE 2000 (2 μl, Life Technologies, Rockville, MD). For apoptosis assays, an enhanced green fluorescent protein vector (pEGFP-N1, 1 μg) was coexpressed to identify transfected cells. Fourteen hours after transfection, cells were washed and placed in their original conditioned medium.
Luciferase Reporter Gene Assay and Apoptosis Assessment.
To induce apoptosis, transfected cerebrocortical cultures were exposed to NMDA (50–100 μM plus 5 μM glycine for 20 min) in Earle's balanced salt solution (24, 31). After NMDA exposure, cells were rinsed and replaced in their original conditioned medium. Luciferase reporter gene assays were performed 4 h after NMDA exposure by using the Luciferase Assay System (Promega). This time point was chosen because it was adequate for transcriptional alterations but preceded neuronal apoptosis. For assessment of apoptosis, cells were fixed 12–24 h after NMDA exposure. Only neurons were susceptible to NMDA-induced caspase activation, gene regulation, and apoptosis in this culture system (24, 29–31). Nuclear apoptotic morphology was evaluated after staining with Hoechst 33342 dye (1 μg/ml) under epifluorescence microscopy.
Electrophoretic Mobility-Shift Assays (EMSA).
Nuclear extracts were obtained from cerebrocortical cultures. Binding to DNA of MEF2A-D, or their caspase-cleaved fragments that were present in the nuclear extracts, was assayed by using a double-stranded probe labeled with 32P-dCTP that represents the consensus sequence corresponding to the brain creatine kinase MEF2 site. The nuclear lysates are incubated with the labeled probe for 20 min at room temperature, resolved on a 6% native polyacrylamide gel, and exposed to x-ray film. Mutated probe was used as a control. Specific anti-MEF2A-D antibodies were used for supershift analysis.
In Vitro Translation of MEF2 and Caspase Reactions.
Recombinant caspase-3, -6, -7, and -8 were purified from Escherichia coli, and their active concentrations were determined by titration with zVAD.fmk (32, 33). In vitro transcription/translation of each MEF2 isoform was performed in a total reaction volume of 25 μl with [35S]methionine and 0.5 μg of each construct by using the TNT Quick Coupled Transcription/Translation System (Promega). One microliter of this reaction mixture and 100 nM purified caspase were incubated in 20 mM Hepes (pH 7.0)/100 mM NaCl/10 mM DTT/1 mM EDTA/0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/10% sucrose for 2 h at 37°C. The reaction mixtures were then subjected to NuPAGE (Invitrogen) and autoradiography.
Cell Lysate Preparation and Western Analysis.
Four hours after an apoptosis-inducing NMDA exposure, cells were harvested with a rubber policeman, and the cultures were centrifuged at 1,000 × g at 4°C. In some experiments, zVAD.fmk (50 μM) was added to the culture medium 2 h before exposure to NMDA. The pellets were resuspended in M-PER Mammalian Protein Extraction Reagent (Pierce), composed of Complete Protease Inhibitor Mixture (Roche Molecular Biochemicals) and NaVO4. Protein concentrations of supernatants were determined with the Pierce BCA protein assay with BSA as the standard. Proteins were resolved on NuPAGE gels (Invitrogen) and transferred to Immobilon-P membrane filters (Millipore) by using a XCell SureLock MiniCell Electrophoresis System (Invitrogen) according to the manufacturer's instructions. Anti-MEF2A and -MEF2C antibodies were purchased from Santa Cruz Biotechnology and Cell Signaling, respectively. Anti-MEF2D antibody was obtained from Ron Prywes (Columbia University). Anti-NR1 antibody was purchased from Upstate Biotechnology (Lake Placid, NY). All of the antibodies were used following the suppliers' instructions. After incubation with specific peroxidase-conjugated secondary antibodies, membranes were developed with an enhanced chemiluminescence Western blotting detection system (Amersham Pharmacia Biotech).
Caspase-3 and -7 Immunostaining.
Five to nine hours after NMDA exposure, cell cultures were fixed with 4% paraformaldehyde for 15 min at room temperature. Cells were then washed three times in PBS, permeabilized with 0.15% Triton X-100 for 5 min, and washed again. Nonspecific antibody binding was minimized by incubation in blocking solution (3% FBS, 3% BSA in PBS, pH 7.4) for 1 h at room temperature. Primary antibodies were diluted as follows in culture medium (DMEM with 10% FBS/20 mM Hepes, pH 7.4): rabbit antiactive caspase-3 (Cell Signaling, Beverly, MA, 1:1,000), rabbit antiactive caspase-7 (Cell Signaling, 1:20), and monoclonal anti-MAP-2 (Sigma, 1:100) or anti-NeuN (Chemicon, 1:50). After a 3-hr incubation, cells were washed three times in PBS and incubated with the following secondary antibodies diluted 1:100: Texas red anti-mouse IgG heavy and light chain (H+L) (Vector Laboratories) and FITC-conjugated donkey anti-rabbit IgG (H+L) (Jackson ImmunoResearch). After a 2-hr incubation, cells were washed extensively and incubated in Hoechst 33342 dye (1 μg/ml) to fluorescently stain nuclei and allow assessment of nuclear morphology. Stained preparations were examined under deconvolution microscopy (Zeiss Axiovert 100 M by using Intelligent Imaging Innovations software, Denver).
Filament Model of Focal Stroke.
Transient ischemia/reperfusion was performed in vivo by using the intravascular filament model to occlude the middle cerebral artery unilaterally for 2 h in C57BL/6 mice, as detailed elsewhere (34). After a 1-h reperfusion period, the animals were killed under anesthesia and nuclear extracts prepared.
Results
Purified Caspases Cleave Recombinant MEF2A, -C, and -D in Vitro.
In the initial experiments, we tested whether recombinant, in vitro synthesized MEF2A, -C, and -D, representing the MEF2 proteins found in cerebrocortical neurons, could be cleaved by purified caspases in vitro (32, 33). Empirically, by using [S35]-labeled MEF2 proteins analyzed on autoradiograms, we found that MEF2A could be cleaved by equivalent concentrations (100 nM) of caspase-3, -6, -7, and -8; MEF2C by caspase-3 and -7; and MEF2D by caspase-7 (Figs. 1 and 2). The predominant caspase activity in each case, as judged by the density of the cleavage bands, was caspase-3 or -7. Mutation of each site (aspartate to alanine) completely blocked cleavage of the MEF2 protein, confirming the site of enzymatic action (Fig. 2). Importantly, in each case, cleavage occurred in the transactivation domain of MEF2 (Figs. 1 and 2), suggesting that the remaining N-terminal cleavage products containing the DNA-binding domain might act as dominant-interfering forms of the protein for transcriptional regulation.
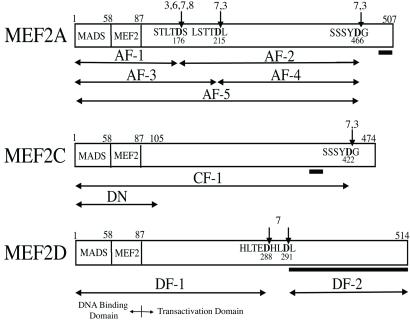
Figure 1.
Caspase cleavage sites of MEF2 proteins. MEF2 proteins contain a conserved N-terminal DNA-binding domain (with MADS and MEF2 domains; amino acids 1–87) and a transactivation domain. Observed caspase cleavage sites on MEF2 family proteins are indicated by arrows (caspases are listed in order of preference for substrate, as judged by density of bands in the autoradiograms of Fig. 2). The caspase-cleaved fragments are designated AF-1 to -5 for MEF2A, CF-1 for MEF2C, and DF-1 and -2 for MEF2D. The MEF2C-DN construct (DN) contains the first 105 amino acids (pcDNA1-MEF2C-1–105flag) and acts as a dominant negative because it lacks the full transactivation domain. The bar underneath the C-terminal segment of each transactivation domain indicates the region that is recognized by isoform-specific MEF2 antibodies.
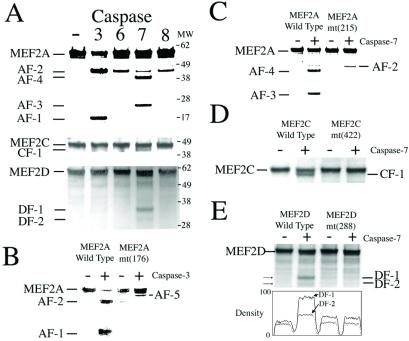
Figure 2.
Identification of caspase cleavage sites on MEF2 proteins. (A) In vitro translated, [35S]methionine-labeled MEF2A, -C, or -D proteins were incubated with purified caspase-3, -6, -7, or -8. Cleaved fragments (corresponding to those shown in Fig. 1) were resolved on NuPAGE and visualized by autoradiography. (B) Mutation of residue 176 of MEF2A from Asp to Ala [MEF2A mt(176)] prevented formation of AF-1 and -2 but enhanced AF-5 formation by caspase-3. (C) Mutation of residue 215 of MEF2A from Asp to Ala [MEF2A mt(215)] prevented formation of AF-3 and -4 by caspase-7. (D) Mutation of residue 422 of MEF2C from Asp to Ala [MEF2C mt(422)] prevented formation of CF-1 by caspase-7. (E) Mutation of residue 288 of MEF2D from Asp to Ala [MEF2D mt(288)] prevented formation of DF-1 and -2. This effect was quantified by densitometry.
NMDA Activates Neuronal Caspase-3 and -7 in Cerebrocortical Neurons.
Because caspase-3 and -7 cleaved the predominant fragments of MEF2 in vitro, it was important to establish whether these caspase enzymes were activated during apoptotic insults in cerebrocortical neurons. Previously, we and others had demonstrated activation of caspase-3 in cerebrocortical neurons as well as other neuronal cell types after apoptosis-inducing stimulation with NMDA or glutamate (24–30). Caspase-3 and -7 manifest similar substrate preferences, so until the recent development of a specific antibody that recognizes active caspase-7, it has been difficult to demonstrate the existence of active caspase-7 in neurons after exposure to NMDA. Using this antibody (which recognizes the new C terminus produced during activation), we found that after NMDA exposure, active caspase-7 is present in cerebrocortical neurons in addition to active caspase-3 (Fig. 3). This is the first demonstration, to our knowledge, of active caspase-7 in cerebrocortical neurons triggered by excitotoxic stress.
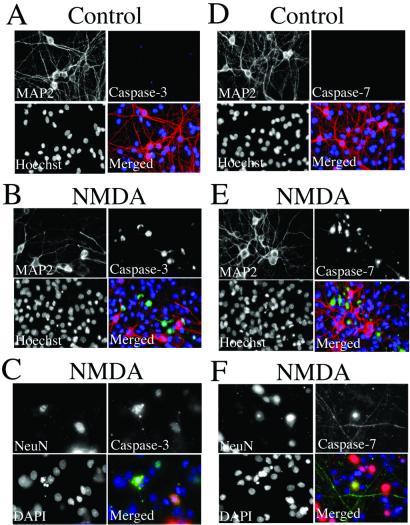
Figure 3.
Activation of neuronal caspase-3 and -7 after NMDA stimulation. Sham-treated (A and D) and NMDA-exposed (B, C, E, F) cerebrocortical cells were fixed and stained with Hoechst or 4′,6-diamidino-2-phenylindole (DAPI) nuclear dye (blue in merged images), anti-MAP-2 or anti-NeuN antibody (red), and antiactivated caspase-3 antibody (green in A–C) or antiactivated caspase-7 antibody (green in D–F). Stained preparations were examined under deconvolution microscopy. Note that images obtained 9 h after exposure to NMDA, a time when neurons destined to undergo apoptosis manifest caspase activity, have often lost their MAP-2 staining (29, 30). Hence, to confirm that these caspases were located specifically in neurons, we used a second neuronal marker more resistant to the initial phases of apoptosis, NeuN.
NMDA-Activated Caspases Cleave Endogenous MEF2 in Cerebrocortical Neurons.
Next, we asked whether endogenous MEF2 proteins could be cleaved by caspases that are activated after NMDA exposure in cerebrocortical neurons. Previously, we had shown that mild insults with NMDA initiated an apoptotic cascade in cultured cerebrocortical neurons (31). This cascade consists of Ca2+ overload of mitochondria, free radical formation, permeability transition, cytochrome c release, and activation of caspase-9 and subsequently caspase-3 (24, 29, 30). Additionally, we and others had demonstrated that excessive NMDA receptor activity could also activate p38 MAPK (1–3). Therefore, initially we sought evidence in our culture system that an apoptosis-inducing exposure to NMDA could activate transcription factors in the MEF2 family, located downstream from p38. Instead, using specific anti-MEF2 antibodies, we found by Western blot that MEF2A, -C, and -D proteins were cleaved within 3 h of NMDA exposure, well in advance of the time of apoptosis, which was observed at 12–24 h (24, 29, 30). The band representing full-length MEF2A was decreased most predominantly, consistent with the multiple sites of cleavage observed in vitro and the fact that the anti-MEF2A-specific antibody recognition site was at the extreme C-terminal end of the molecule (Figs. 1, 2, and 4A). Decrements in MEF2C and -D were also consistently observed in three separate experiments after short exposure to NMDA. As shown in Figs. 1 and 2, MEF2C is expected to be cleaved into a slightly smaller cleavage fragment (designated CF-1; the antibody recognition site is indicated by a bar in Fig. 1). To visualize the CF-1 fragment on Western blots (arrow in Fig. 4A), the blots had to be overexposed such that the decrement in full-length MEF2C was no longer obvious. In the case of MEF2D, two potential cleavage sites are located close to one another and N-terminal to the antibody-binding site (Fig. 1); hence, the larger fragment would not be expected to react with the antibody. In fact, by Western blot, we found a decrease in full-length MEF2D and the expected cleavage fragment DF-2 (arrowhead in Fig. 4A). The addition of the polycaspase inhibitor zVAD.fmk abrogated the NMDA-induced decrement in each MEF2 protein, whereas neither NMDA exposure nor zVAD treatment affected the level of a control protein, NR1, the major subunit of the NMDA receptor.
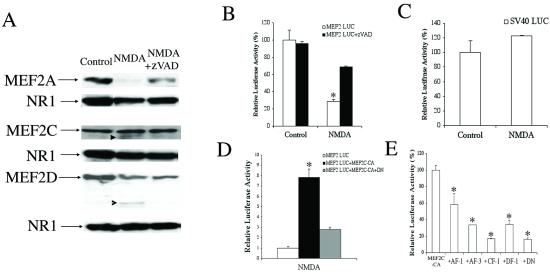
Figure 4.
Caspase-mediated MEF2 cleavage inhibits MEF2 transcriptional activity after NMDA insult. (A) Western blots of cell lysates from control, NMDA-exposed, and NMDA plus zVAD.fmk-treated cultures probed with specific antibodies to MEF2A, -C, or -D. The blots were stripped and incubated with anti-NR1 antibody (the principal subunit of the NMDA receptor) as a loading control. Arrowheads indicate cleaved MEF2C or -D; these cleavage fragments correspond by molecular weight to CF-1 and DF-2, respectively (see Figs. 1 and 2). (B) NMDA insult reduces MEF2 transcriptional activity in luciferase reporter gene assays. The decrement in luciferase activity was prevented by zVAD.fmk (in other experiments, the control compound zFA.fmk had no effect). Cerebrocortical neurons were transfected with a MEF2 reporter gene (MEF2 LUC) before exposure to NMDA ± zVAD. Luciferase activity was measured 4 h after NMDA insult (NMDA-induced killing did not occur until 12–24 h). Baseline activity from untreated cells was assigned a value of 100% (values are mean ± SE; *, P < 0.001 by ANOVA). (C) As a control, transcriptional activity monitored in cells transfected with the simian virus 40 (SV40) enhancer-driven luciferase construct (SV40 LUC) was unaffected after NMDA exposure. (D) Constitutively active MEF2 (MEF2C-CA) prevented the decrease in MEF2 transcriptional activity engendered by NMDA exposure, but this effect was blocked by coexpression of a dominant-interfering form of MEF2 (MEF2C-DN, representing cleavage of MEF2 in the transactivation domain). (E) Truncated MEF2 proteins inhibit MEF2 transcriptional activity. Neurons were transfected with a MEF2 reporter gene (MEF2 LUC), MEF2C-CA, and expression vectors for the caspase cleavage fragments of MEF2 proteins (AF-1, AF-3, CF-1, DF-1) or dominant-negative MEF2 (DN). One day after transfection, luciferase activity was measured. In this case, activity observed after MEF2C-CA transfection was assigned a value of 100%.
NMDA-Mediated Insults Decrease MEF2 Transcriptional Activity in Cerebrocortical Neurons.
The next question we asked was whether the caspase-cleaved MEF2 proteins produced in response to an NMDA insult would affect the transcriptional activity of endogenous MEF2. We predicted that, because the cleavage sites were in the transactivation domain, MEF2 transcriptional activity would be lost by these cleavage fragments. In fact, the fragments containing the DNA-binding domain but without a full-length transactivation domain would be expected to function as dominant-interfering forms of MEF2. To begin to test this premise, we performed reporter gene assays and found that induction of MEF2-dependent luciferase activity was greatly reduced within 4 h of NMDA exposure; zVAD.fmk prevented the decrease in MEF2 transcriptional activity engendered by NMDA exposure (Fig. 4B). As a control, the transcriptional activity of the simian virus 40 promoter was unaffected by NMDA exposure under these conditions, indicating that a nonspecific effect such as cell toxicity was not responsible for this finding (Fig. 4C). Transfection with constitutively active MEF2C-CA (pcDNA1-MEF2C-1–117/VP16) also blocked the decrease in MEF2-dependent luciferase activity produced by NMDA (Fig. 4D). We also found on Western blots that the overexpressed MEF2C-CA was not substantially cleaved by endogenous caspases (data not shown), presumably because of the high levels of MEFC-CA present after transfection. Conversely, cotransfection with a dominant-interfering form, MEF2C-DN (pcDNA1-MEF2C-1–105flag), abrogated the effect of MEF2C-CA, indicating the specificity of the effect. These results suggested that caspase cleavage of the MEF2 proteins resulted in their decreased transcriptional activity. Moreover, the inhibitory effect of pcDNA1-MEF2C-1–105flag, which like each caspase-cleaved MEF2 protein was truncated in the transactivation domain, was consistent with the notion that cleavage in the transactivation domain produces dominant-interfering forms. To further test this premise, cerebrocortical cells were transfected with constructs encoding the caspase-induced cleavage fragments that retained their DNA-binding domains. These fragments of MEF2A, -C, and -D were designated AF-1, AF-3, CF-1 (closely resembling AF-5), and DF-1 (Fig. 1). We found that these MEF2 fragments functioned as dominant-interfering forms by inhibiting the transcriptional activity of MEF2-CA in a similar fashion to MEF2C-DN (Fig. 4E).
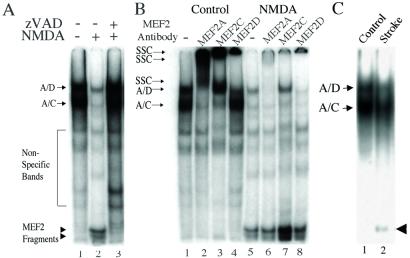
After NMDA-Mediated Insult or Stroke, MEF2 Cleavage Fragments Bind to DNA.
To determine whether the DNA-binding region was indeed present in the MEF2 cleavage fragments, we performed EMSAs. In these experiments, cleaved fragments of MEF2 isoforms in nuclear lysates were shown to bind to the radiolabeled MEF2 site DNA probe, as evidenced by the presence of lower molecular weight bands in the EMSA (Fig. 5A). Specific anti-MEF2A, -C, and -D antibodies supershifted individual bands, indicating that particular MEF2 isoforms were indeed bound to DNA as dimers (Fig. 5B). We did not find evidence for the presence of MEF2B by supershift analysis (data not shown). Note that the vast majority of MEF2 cleavage fragments that bound to DNA did not contain the antibody recognition site for their cognate antibodies. Hence, these small MEF2 cleavage fragments were not supershifted; nonetheless, the cleaved MEF2 fragments that bound to DNA were apparent at the bottom of gel and were observed only coincident with the disappearance of their full length counterparts. Additionally, we found evidence for the production of MEF2 cleavage fragments in vivo within 3 h of the onset of insult in a mouse model of focal middle cerebral artery ischemia/reperfusion (Fig. 5C) (34). Neuronal damage in this stroke model is known to be NMDA receptor mediated.
Figure 5.
EMSA for MEF2 full-length protein and cleavage fragment binding to the MEF2 site on DNA. (A) EMSA for MEF2. Cerebrocortical cultures were incubated for 2 h with vehicle (DMSO, lanes 1 and 2) or zVAD.fmk (lane 3) before stimulation with 100 μM NMDA for 20 min (lanes 2 and 3). Nuclear extracts were prepared 4 h after NMDA insult. EMSAs were performed by using 10 μg of nuclear cell extracts and 32P-labeled oligonucleotide representing the MEF2-binding site. Arrows: A/D, heteromer of full-length MEF2A and MEF2D binding to DNA (as determined by supershift assay; see below); A/C, heteromer of full-length MEF2A and MEF2C binding to DNA (as determined by supershift assay; see below). Arrowheads: low mobility complexes of MEF2 cleavage fragments and DNA. (B) Supershift analysis. Nuclear extracts were prepared from untreated (lanes 1–4) or NMDA exposed (lanes 5–8) cerebrocortical cultures. The nuclear lysates were preincubated with specific anti-MEF2 antibodies and then incubated with 32P-labeled oligonucleotide representing the MEF2-binding site. Arrows: SSC, supershifted complex of full-length MEF2 and DNA; A/D, heteromer of full-length MEF2A and MEF2D binding to DNA; A/C, heteromer of full-length MEF2A and MEF2C binding to DNA. Bottom of gel: low mobility complexes of MEF2 cleavage fragments and DNA. (C) MEF2 cleavage in vivo after stroke. Nuclear extracts were prepared 3 h after focal ischemia/reperfusion to one hemisphere or from the control undamaged hemisphere (n = 6 mice). Representative EMSA shows the presence of MEF2 cleavage fragments that bind to DNA only from the side with stroke damage (arrowhead).
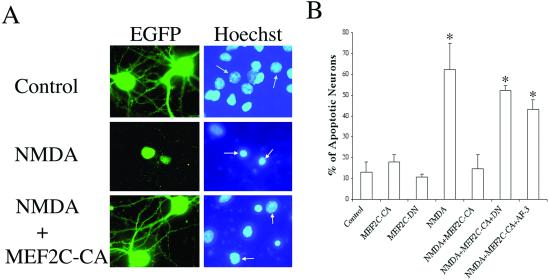
Constitutively Active MEF2C Prevents NMDA-Induced Neuronal Apoptosis, an Effect Reversed by Dominant-Interfering Forms of MEF2C.
We had previously shown that caspase inhibition (with the polycaspase inhibitor zVAD.fmk or the caspase-3, -7 inhibitor DEVD.CHO) could ameliorate NMDA-induced neuronal apoptosis in our cerebrocortical culture system (24, 29, 30). In the present study, we found that transfection with MEF2C-CA (assessed by cotransfection with EGFP) abrogated the apoptotic effect of NMDA (Fig. 6 A and B). Additionally, overexpression of MEF2C-DN or a caspase-generated fragment of MEF2 (e.g., AF-3 in Fig. 1) was able to overcome the antiapoptotic action of MEF2C-CA, arguing against a nonspecific effect (Fig. 6B). Taken together, these results suggest that constitutively active MEF2 could prevent NMDA-induced neuronal apoptosis. Conversely, expression of a dominant-interfering form of MEF2 resulting from cleavage in the transactivation domain was proapoptotic and capable of inhibiting the antiapoptotic effect of active MEF2.
Figure 6.
Dependence of NMDA-induced neuronal apoptosis on MEF2 transcriptional activity. (A) NMDA-induced neuronal apoptosis was prevented by constitutively active MEF2. Cerebrocortical neurons were transfected with an empty vector or an expression vector encoding MEF2C-CA in conjunction with an EGFP expression vector to label transfected cells (green). One day after exposure to NMDA, apoptotic cells were scored by morphological changes with Hoechst nuclear staining (blue); neurons labeled by EGFP are marked by an arrow. Cells were verified to be neurons with MAP-2 staining (not shown). (B) Dominant-interfering forms of MEF2 prevent the neuroprotective effect of constitutively active MEF2. Neurons were transfected with an EGFP expression vector plus expression vectors for control (empty vector), MEF2C-CA, MEF2C-DN, or a caspase-cleaved form of MEF2A (AF-3). Apoptosis in transfected neurons was as-sessed with Hoechst nuclear staining. MEF2C-CA ameliorated NMDA-induced neuronal apoptosis, whereas dominant-interfering forms of MEF2 (MEF2-DN and AF-3) blocked this neuroprotective effect. Values are mean ± SE; *, P < 0.001 by ANOVA.
Discussion
In the developing brain, MEF2 proteins are expressed at the time of initial neuronal differentiation (7, 11, 12, 17, 35, 36). Early in development, MEF2C is found in the cortical plate and later is observed in neurons migrating preferentially to layers II, IV, and VI of the mature cerebrocortex. Because of this distribution, we had previously suggested that MEF2C might be involved in neuronal differentiation (7). Along these lines, more recent work has shown that MEF2C serves an antiapoptotic function and can promote the neurogenic phenotype, as can upstream activation with p38 MAPK (4, 16). In apparent contrast to these findings, in mature neurons, activation of the same p38 MAPK pathway under periods of stress, e.g., induced by excitotoxic insults, has been found to trigger apoptosis, and inhibition of the pathway can prevent this form of neuronal cell death (1–3).
Here we describe a mechanism that can account for a switch from the proneurogenic antiapoptotic effects of the p38 MAPK-MEF2 transcription factor pathway into a proapoptotic cascade in mature neurons. The events entail the activation of neuronal caspases, e.g., by mild NMDA insults, which in turn cleave MEF2 proteins in their transactivation domains. Such cleavage not only renders inactive the transcriptional regulation of MEF2 but also produces dominant-interfering forms that can block the transcriptional activity of MEF2. Under these circumstances, apoptosis ensues after NMDA exposure. These proapoptotic effects of caspase-cleaved MEF2 fragments can be abrogated by inhibition of caspases or by expression of a constitutively active form of MEF2 (MEF2C-CA). Conversely, dominant-interfering forms of MEF2 transcriptional regulation, produced by truncation or caspase cleavage of MEF2 in the transactivation domain while preserving the DNA-binding domain, can reverse the antiapoptotic effect of MEF2C-CA. Taken together, our results show that caspase cleavage of the various MEF2 isoforms found in cerebrocortical neurons renders them transcriptionally inactive, produces dominant-interfering forms of MEF2 that interfere with transcriptional regulation by any remaining intact MEF2, and contributes to apoptotic neuronal cell death.
One of the features of the cerebrocortical culture system used in the present study is that it embodies all of the cell types found in the intact brain (e.g., glia and neurons). However, in this culture system, functional NMDA receptors are found only on neurons (31, 37). Because the NMDA insult is specific for neurons (24, 29–31), caspase activation, gene regulation, and apoptosis in response to NMDA can be studied specifically in this cell type. Importantly, in control experiments, transfection with MEF2C-DN alone did not affect neuronal cell survival under basal conditions. Nonetheless, dominant-interfering forms of MEF2 abrogated the protective effect from NMDA neurotoxicity engendered by MEF2C-CA. Because interference with the p38-MEF2 pathway under basal conditions does not affect neuronal survival, but inhibition of MEF2 transcriptional activity (e.g., with MEF2C-DN or AF-3) during insults with NMDA does invoke cell death, then the p38-MEF2 pathway is apparently necessary only for survival under periods of neuronal cell stress.
In summary, we describe a pathway for neuronal apoptosis involving caspase-catalyzed cleavage of MEF2 transcription factor proteins. This reaction disrupts the normal prosurvival effects of the p38-MEF2 pathway and contributes to neuronal cell death, e.g., after excessive NMDA receptor activation. That caspase inhibitors or constitutively active MEF2 constructs can prevent apoptotic neuronal cell death after NMDA exposure may have therapeutic implications, because a large number of acute and chronic neurodegenerative disorders, ranging from stroke and spinal cord trauma to dementia and glaucoma, involve excessive NMDA receptor stimulation (38, 39).
Acknowledgments
We thank Dr. J. Han (The Scripps Research Institute) for MEF2 constructs and Scott Snipas (The Burnham Institute) for producing the recombinant caspases and performing sequence analyses. This work was supported in part by the Parkinson's Disease Association, San Diego Chapter (S.-i.O.) and National Institutes of Health Grants P01 HD29587, R01 EY05477, and R01 EY09024 (S.A.L.).
Abbreviations
- MAPK
mitogen-activated protein kinase
- NMDA
N-methyl-D-aspartate
- MEF2
myocyte enhancer factor-2
- MADS
MCM1-agamous-deficiens-serum response factor
- EGFP
enhanced green fluorescent protein
- EMSA
electrophoretic mobility-shift assays
References
- 1.Kawasaki H, Morooka T, Shimohama T, Gotoh Y, Nishida E. J Biol Chem. 1997;272:18518–18521. doi: 10.1074/jbc.272.30.18518. [DOI] [PubMed] [Google Scholar]
- 2.Kikuchi M, Tenneti L, Lipton S A. J Neurosci. 2000;20:5037–5044. doi: 10.1523/JNEUROSCI.20-13-05037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee P K, DeCoster M A, Campbell F Z, Davis R J, Bazan N G. J Biol Chem. 1999;274:6493–6498. doi: 10.1074/jbc.274.10.6493. [DOI] [PubMed] [Google Scholar]
- 4.Okamoto S-i, Krainc D, Sherman K, Lipton S A. Proc Natl Acad Sci USA. 2000;97:7561–7566. doi: 10.1073/pnas.130502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Nature (London) 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 6.Zhao M, New L, Kravchenko V V, Kato Y, Gram H, di Padova F, Olson E N, Ulevitch R J, Han J. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leifer D, Krainc D, Yu Y-T, McDermott J, Breitbart R E, Heng J, Neve R L, Kosofsky, Nadal-Ginard B, Lipton S A. Proc Natl Acad Sci USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molkentin J D, Black B L, Olson E. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 9.Pollock R, Treisman R. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 10.Yu Y T, Breitbart R E, Smoot L B, Lee Y, Mahdavi V, Nadal-Ginard B. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 11.Lin X, Shah S, Bulleit R F. Mol Brain Res. 1996;42:307–316. doi: 10.1016/s0169-328x(96)00135-0. [DOI] [PubMed] [Google Scholar]
- 12.Lyons G E, Micales B K, Schwarz J, Martin J F, Olson E N. J Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molkentin J D, Olson E N. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naya F S, Olson E. Curr Opin Cell Biol. 1999;11:683–688. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- 15.Ornatsky O I, Andreucci J J, McDermott J C. J Biol Chem. 1997;272:33271–33278. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- 16.Skerjanc I S, Wilton S. FEBS Lett. 2000;472:53–56. doi: 10.1016/s0014-5793(00)01438-1. [DOI] [PubMed] [Google Scholar]
- 17.Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg M E. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- 18.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A. Nature (London) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 19.Salvesen G S, Dixit V M. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 20.Tewari M, Quan L T, O'Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 21.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt P. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 22.Yuan J, Shaham S, Ledouz S, Ellis H M, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Linseman D A, Allen M P, Meintzer J K, Wang X, Laessig T, Wierman M E, Heidenreich K A. J Neurosci. 2001;21:6544–6552. doi: 10.1523/JNEUROSCI.21-17-06544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Budd S L, Tenneti L, Lishnak T, Lipton S A. Proc Natl Acad Sci USA. 2000;97:6161–6166. doi: 10.1073/pnas.100121097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Y, Bales K R, Dodel R C, Hamilton-Byrd E, Horn J W, Czilli D L, Simmons L K, Ni B, Paul S M. Proc Natl Acad Sci USA. 1997;94:11657–11662. doi: 10.1073/pnas.94.21.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan J, Galindo M F, Miller R J. J Neurochem. 1997;68:1612–1621. doi: 10.1046/j.1471-4159.1997.68041612.x. [DOI] [PubMed] [Google Scholar]
- 27.Mattson M P, Keller J N, Begley J G. Exp Neurol. 1998;153:35–48. doi: 10.1006/exnr.1998.6863. [DOI] [PubMed] [Google Scholar]
- 28.Tenneti L, D'Emilia D M, Lipton S A. Neurosci Lett. 1997;236:139–142. doi: 10.1016/s0304-3940(97)00780-5. [DOI] [PubMed] [Google Scholar]
- 29.Tenneti L, D'Emilia D M, Troy C M, Lipton S A. J Neurochem. 1998;71:946–959. doi: 10.1046/j.1471-4159.1998.71030946.x. [DOI] [PubMed] [Google Scholar]
- 30.Tenneti L, Lipton S A. J Neurochem. 2000;74:134–142. doi: 10.1046/j.1471-4159.2000.0740134.x. [DOI] [PubMed] [Google Scholar]
- 31.Bonfoco E, Krainc D, Ankarcrona M, Nicotera P, Lipton S A. Proc Natl Acad Sci USA. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stennicke H R, Salvesen G S. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 33.Stennicke H R, Salvesen G S. Methods Enzymol. 2000;322:91–100. doi: 10.1016/s0076-6879(00)22010-7. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y F, Tsirka S E, Strickland S, Stieg P E, Soriano S G, Lipton S A. Nat Med. 1998;4:228–231. doi: 10.1038/nm0298-228. [DOI] [PubMed] [Google Scholar]
- 35.Ikeshima H, Imai S, Shimoda K, Hata J, Takano T. Neurosci Lett. 1995;200:117–120. doi: 10.1016/0304-3940(95)12092-i. [DOI] [PubMed] [Google Scholar]
- 36.McDermott J C, Cardoso M C, Yu Y T, Andres V, Leifer D, Krainc D, Lipton S A, Nadal-Ginard B. Mol Cell Biol. 1993;13:2564–2577. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei S Z, Pan Z-H, Aggarwal S K, Chen H-S V, Hartman J, Sucher N J, Lipton S A. Neuron. 1992;8:1087–1099. doi: 10.1016/0896-6273(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 38.Dreyer E B, Lipton S A. J Am Med Assoc. 1999;281:306–308. doi: 10.1001/jama.281.4.306. [DOI] [PubMed] [Google Scholar]
- 39.Lipton S A, Rosenberg P A. N Engl J Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]