Abstract
Polyadenylation of synthetic RNAs stimulates rapid degradation in vitro by using either Chlamydomonas or spinach chloroplast extracts. Here, we used Chlamydomonas chloroplast transformation to test the effects of mRNA homopolymer tails in vivo, with either the endogenous atpB gene or a version of green fluorescent protein developed for chloroplast expression as reporters. Strains were created in which, after transcription of atpB or gfp, RNase P cleavage occurred upstream of an ectopic tRNAGlu moiety, thereby exposing A28, U25A3, [A+U]26, or A3 tails. Analysis of these strains showed that, as expected, polyadenylated transcripts failed to accumulate, with RNA being undetectable either by filter hybridization or reverse transcriptase–PCR. In accordance, neither the ATPase β-subunit nor green fluorescent protein could be detected. However, a U25A3 tail also strongly reduced RNA accumulation relative to a control, whereas the [A+U] tail did not, which is suggestive of a degradation mechanism that does not specifically recognize poly(A), or that multiple mechanisms exist. With an A3 tail, RNA levels decreased relative to a control with no added tail, but some RNA and protein accumulation was observed. We took advantage of the fact that the strain carrying a modified atpB gene producing an A28 tail is an obligate heterotroph to obtain photoautotrophic revertants. Each revertant exhibited restored atpB mRNA accumulation and translation, and seemed to act by preventing poly(A) tail exposure. This suggests that the poly(A) tail is only recognized as an instability determinant when exposed at the 3′ end of a message.
Modulation of RNA stability plays a variety of roles in gene regulation during organism development and environmental responses. Moreover, mRNA turnover can eliminate aberrant transcripts and thereby contribute to accurate translation (1, 2). Studies in yeast have defined the major steps for turnover of nucleus-encoded mRNAs, which initiates with 3′ end deadenylation followed by 5′ end decapping (3). These products are then degraded by a 5′- to 3′-exoribonuclease, or by the exosome complex of 3′- to 5′-exonucleases (4). A multiprotein complex may also be involved in RNA degradation in Escherichia coli. This complex, termed the degradosome, consists of polynucleotide phosphorylase (PNPase), a DEAD-box RNA helicase, RNase E, enolase, and possibly other proteins. In contrast to eukaryotic mRNA degradation, however, 3′-polyadenylation stimulates decay in E. coli through its recognition by PNPase (5, 6). Thus, although both prokaryotic and eukaryotic cells have exonuclease-containing degradation complexes, one prefers polyadenylated substrates and the other acts on deadenylated molecules.
The chloroplast, an endosymbiotic organelle, can be viewed as a prokaryotic compartment in a eukaryotic cell. In the chloroplast, polyadenylated mRNAs have been found by using reverse transcriptase–PCR (RT-PCR), and their incubation in soluble protein extracts results in rapid degradation (7–9). Chloroplasts contain a nucleus-encoded form of PNPase (10), which has a strong affinity for poly(A) (11), and likely recognizes poly(A) tails in vivo. Although the existence of a chloroplast degradosome was reported (10), it now seems that chloroplast PNPase is not part of a multiprotein complex (12), and that it both synthesizes and degrades poly(A) tails (13). E. coli, by contrast, encodes a poly(A) polymerase. These differences may reflect the evolution of the chloroplast to a compartment whose gene regulation is controlled by the nucleus.
Unlike eukaryotic poly(A) tails, the roles of 3′-UTR tails in prokaryotic gene expression are largely unexplored, although they are widely distributed in microorganisms including cyanobacteria (14). Polyadenylated mRNAs have also been found in both plant (15, 16) and animal mitochondria (17). The functions of these tails have mostly been tested in vitro, which has highlighted RNA degradation but not interactions with other RNA processing or expression pathways. In particular, the site and composition of the 3′-UTR tail has not been manipulated in vivo, which might reveal important mechanisms.
Our studies with Chlamydomonas have detected polyadenylated forms of mRNA, tRNA, and rRNA by RT-PCR, and shown that an A25 tail is sufficient to confer marked instability in vitro (9). Here, we have manipulated the endogenous atpB gene, which encodes the β-subunit of ATP synthase, and a green fluorescent protein (GFP) reporter gene, to examine the consequences of adding specific 3′ end tails in vivo. Our data suggest that both poly(A) and poly(U) destabilize the upstream mRNA. We also demonstrate a genetic approach to identifying nuclear factors involved in RNA processing and the polyadenylation/degradation pathways.
Materials and Methods
Strains and Culture Conditions.
Extracts for in vitro assays were obtained from strain CC406. Strain CC373 (ac-u-c-2–25), which carries a deletion in the atpB gene and downstream region (18), was used for chloroplast transformations. Cells were grown under constant light in Tris-acetate-phosphate medium (19).
Plasmids, DNA Constructs, and Chloroplast Transformation.
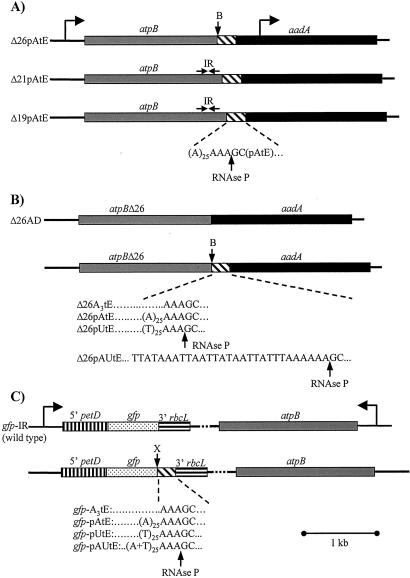
The insert of the atpX-AAD selectable marker cassette (20) was amplified by PCR and inserted into pGEM-T (Promega) after adding a BglII site to the 5′ primer. The cassette was excised as a BglII-BamHI fragment, and inserted into the BglII sites of plasmids atpBΔ19, atpBΔ21, and atpBΔ26; these sites lie immediately downstream of deletion endpoints ranging from several base pairs after the stop codon to within the 3′-UTR stem-loop structure (21) (see Fig. 2). Clones were identified in which the cassette was transcribed in tandem with atpB (Fig. 1A), yielding constructs atpBΔ19AD, atpBΔ21AD, and atpBΔ26AD; these retained a BglII site between the modified atpB gene and the aadA cassette. The trnE gene was cloned by using PCR, with the amplified fragment containing a BamHI site plus 3 bp upstream of the mature tRNA sequence, and 148 bp downstream plus a BglII site. Modified trnE genes were created by PCR with an upstream primer containing a BglII site and one of following: (A)3, (A)25, (T)25, or the arbitrary [A+U]26 sequence (Fig. 1B), immediately upstream of the RNase P site; and a downstream primer containing a BamHI site. These modified trnE genes were inserted as BglII-BamHI fragments into the BglII sites of atpBΔ19AD, atpBΔ21AD, and atpBΔ26AD [Fig. 1 A and B; only the (A)25 cassette was inserted into atpBΔ19AD and atpBΔ21AD]. For GFP constructs, plasmid MR220 containing cpgfp1 (GenBank accession no. AF303131), which is optimized for tobacco chloroplast expression, was obtained from M. Hanson (Cornell University). The gfp gene was excised with NcoI and XbaI, and inserted into NcoI- and XbaI-digested pDAAD (22), creating plasmid pDGFP. pDAAD contains the cassette petD promoter-aadA-coding region-rbcL 3′-UTR, inserted downstream of the atpB gene, and in pDGFP the aadA-coding region is replaced by that of gfp. Modified trnE genes were created as above, except both PCR primers contained SpeI sites, because the trnE gene contains compatible XbaI sites. Modified trnE gene fragments were digested with SpeI and inserted into XbaI-digested pDGFP, placing the tRNA and flanking sequences between the gfp-coding region and the rbcL 3′-UTR, as shown in Fig. 1C. Each construct was verified by DNA sequence analysis. Chloroplast transformation was performed as described (21).
Figure 2.
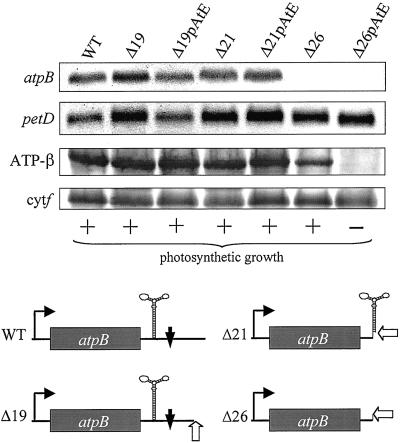
Effect of an IR upstream of the polyadenylation position in atpB transcripts. RNA filter hybridization analysis (upper two panels; petD is a loading control) showed that discrete atpB transcripts were not detectable in Δ26 and Δ26pAtE. The bottom two panels show immunoblot analysis, with cytochrome f as a loading control. The photosynthetic growth phenotype, determined by plating on medium lacking acetate, is shown below the blots. The lower part of the figure shows representations of the atpB genes in each transformant. The open arrow denotes the site of poly(A) tail addition, the bent arrow the atpB promoter, and the filled vertical arrow the ECS.
Figure 1.
Gene organization in chloroplast transformants to test 3′-UTR modifications. (A) Addition of a poly(A) sequence downstream of different versions of atpB. The gray box represents the atpB-coding region, which in strain Δ26 is followed by a BglII site (B) in lieu of the normal stem-loop-forming inverted repeat (IR). The hatched box represents a sequence of 25 adenosines immediately followed by an RNase P cleavage site (AAA↓GC), and then the sequence corresponding to mature trnE and some downstream sequences (see Materials and Methods). In Δ21 and Δ19, most or all, respectively, of the atpB 3′ IR remains, as indicated by the horizontal arrows. The aadA selectable marker cassette, transcribed in the same orientation as atpB, is shown as a filled box. Construct designations are shown at the left. (B) Derivatives of the atpBΔ26 gene. Δ26AD is a control which contains only the 3′ IR-deleted atpB gene and the aadA cassette. The four derivatives shown below produce modified atpB transcripts with a three adenines, A28, U25A3, or (AU)26 as atpB 3′-UTRs after cleavage at the RNase P site. The precise sequence of the (AU)26 tail is shown. (C) Transformants containing the GFP reporter gene. The gfp-coding region is flanked by the petD promoter and 5′-UTR (vertical stripes), and the rbcL 3′-UTR (horizontal stripes). The cassette is transcribed convergently to the atpB gene, which was used as a selectable marker. The atpB gene is shown in the opposite orientation as compared with A and B, so that the modified cassettes are at left in all cases. In gfp IR, gfp mRNA contains a stem-loop forming inverted repeat at its 3′ end (derived from rbcL), and its expression was considered as a wild-type or baseline level. Insertions conferring A3, A28, U25A3, or (AU)26 tails, flanked by an RNase P site and trnE, were inserted at an XbaI site (X) immediately upstream of the rbcL 3′-UTR. The tail sequence in gfp-pAUtE is identical with that in Δ26pAUtE.
The cpDNA configuration in the spa revertants (Fig. 6) was determined by sequence analysis with PCR amplification from total DNA with primers YK03 and atpX-3′. YK03 anneals 60 bp upstream of the atpB stop codon, and atpX-3′ anneals immediately upstream of the atpA translation initiation codon fused to the aadA-coding region. When used to amplify DNA from Δ26pAtE, the product obtained is 984 bp, which was also the case for each spa mutant except spa2. In spa2, a deletion begins 37 bp into the trnE cassette, within the mature tRNA-coding region, and ends 74 bp upstream of the atpA translation initiation codon, a total of 765 bp.
Figure 6.
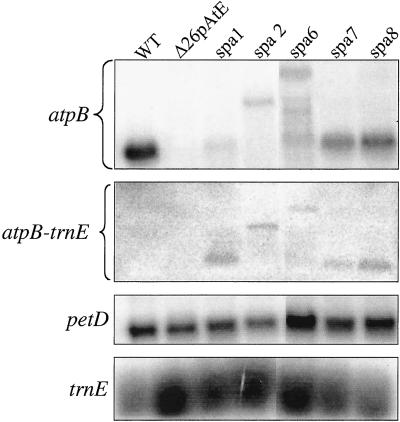
RNA analysis of spontaneous photoautotrophic revertants obtained from Δ26pAtE. Replicate RNA filter blots were probed, from top to bottom, with the atpB-coding region, trnE, the petD coding region, and trnE (the second and bottom panels are parts of the same gel).
RNA and Protein Analysis.
Total RNA and protein were isolated from early-log-phase cultures (1–2 × 106 cells per ml). Each assay involved at least three independent preparations, and results were averaged. The data shown here are representative of the averaged results. RNA filter hybridizations were performed as described (23).
For RT-PCR, total RNA was precipitated with 2 M LiCl, and 25-μg aliquots were treated with 10 units RQ1 DNase (Promega) for 1 h and purified by using the RNeasy Plant Mini Kit (Qiagen, Chatsworth, CA). Negative controls were identical reactions except reverse transcriptase was omitted, and no product was generated (data not shown). For the competitive RT-PCR shown in Figs. 3B and 5A, RNA competitors were generated by using a RT-PCR competitor construction kit (Ambion, Austin, TX). The primers used to amplify PCR products for generating competitors (primers a and b in Fig. 3B) were: (T7)TGGCTGAATATTTCCGTGATGTΔ24CATTGATAACATTTTCCGTTTCGTAC and GTACTGTAGTAGCATCTAAGTGAG for atpB, and (T7)GGTTCAGTACAATTAGCAGATCΔ17CCTATCGGGTGATGGTCCTG for gfp. The Δ24 in the upstream atpB primer represents a deletion of 24 bp of atpB sequences, as shown schematically in Fig. 3B; similarly, 17 bp are deleted from gfp. (T7) indicates the T7 RNA polymerase promoter. The purified PCR products were used as templates for T7 transcription. The primers used for RT-PCR (primers c and d in Fig. 3B) were for primers c, the underlined part of primers a; and for primers d, GCCTGAACTGATGTGATAG for atpB and GTGTGATACCAGCAGCTG for gfp. Reverse transcriptase reactions (20 μl) were performed by using Superscript II (Invitrogen) with 500 ng of total RNA prepared as described above, RNA competitor (1 ng for atpB and 0.04 ng for gfp) and the 3′ end primer for each gene (0.5 mM final concentration) at 50°C for 45 min. PCR was performed with 2 μl of the RT reaction products for 25 cycles (94°, 50°, 72° each for 30 sec). Conditions were identical for petD except no competitor was added. Controls (not shown) verified that 25 cycles did not saturate the PCR reactions. PCR products were resolved in 6% polyacrylamide gels and stained with SYBR Green I (Molecular Probes, 10,000× dilution) for 30 min. The products were detected and quantified by using a Storm System (Molecular Dynamics) in blue fluorescence mode. For atpB and gfp, relative product accumulations were estimated by comparing their levels to PCR products from wild-type cells (atpB) or gfp flanked by the rbcL 3′-UTR.
Figure 3.
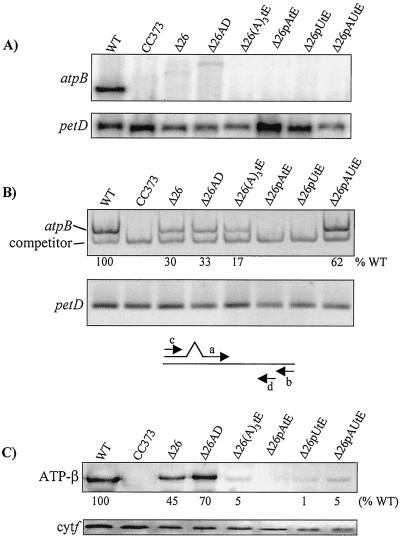
Analysis of transformants containing modified atpBΔ26 genes (see Fig. 1B). CC373 is an atpB deletion mutant and was the transformation recipient. (A) RNA filter hybridizations, with petD as a loading control. The weak signals in the Δ26 and Δ26AD lanes correspond to low-abundance atpB transcripts, most likely stabilized by sequences in the downstream aadA cassette. (B) RT-PCR analysis of transcript accumulation in the same strains as in A. Equal amounts of RNA were analyzed by using competitive RT-PCR for atpB, or RT-PCR under the same conditions for petD-coding regions (see Materials and Methods). Relative atpB RNA accumulation is an average of two experiments. Control PCR reactions were also conducted on RNA samples with RT being omitted. In these cases, no amplification product was observed (data not shown). Below the gel pictures, a schematic of the competitive RT-PCR strategy is shown (see text). Primers a and b were used to create a template for synthesis of competitor RNA, and primers c and d were used for RT-PCR. (C) Immunoblot analysis of the same strains, with cytochrome f as a loading control. The estimated accumulation of the atpB gene product, relative to the wild-type control, is shown beneath the blot, and is an average of at least three experiments. The basis for quantitation is given in Materials and Methods.
Figure 5.
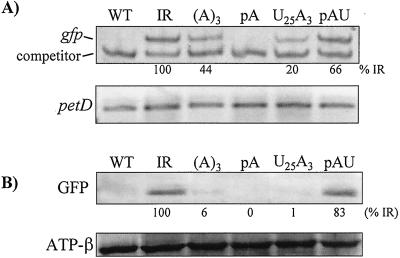
Analysis of GFP expression in the strains shown in Fig. 5. (A) RT-PCR analysis of gfp transcripts, with petD as a control. Competitive RT-PCR was used for gfp, and simple RT-PCR for petD. See Fig. 3B and legend for strategy. Relative RNA amounts for gfp are an average of two experiments. Omission of RT resulted in no PCR product (data not shown). (B) Immunoblot analysis of GFP with use of a monoclonal antibody. An estimated amount of GFP is shown as percentage relative to strain IR, and is the average of at least three experiments. The ATPase β-subunit was used as a loading control.
Total protein was isolated from 3-ml cultures, with pelleted cells resuspended in 50 μl of 0.1 M DTT, 0.1 M sodium carbonate after dissolving a protease inhibitor tablet (1 per 50 ml), and frozen in liquid N2. An equal amount of 2× SDS sample buffer was added to thawed samples which were microfuged for 10 min at 4°C. The protein concentrations of supernatants were measured by using the Bio-Rad protein assay. Proteins were resolved in SDS-12% polyacrylamide gels. Immunoblots were incubated in 0.1 M maleic acid/0.15 M NaCl, pH 7.5, using antibodies raised against the Chlamydomonas ATPase β-subunit, cytochrome f or a monoclonal anti-GFP antibody (Boehringer Mannheim), and detection was achieved with either alkaline phosphatase or enhanced chemiluminescence (ECL+; Amersham Pharmacia).
Fluorescence Microscopy.
Cells were harvested in late-log phase (1–2 × 107 cells per ml) to reduce mobility, observed under a Olympus BX50 microscope (Olympus, New Hyde Park, NY), and analyzed with metamorph software (Universal Imaging Corp., West Chester, PA).
Results
A Poly(A) Tail Does Not Affect RNA Accumulation in Vivo When Added Downstream of a 3′-UTR Structure.
We have shown that a 3′-UTR stem-loop in Chlamydomonas atpB mRNA serves as an RNA stability determinant (21), by impeding 3′→5′ exonuclease digestion following cleavage at a nearby downstream primary processing site [endonuclease cleavage site (ECS)]. When this stem-loop is destroyed through deletions, discrete atpB mRNA no longer accumulates; however, cells remain photoautotrophic because the remaining heterodisperse transcripts are translatable. Even cells totally lacking the normal atpB 3′-UTR (e.g., strain atpBΔ26, hereafter referred to as Δ26) accumulate 10–20% of the wild-type level of the β-subunit. Two other deletion strains, Δ19 and Δ21, still possess all (Δ19) or most of (Δ21) the 3′ stem-loop, but Δ21 lacks the ECS. In both cases, wild-type levels of atpB mRNA and protein accumulate.
In our studies of polyadenylation in Chlamydomonas chloroplasts, we found atpB polyadenylation both at the mature mRNA 3′ end and at the ECS. If polyadenylation were to stimulate rapid RNA degradation in vivo, we suggested that the processing machinery and this degradation machinery might be in competition (9). One way to test this was to create precursor mRNAs in vivo that had poly(A) tails at defined sites, and then measure transcript accumulation. To do this, we used the strategy shown in Fig. 1A. The deletion endpoints of Δ26, Δ21, and Δ19 contain BglII sites into which we inserted an ectopic copy of the trnE gene, preceded by sequences encoding a tail of 25 adenines (A25). Because the natural 5′ processing (RNase P) site of tRNAGlu is at the sequence AAA↓GC, we reasoned that after atpB transcription, pre-mRNAs would be generated containing atpB-A25-trnE, and these would be cleaved to yield atpB mRNA with a particular version of its 3′-UTR followed by an A28 tail. In the cases of Δ21 and Δ19, it should be noted that the 3′-UTR stem-loop is not an effective transcription termination signal (24), nor is transcription rate affected by manipulating the atpB 3′-UTR (21).
These constructs were introduced into chloroplasts by biolistic transformation, and Fig. 2 shows accumulation of RNA and protein in the transformants, compared with control strains lacking the A25 motif. At the RNA level, strains Δ19 and Δ21 accumulated essentially wild-type levels of the atpB transcript, irrespective of the presence of the A25 sequence. On the other hand, neither version of Δ26 accumulated detectable atpB mRNA; this was expected because Δ26 lacks the 3′ stem-loop that defines the normal 3′ end. However, the Δ26 strain producing an A28 tail (Δ26pAtE) could be differentiated from Δ26 in two ways. First, Δ26 but not Δ26pAtE could grow photoautotrophically, and second, only Δ26 accumulated detectable β-subunit (Fig. 2, third row). This result suggested that the A28 tail might be responsible for the absence of atpB mRNA and its gene product, whereas constructs producing at least 20% of wild-type mRNA were unaffected.
Influence of 3′-UTR Tail Composition on RNA Accumulation in Vivo.
Having established that atpB Δ26 mRNA failed to accumulate when flanked by an A28 tail, presumably because of RNA instability, we created a series of modified Δ26 transcripts to examine the specificity of this effect. The following strains were obtained, as diagrammed in Fig. 1B: Δ26AD, which is the Δ26 deletion followed by the aadA cassette; Δ26(A)3tE, predicted to produce an A3 tail after RNase P processing; Δ26pAtE, which produces a A28 tail; Δ26pUtE, which produces a U25A3 tail; and Δ26AUtE, which produces an arbitrary 26-nt tail that is 100% [A+U], as a control for any effect of base composition. Because the trnE RNase P site must be included, it was not possible to produce a homopolymer U-tail. However, we note that both the U25A3 [hereafter referred to as “poly(U)”] and [A+U]26 tails terminate in three A's for this reason, and can thus be compared for the effect of the poly(U) stretch. We also note that the [A+U]26 tail is not predicted to form a stable secondary structure; the Mulfold algorithm (bioinfo.math.rpi.edu/≈mfold/rna/form1.cgi) yields ΔG = −0.3 kcal/mol.
Homoplasmic transformants were obtained for the strains above, and RNA and protein analysis was performed as shown in Fig. 3. Fig. 3A shows that none of the strains producing 3′ tails accumulated discrete atpB mRNA, which was expected because none have strong 3′-UTR secondary structures. We therefore used competitive RT-PCR to estimate relative RNA levels. As shown in the diagram below Fig. 3B, a synthetic atpB RNA competitor template was generated carrying a short deletion (primers a and b), and transcribed in vitro. A defined amount of this RNA was introduced into each RT-PCR reaction (primers c and d) along with an equal amount of total RNA from each strain. By using a titration (not shown), an amount of competitor was chosen so that competition still occurred with RNA from wild-type cells where atpB mRNA accumulation is greatest. This, and control experiments with different numbers of cycles, ensured that the analysis was within linear range.
The competitive RT-PCR assay revealed products from all strains except Δ26pAtE and Δ26pUtE, as well as the transformation recipient CC373, where the coding region is deleted. Strains Δ26 and Δ26AD accumulated about 30% of the wild-type RNA level, in accordance with earlier results for Δ26 obtained by slot-blot analysis (21). Strain Δ26(A)3tE had slightly lower accumulation, consistent with a small destabilizing effect of the A3 tail, but Δ26AUtE accumulated almost the wild-type level, although it also terminates in an A3 motif. One interpretation is that the upstream A+U sequence does not support rapid degradation, i.e., degrading activity would slow after its encounter with A3.
Immunoblot analysis (Fig. 3C) was consistent with the RNA results for Δ26pAtE and Δ26AUtE, which had no RNA according to RT-PCR and accumulated no or only a trace amount of β-subunit, respectively. Also consistent were Δ26, Δ26AD, and Δ26(A)3tE, which accumulated 45%, 70%, and 5% of the wild-type β-subunit amount, respectively. There are many examples where chloroplast RNA and protein accumulation do not display a linear relationship, and in this respect our results were not surprising. On the other hand, Δ26AUtE accumulated ≈5% of the wild-type β-subunit level, but approximately 60% of the wild-type RNA level. One possibility is that the 3′-UTR itself may affect translation; we have shown a correlation between atpB 3′ processing and translation (25). However, the RT-PCR used here does not prove that the target transcript is intact and translatable. For example, if the 5′ portion had been degraded, RT-PCR would be unaffected but the gene product could not be made. For all these reasons, immunoblot assays may not always correlate with RNA accumulation measures.
A GFP Reporter Gives Results Largely Consistent with Those for atpB.
To verify that the results above were not specific to atpB, we used a reporter gene. Although we have previously used β-glucuronidase as a reporter in Chlamydomonas chloroplasts (26), we adapted GFP to this system because it can be observed in living cells. GFP had already been expressed as a nuclear gene in Chlamydomonas (27); however, we elected to use a version that had been modified for tobacco chloroplast expression (28). The control gfp construct (gfp-IR) is driven by the petD promoter/5′-UTR and terminated by the rbcL 3′-UTR (Fig. 1C). Four derivatives were made by placing various 3′-UTR/trnE combinations between the GFP-coding region and the rbcL 3′-UTR. These derivatives were analogous to those for which atpB results were described above, and were expected, apart from GFP-IR, to generate petD-gfp mRNAs with added 3′ tails. The GFP cassettes were introduced into the chloroplast genome, using the intact flanking atpB gene as a selectable marker for photoautotrophic growth.
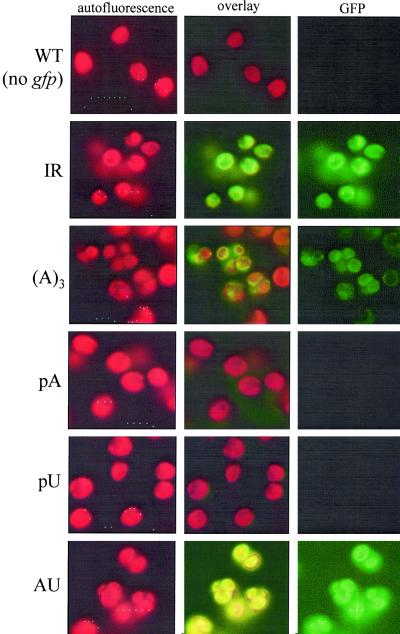
Homoplasmic transformed cells were examined by fluorescence microscopy (Fig. 4). Chlorophyll autofluorescence (Left) revealed the large cell volume occupied by the chloroplast, and GFP fluorescence (Right) was seen for three strains: IR, A3, and AU. In contrast, no GFP fluorescence was observed for the negative control (WT cells), nor for the pA or pU strains. Because the same exposure time was used for each set of images (e.g., autofluorescence or GFP fluorescence), the relative fluorescence should reflect GFP accumulation. In this context, strain A3 clearly had lower fluorescence than IR or AU. To support these observations, GFP transcription and protein accumulation were monitored by competitive RT-PCR and immunoblotting, respectively (Fig. 5). RT-PCR was used in part because the transcripts were undetectable on filter blots (data not shown). RT-PCR measurements (Fig. 5A) showed no accumulation in the pA strain and intermediate amounts in other strains, when compared with strain IR. The results are similar to those for atpB, except we did detect accumulating RNA for strain pU. At the protein level, results were consistent with fluorescence, and also with results for atpB, except that the relative GFP level in strain AU was higher. As discussed above, many factors can influence the relationship between RNA and protein level. Taking the atpB and GFP results together, we propose that poly(A) tails destabilize chloroplast transcripts in vivo, and that poly(U) also stimulates RNA decay. Furthermore, an A3 tail is sufficient to exert a negative effect.
Figure 4.
Differential GFP expression observed by fluorescence microscopy. The left-hand columns are chlorophyll autofluorescence, the right-hand columns are GFP fluorescence, and the center columns are the combined images. Strains shown at the sides (see Fig. 1C) are: WT, negative control (no gfp gene), IR (gfp-IR), (A)3 (gfp-A3tE), pA (gfp-pAtE), pU (gfp-pUtE), and AU (gfp-pAUtE). Each autofluorescence image was exposed for the same amount of time, as was each GFP image. However, longer exposures were necessary for GFP.
Photoautotrophic Revertants of Δ26pAtE Fail to Expose the Poly(A) Tail.
Poly(A) tail-mediated RNA instability is likely to be caused by an interaction with cellular factors, rather than an intrinsic property of the sequence. One way to test this hypothesis is to use a genetic approach, which we initiated by using Δ26pAtE. This strain is acetate-requiring because it does not accumulate the ATP synthase, and we selected spontaneous revertants by plating cells on minimal medium. To date 38 phenotypic revertants have been isolated, and all accumulate the ATP synthase, as expected (data not shown). The RNA analysis of five such revertants is presented here.
When RNA filter hybridizations were performed, we found that the spa (suppressor of polyadenylation) revertants produced stable atpB transcripts of variable sizes (Fig. 6 Top). When DNA from the strains was isolated and sequenced, however, the poly(A) sequence and RNase P site were still present, although spa2 was found to carry a deletion starting further downstream (see below). Thus, the revertants had apparently overcome poly(A)-mediated RNA instability through a mechanism other than stabilizing poly(A) tails. Indeed, upon probing another filter blot with trnE (Upper Center), the same transcripts were identified, suggesting that cleavage at the RNase P site was no longer occurring efficiently. In spa2 and spa6, the transcripts extended downstream into the aadA cassette, which was confirmed by hybridization (data not shown). Because these transcripts still contain the poly(A) sequence, we can tentatively draw two conclusions: first, the sequence is not intrinsically destabilizing; and second, it is not a site for endonuclease cleavage, or at least it is not destabilizing when unexposed at the transcript 3′ end.
Although genetic analysis is still incomplete, the spa mutants fall into two classes, having arisen through both chloroplast (spa6) and nuclear (spa1) mutations. We have not yet found the mutation conferring reversion in spa6, despite having sequenced the atpB 3′–trnE-atpA 5′ region. Other possibilities include a mutation elsewhere in atpB or aadA that influences RNA secondary structure, or an unlinked mutation in the chloroplast genome. On the other hand, spa2 was found to have a deletion beginning within trnE and continuing into the aadA cassette promoter (see Materials and Methods). This deletion probably disrupts processing at the RNase P site (29), although this remains to be confirmed. Taken together, we suggest that both the sequence context and putative trans-acting factors are relevant to the RNA processing and degradation pathways studied here.
Discussion
Taking advantage of Chlamydomonas chloroplast transformation, we have created an in vivo system for studying RNA stability modulation by 3′-UTR tails. To expose a particular sequence at the RNA 3′ end, we used an RNase P site, which seems to have been very effective because atpB-trnE cotranscripts were never observed, except in the spa mutants. One reason for its efficiency is that positions more than 1 nt upstream of prokaryotic RNase P sites do not contribute significantly to catalytic rate or site recognition (29). This contrasts with some other endonuclease cleavage sites; for example, the petD mRNA 5′-processing site is used inefficiently in ectopic locations (30, 31). Another advantage of the trnE RNase P site is that it naturally leaves an A tail. Although we had to begin with a relatively unstable (atpBΔ26) mRNA to test added 3′-UTR sequences, the results clearly showed a destabilizing effect of poly(A), which was expected on the basis of previous in vitro results, and a probable stimulatory effect of poly(U), which was unexpected. Although we did not measure transcription rates here, we have shown (21, 26, 32) that 3′-UTR manipulations do not affect the atpB or petD promoters. Thus, the absence or decrease in RNA levels must be caused by instability. By using the obligate heterotroph Δ26pAtE, we were able to obtain revertants to photoautotrophy. Their properties suggest that the 3′ end exposure of the poly(A) tail is necessary for rapid RNA turnover, and that nucleus-encoded factors act at some level in the degradation pathway. Both are consistent with known properties of chloroplast PNPase, a nucleus-encoded enzyme that does not recognize internal poly(A) stretches (11).
When a poly(A) tail was added downstream of an inverted repeat sequence (Fig. 1A), we found that the IR interfered with poly(A)-promoted degradation (Fig. 2). In fact, the atpB transcripts in Δ19 and Δ21 seemed to be processed correctly, consistent with competition between RNA processing and decay pathways. Indeed, cleavage at the atpB 3′-UTR ECS is accompanied by rapid degradation of downstream sequences (24, 33), which in this case would be the poly(A) moiety. Processing can also influence degradation in other systems. For example, the human phlp mRNA has alternative polyadenylation sites, and if the distal site is used the mRNA is less stable because of the inclusion of a canonical AU-rich element (34).
Δ21pAtE RNA, however, lacks an ECS between the atpB-coding region and the poly(A) signal. This suggested another interpretation, namely that the enzyme recognizing the poly(A) tail, presumably PNPase, could not efficiently degrade the IR. These results contrast with expectations based on E. coli, where the RNA helicase subunit of the degradosome can unwind stem-loops after PNPase has initiated degradation (5). Although plant chloroplasts certainly have RNA helicase (35), recent data suggest that they do not have a degradosome (12). Thus, in chloroplasts RNA unwinding may be uncoupled from PNPase activity. In vivo, however, RNAs that terminate in stem-loops are poorly polyadenylated, most likely for steric reasons (36), which affords another level of protection for correctly matured transcripts.
We have adapted GFP to Chlamydomonas chloroplasts, and our data suggest that it is a sensitive reporter. We used a version that yielded high protein levels in tobacco chloroplasts (28), and were able to estimate relative RNA stabilities by monitoring GFP fluorescence. However, GFP accumulation was 100-fold lower in Chlamydomonas, on a total protein basis, as compared with tobacco (data not shown). We speculate that codon usage or other sequences affected gfp mRNA stability. Results with GFP largely paralleled those obtained with atpB. No RNA was detected when a poly(A) tail was added, but RNA did accumulate with A3 or arbitrary AU tails. This suggests that the overall properties we observed were not caused by particular combinations of reporter genes and 3′-UTRs. On the other hand, some subtle differences can be noted. In U25A3, RNA was observed for gfp but not for atpB. In both instances, however, 1% of protein was measured relative to the control, implying that atpB mRNA also accumulated, albeit to a very low level. With the AU tail, RNA accumulations were similar relative to the control; however, relative protein accumulation was higher for GFP, which suggests that translational efficiency and/or protein stability is reflected in the final data.
Our results suggest that not only poly(A), but also poly(U) sequences are associated with RNA instability. In contrast, a tail of an arbitrary AU sequence did not confer instability, even though like the poly(U) tail it terminated in A3. We infer that the homopolymers are recognized by PNPase or another enzyme. One nucleus-encoded PNPase homologue has been identified by the Chlamydomonas expressed sequence tag project (37); however, its subcellular localization is unknown. The results with poly(U) are consistent with the known affinity of both chloroplast (11) and E. coli (6) PNPases for poly(U). Alternatively, a UTP-dependent RNA decay pathway known in trypanosome mitochondria (38) could exist in Chlamydomonas, although our results may rather suggest that degradation begun by recognition of the terminal A3 motif is stimulated by the upstream U25 stretch.
Because both Δ26pAtE and Δ26pUtE are obligate heterotrophs, they open the door to studying factors involved in their common or respective RNA degradation pathways. Here we have shown data for several photoautotrophic revertants of Δ26pAtE. All accumulated stable atpB transcripts, apparently resulting from the prevention of RNase P cleavage and thus poly(A) tail exposure. These observations indicated that reversion was caused by alteration(s) in RNA processing rather than in the degradation pathway. Because RNase P is essential for tRNA processing and thus translation, we speculate that the lack of RNase P processing is a secondary effect caused by chloroplast genome alterations or nuclear mutations affecting other endonuclease activities.
Acknowledgments
We thank members of the Stern laboratory for helpful comments, and M. Hanson for the GFP construct. This work was supported by Binational Agricultural Research and Development Grant US-3177–99C (to G.S. and D.S.).
Abbreviations
- RT-PCR
reverse transcriptase–PCR
- UTR
untranslated region
- GFP
green fluorescent protein
- ECS
endonuclease cleavage site
- PNPase
polynucleotide phosphorylase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jacobson A, Peltz S W. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 2.Hilleren P, Parker R. Annu Rev Genet. 1999;33:229–260. doi: 10.1146/annurev.genet.33.1.229. [DOI] [PubMed] [Google Scholar]
- 3.Herrick D, Parker R, Jacobson A. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell P, Tollervey D. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 5.Carpousis A J, Vanzo N F, Raynal L C. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 6.Lisitsky I, Schuster G. Eur J Biochem. 1999;261:468–474. doi: 10.1046/j.1432-1327.1999.00285.x. [DOI] [PubMed] [Google Scholar]
- 7.Kudla J, Hayes R, Gruissem W. EMBO J. 1996;15:7137–7146. [PMC free article] [PubMed] [Google Scholar]
- 8.Lisitsky I, Klaff P, Schuster G. Proc Natl Acad Sci USA. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komine Y, Kwong L, Anguera M C, Schuster G, Stern D B. RNA. 2000;6:598–607. doi: 10.1017/s1355838200992252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes R, Kudla J, Schuster G, Gabay L, Maliga P, Gruissem W. EMBO J. 1996;15:1132–1141. [PMC free article] [PubMed] [Google Scholar]
- 11.Lisitsky I, Kotler A, Schuster G. J Biol Chem. 1997;272:17648–17653. doi: 10.1074/jbc.272.28.17648. [DOI] [PubMed] [Google Scholar]
- 12.Baginsky S, Shteiman-Kotler A, Liveanu V, Yehudai-Resheff S, Bellaoui M, Settlage R E, Shabanowitz J, Hunt D F, Schuster G, Gruissem W. RNA. 2001;7:1464–1475. [PMC free article] [PubMed] [Google Scholar]
- 13.Yehudai-Resheff S, Hirsh M, Schuster G. Mol Cell Biol. 2001;21:5408–5416. doi: 10.1128/MCB.21.16.5408-5416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grunberg-Manago M. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 15.Gagliardi D, Leaver C J. EMBO J. 1999;18:3757–3766. doi: 10.1093/emboj/18.13.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupold D S, Caoile A G F S, Stern D. J Biol Chem. 1999;274:3897–3903. doi: 10.1074/jbc.274.6.3897. [DOI] [PubMed] [Google Scholar]
- 17.Ojala D, Montoya J, Attardi G. Nature (London) 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 18.Shepherd H S, Boynton J E, Gillham N W. Proc Natl Acad Sci USA. 1979;76:1353–1357. doi: 10.1073/pnas.76.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris E H, Boynton J E, Gillham N W. Microbiol Rev. 1994;58:700–754. doi: 10.1128/mr.58.4.700-754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldschmidt-Clermont M. Nucleic Acids Res. 1991;19:4083–4090. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stern D B, Radwanski E R, Kindle K L. Plant Cell. 1991;3:285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rott R, Drager R G, Stern D B, Schuster G. Mol Gen Genet. 1996;252:676–683. doi: 10.1007/BF02173973. [DOI] [PubMed] [Google Scholar]
- 23.Church G, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stern D B, Kindle K L. Mol Cell Biol. 1993;13:2277–2285. doi: 10.1128/mcb.13.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rott R, Levy H, Drager R G, Stern D B, Schuster G. Mol Cell Biol. 1998;18:4605–4611. doi: 10.1128/mcb.18.8.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto W, Kindle K L, Stern D B. Proc Natl Acad Sci USA. 1993;90:497–501. doi: 10.1073/pnas.90.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuhrman M, Oertel W, Hegemann P. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- 28.Reed M L, Wilson S K, Sutton C A, Hanson M R. Plant J. 2001;27:257–265. doi: 10.1046/j.1365-313x.2001.01088.x. [DOI] [PubMed] [Google Scholar]
- 29.Kirsebom L A, Vioque A. Mol Biol Rep. 1996;22:99–109. doi: 10.1007/BF00988713. [DOI] [PubMed] [Google Scholar]
- 30.Lisitsky I, Rott R, Schuster G. Planta. 2001;212:851–857. doi: 10.1007/s004250000449. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto W, Sturm N R, Kindle K L, Stern D B. Mol Cell Biol. 1994;14:6180–6186. doi: 10.1128/mcb.14.9.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drager R G, Girard-Bascou J, Choquet Y, Kindle K L, Stern D B. Plant J. 1998;13:85–96. doi: 10.1046/j.1365-313x.1998.00016.x. [DOI] [PubMed] [Google Scholar]
- 33.Hicks A J, Drager R G, Higgs D C, Stern D B. J Biol Chem. 2002;277:3325–3333. doi: 10.1074/jbc.M108979200. [DOI] [PubMed] [Google Scholar]
- 34.Lazarov M E, Martin M M, Willardson B M, Elton T S. Biochim Biophys Acta. 1999;1446:253–264. doi: 10.1016/s0167-4781(99)00098-6. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Duby G, Purnelle B, Boutry M. Plant Cell. 2000;12:2129–2142. doi: 10.1105/tpc.12.11.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuster G, Lisitsky I, Klaff P. Plant Physiol. 1999;120:937–944. doi: 10.1104/pp.120.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S. DNA Res. 1999;6:369–373. doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- 38.Militello K T, Read L K. Mol Cell Biol. 2000;20:2308–2316. doi: 10.1128/mcb.20.7.2308-2316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]