Abstract
We have designed a synthetic tetracationic metallo-supramolecular cylinder that targets the major groove of DNA with a binding constant in excess of 107 M−1 and induces DNA bending and intramolecular coiling. The two enantiomers of the helical molecule bind differently to DNA and have different structural effects. We report the characterization of the interactions by a range of biophysical techniques. The M helical cylinder binds to the major groove and induces dramatic intramolecular coiling. The DNA bending is less dramatic for the P enantiomer.
While DNA encodes the essential blueprint for life, within biological systems its structure and function is regulated by proteins. In the postgenomic environment the goal is to understand the processing of the genetic code and how to stimulate or prevent this processing. To achieve this end a variety of different types of molecular tools will be required that recognize the genetic code in a sequence-selective fashion. Within biological systems, sequence-specific code recognition is generally achieved by the surface motifs of proteins, which generally interact noncovalently with the major groove of DNA (1–3). The major groove is particularly attractive for DNA recognition because the size and shape of the major groove of B-DNA varies most with base sequence. For example, transcriptional regulators often involve cylindrical binding units, such as α-helices or zinc fingers, that insert into the major groove and may kink the DNA (1–3).
Oligonucleotides (synthetic and natural) can selectively recognize DNA by forming triplexes through binding in the major groove (4). Neutral oligonucleotide analogues (e.g., peptide nucleic acids) can achieve similar effects, although more commonly they achieve sequence selectivity through strand displacement (5).
Such biomacromolecule recognition of DNA contrasts with synthetic small molecule recognition agents. Synthetic molecules that achieve sequence selectivity include well-known minor groove binders such as amide-linked imidazole/pyrole oligomers and polymers (6), which in common with many small-molecule DNA-binding agents target the minor groove (7). Alternatively small molecules can act by means of intercalation (8, 9). Synthetic agents that target the major groove of DNA with recognition through noncovalent surface motifs have the potential to be a powerful new tool in the armory of reagents that is being developed to tackle the postgenomic challenge. However, to date little progress has been made in this direction, caused in large part by the size of the molecular surfaces required.
DNA binding by metal complexes through formation of metal-ligand bonds (e.g., cisplatin) has been extensively studied and usually focuses on binding to N7 of G and A residues (ref. 10 and references therein). By contrast, noncovalent binding of metal complexes to DNA is a less well-developed area and has primarily centered around approximately spherical ruthenium polypyridyl complexes, complexes bearing planar intercalating units, or combinations thereof (refs. 11–13 and references in refs. 12 and 13). The size of these compounds means they often bind in the minor, rather than the major, groove. Moreover their small size means that they cannot target more than 2–3 bp and consequently are not suitable scaffolds for sequence-specific recognition. There have been a very few studies, primarily from two American groups, in which these noncovalent binding metal complexes are conjugated to other biomolecular recognition motifs to create hybrid systems (14–16), and to date these have not shown enhanced sequence selectivity over the parent recognition motifs because of the poor recognition properties of the small metallo-units used.
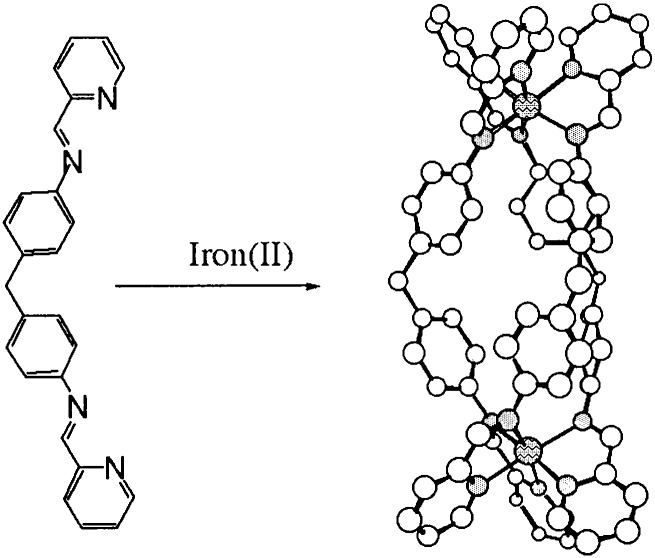
Supramolecular chemistry (17, 18) provides an excellent methodology for designing large synthetic arrays and consequently has the potential to allow us synthetically to bridge the size gap between traditional small-molecule and larger-biomolecule DNA recognition motifs. Within this field, metallo-supramolecular assembly is particularly attractive for design of noncovalent DNA recognition agents (19) because of the cationic charge the metallo-centers impart, and which will afford a substantial energetic contribution to the noncovalent binding to anionic DNA. This hypothesis has been confirmed through preliminary studies on the racemic mixture of the cylindrical tetra cationic dimetallo triple-helicate illustrated in Fig. 1 (20). The effect of the racemic mixture is to induce a dramatic intramolecular bending that results in coils of DNA. Such an effect is unprecedented with a synthetic DNA binder. The NMR data collected for the racemate confirm major groove binding, but were intriguing in that only the M enantiomer (ΛΛ) was present in the refined structure. Herein we explore the enantio-selectivity of the binding of this molecule. This work forms a step in the program to design groove and sequence-selective DNA-binding agents.
Figure 1.
Schematic illustrating the design and structure of the molecular cylinders.
Materials and Methods
Ultrapure water (18.2 ΩM) was used in all experiments. Calf thymus (ct)-DNA was purchased from Sigma/Aldrich. Synthetic double-stranded DNA copolymers, poly[d(A-T)2] (AT) and poly[d(G-C)2] (GC) were obtained from Pharmacia Biochemicals. All polynucleotides were dissolved in water without any further purification and kept frozen until the day of the experiment. The DNA concentrations (moles of bases per liter) of all polynucleotides were determined spectroscopically by using the molar extinction coefficients at the maximum of the long wavelength absorbance: poly[d(A-T)2] ɛ262 = 6,600 cm−1⋅mol−1⋅dm3; poly[d(G-C)2] ɛ254 = 8,400 cm−1⋅mol−1⋅dm3; ct-DNA ɛ258 = 6,600 cm−1⋅mol−1⋅dm3.
[Fe2L3]Cl4 was synthesized following literature methods (21). A sample was freeze-dried, and CHN data (Warwick Analytical Service, Coventry, U.K.: 63.0% C, 4.5% H, 11.6% N) were consistent with the complex having 2½ waters of crystallization: [Fe2(C25H20N4)3]Cl4 (H2O)2.5 (theoretical: 63.1% C, 4.6% H, 11.8% N). Concentrations of stock solutions of the racemic metal complex were determined from accurately weighed samples of this material. ɛ320 was found to be 32,980 mol−1⋅dm3⋅cm−1. Enantiomerically pure material was obtained by using a cellulose (≈20 μm, Aldrich) column and eluting with aqueous NaCl (20 mM) (22). After the first band had eluted, pressure was applied to get the second band off the column. The UV and CD spectra of each band were collected. The compound was concluded to be enantiomerically pure if the per-mol CD spectra were equal and opposite. The first eluting band was identified to be the M enantiomer (for (ΛΛ)-(−)-[Fe2L3]4+) with the longest wavelength negative CD signal from x-ray diffraction and CD theory and the second compound to be P for (ΔΔ)-(+)-[Fe2L3]4+.
A 100 mM stock sodium cacodylate buffer was prepared by mixing 50 ml of 0.2 M solution of sodium cacodylate [42.8 g of Na(CH2)2AsO2⋅3H2O in 100 ml] with 9.3 ml of 0.2 M hydrochloric acid, and diluting to a total of 200 ml. Stock solutions (500 μM) of metal complex were prepared and the salt, sodium chloride, and all DNA experiments were conducted in sodium cacodylic buffer (pH = 6.8, 1 mM) and 20 mM NaCl.
Spectroscopic titration series experiments keeping either the DNA or metal complex concentration constant were undertaken by adding the salt, buffer, water, and metal complex (in that order) to the DNA. A stock solution of DNA or metal complex at the set concentration also was prepared and then used to dilute, respectively, the metal complex or DNA. The CD spectra were run on a Jasco J-715 spectropolarimeter. For each dilution a UV-visible spectrum was recorded on a Jasco V-550 spectrophotometer. Linear dichroism (LD) experiments were performed on a J-715 spectropolarimeter adapted for LD in a 1-mm pathlength couette flow cell (23).
Ethidium bromide (EB) displacement by the metallo helicates was measured by measuring the quenching of the EB fluorescence as it leaves the protection of the DNA. A DNA/EB ratio of 4:5 (12 μM:15 μM) was used and the emission spectra were recorded as a function of triple-helix concentration by using a Perkin–Elmer LS 50b (parameters: excitation, 540 nm; excitation slit, 10.0; emission slit, 15.0).
The heat denaturing of DNA from a double-stranded helix to single-stranded coils, also called melting, was monitored experimentally by observing the absorption of UV light at 260 nm. The midpoint (strictly where 50% of the DNA is duplex and 50% is single-stranded) on the melting curve, termed the melting temperature, Tm, gives an indication of the DNA's thermal stability. The melting curve experiments were run simultaneously on six masked cuvettes of 1.2-ml volume by using a thermostatic cell-changer UV-visible Cary 1E spectrophotometer with parameters: wavelength, 260 nm; signal band width, 1.0 nm; average time, 10.0 s; data interval, 0.2°C; temperature scan, 20−95°C; ramp rate, 0.2°C/min; hold time, 10.0 min.
The high-temperature kinetic CD experiment (85°C) was performed
on solutions that were first equilibrating at 85°C a solution
containing  of the total amount of water, the total salt, and
the total buffer in the Peltier cell holder before adding a room
temperature solution made of the DNA, the remaining water, and the
metal complex (in that order). The metal complex was carefully added
under stirring to avoid precipitation. Single wavelength CD data were
then recorded at 575 nm [metal-to-ligand-charge-transfer (MLCT) band]
every 3 min as well as wavelength scans every 20 min for 4 h. A
parallel UV temperature kinetic experiment was run following the same
sample preparation.
of the total amount of water, the total salt, and
the total buffer in the Peltier cell holder before adding a room
temperature solution made of the DNA, the remaining water, and the
metal complex (in that order). The metal complex was carefully added
under stirring to avoid precipitation. Single wavelength CD data were
then recorded at 575 nm [metal-to-ligand-charge-transfer (MLCT) band]
every 3 min as well as wavelength scans every 20 min for 4 h. A
parallel UV temperature kinetic experiment was run following the same
sample preparation.
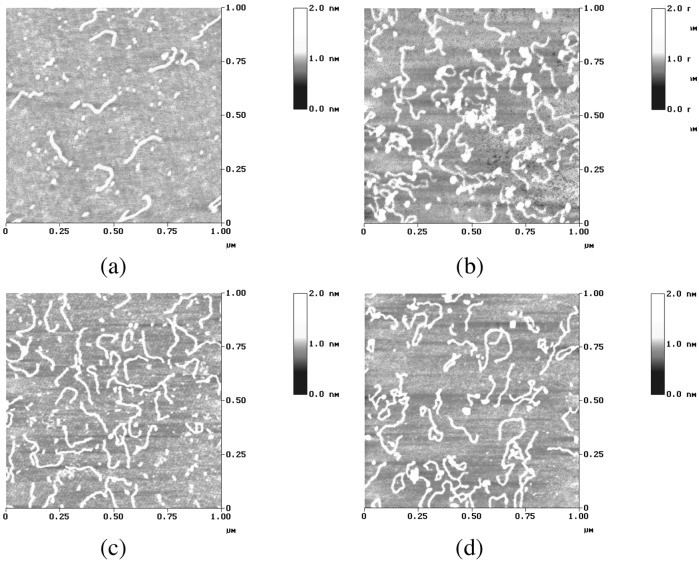
Atomic force microscopy (AFM) images were collected of a fragment of linear DNA 200 bp with [Fe2L3]4+, free DNA, [Fe2L3]4+ enantiomer M, [Fe2L3]4+ enantiomer P, and racemic [Fe2L3]4+ (DNA base pair to metal complex ratio was 10:3 for all of the samples). DNA-metal complex adducts were prepared as follows: the linear DNA was incubated in an appropriate volume with the metal complex in Hepes buffer (4 mM Hepes, pH 7.4/2 mM MgCl2). All solutions were made with 18.2 MΩ water filtered through 0.2-nm FP030/3 filters (Scheicher & Schuell) and centrifuged at 4,000 × g several times to avoid salt deposits and provide a clear background when they were imaged by AFM. The samples were left to equilibrate at 37°C for 5 h in the dark. Samples were prepared for AFM by placing a drop (6 μl) of DNA solution or DNA-metal complex solution onto Ni-treated green mica (Ashville-Schoonmaker Mica, Newport News, VA). After adsorption for 5 min at room temperature, the samples were rinsed for 10 s in a jet of deionized water of 18.2 MΩ⋅cm−1 Milli-Q water directed onto the surface with a squeeze bottle. The samples were blow-dried with compressed argon over silica gel and then imaged by using a Nanoscope III Multimode AFM (Digital Instrumentals, Santa Barbara, CA) operating in tapping mode in air at a scan rate of 1−3 Hz. The AFM probes were 125-nm-long monocrystalline silicon cantilevers with integrated conical-shaped Si tips (Nanosensors, Norderfried Richskoog, Germany) with an average resonance frequency of 330 kHz and spring constant K = 50 N/m. The cantilever is rectangular and the tip radius given by the supplier is 10 nm with a cone angle of 35° and high aspect ratio. The images were obtained at room temperature (T = 20 ± 2°C) and typical relative humidity of >55%. Four different samples from each reaction were imaged in several places and many times to obtain reliable measurements.
Results
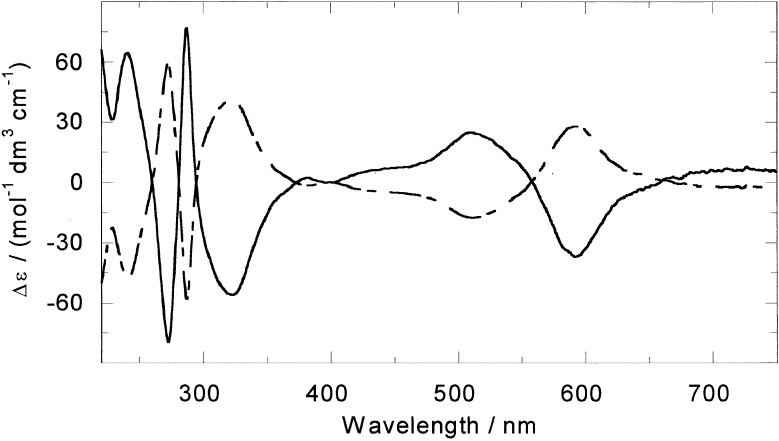
The key discovery that has enabled us to work with the resolved enantiomers of [Fe2L3]4+ (Fig. 1) is that enantiomerically pure samples in milligram quantities can be obtained by loading the racemic mixture on a cellulose column and eluting with 20 mM NaCl (22). The M enantiomer elutes first. The CD spectra of the two enantiomers in aqueous NaCl (20 mM) solution are given in Fig. 2. Both enantiomers were extremely effective at displacing EB from DNA in an EB displacement assay, indicating that both enantiomers have binding constants at 20 mM NaCl well in excess of 107 M−1. It therefore can be assumed that mixing ratios in solution correspond to binding ratios for the subsequent experiments.
Figure 2.
CD spectra of M-[Fe2L3]4+ (solid line) and P-[Fe2L3]4+ (dashed line).
Absorption Spectroscopy.
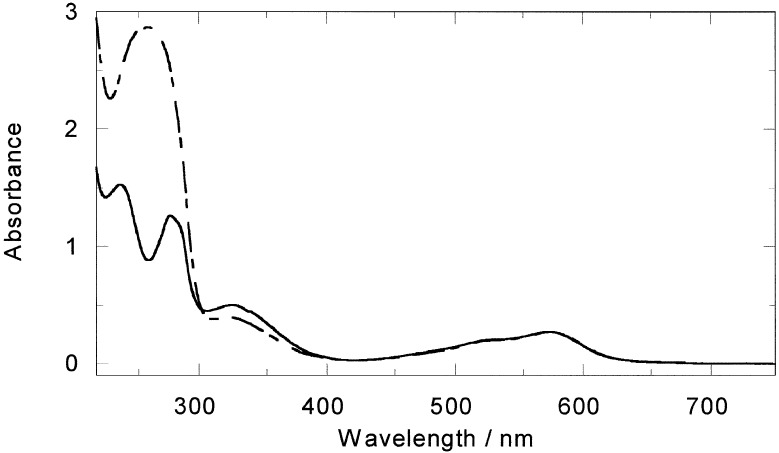
In the presence of ct-DNA a decrease in the absorbance intensity of the in-ligand band of the M enantiomer together with a small (2 nm) wavelength shift toward longer wavelength of the MLCT maximum (Fig. 3) was observed. The P enantiomer (not shown) gave only a small decrease in intensity in the in-ligand region and had no effect on the MLCT region.
Figure 3.
Absorbance spectra of M-[Fe2L3]4+ (20 μM) alone and in the presence of ct-DNA (10:1 base/metal complex mixing ratio).
CD.
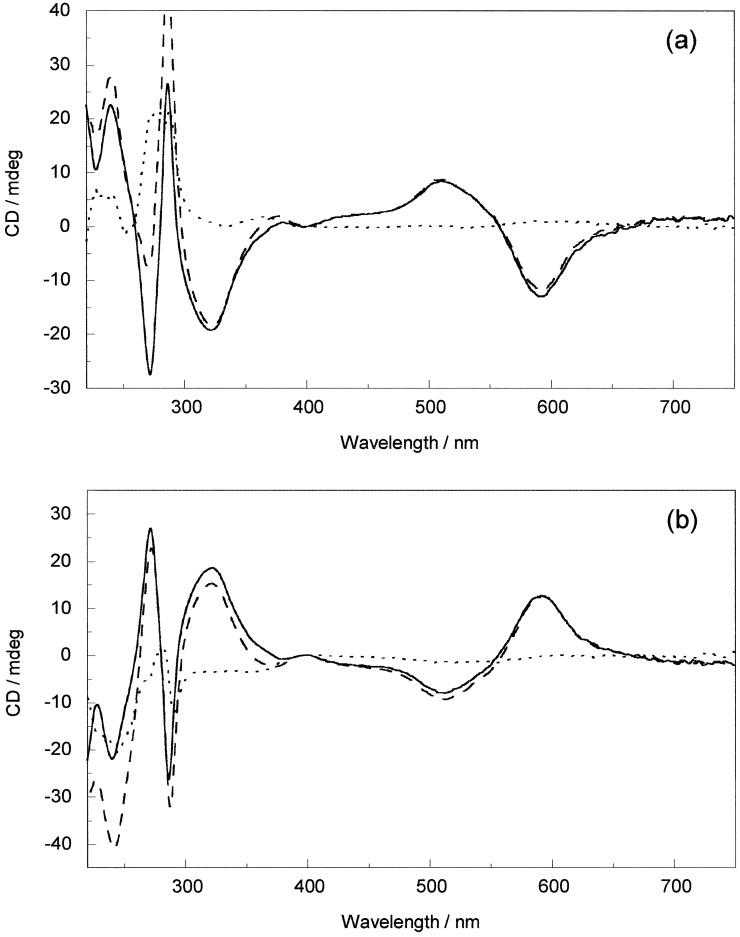
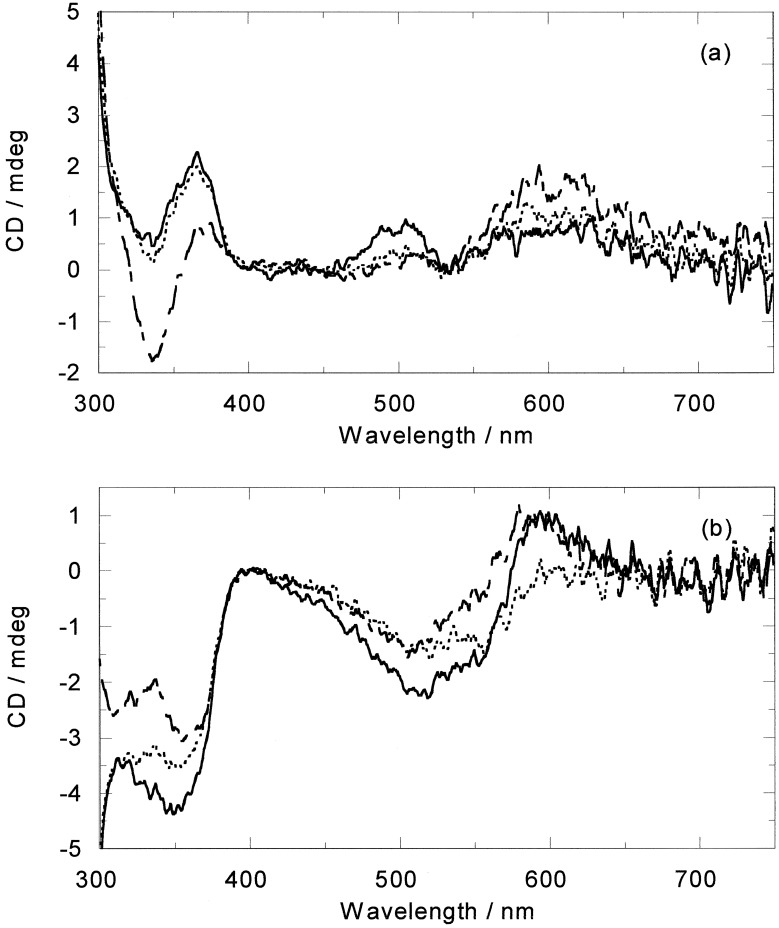
CD is the difference in absorption of left and right circularly polarized light. As such it is uniquely sensitive to any asymmetric interaction such as that between the chiral DNA and the chiral metallo helicate. Compared with the absorption spectra, the CD spectra of both enantiomers show more dramatic changes upon addition of ct-DNA. In the short wavelength region of M, a small wavelength shift is noticed, whereas in the longer wavelength part of the MLCT wavelength region a decrease in intensity is apparent. Upon subtracting the M CD from the DNA−M CD, the induced CD (ICD) of Fig. 4a is obtained. Intriguingly, on addition of P to ct-DNA, as well as slightly larger in-ligand spectra changes, the MLCT band is perturbed only in the shorter wavelength part, whereas the longer wavelength ICD stays constant (Fig. 4b). This result is extremely useful as it means we have independent probes of the binding of the two enantiomers to ct-DNA.
Figure 4.
CD spectra of (a) M and (b) P (20 μM) alone (solid line) and in the presence of ct-DNA (10:1 base/metal complex mixing ratio) (dashed line) and the resulting ICD (dotted line).
To get an indication of any sequence variations, the ICD obtained for both enantiomers with AT-, GC-, and ct-DNA were compared (Fig. 5). The longer wavelength part of the ICDs of the M enantiomer overlay, whereas the shorter wavelength MLCT band has the ct-DNA intermediate between the two alternating homopolymers, suggesting different spectroscopic interactions but little sequence preference for this enantiomer. By way of contrast, the P enantiomer shows bigger MLCT ICD signals with both AT- and GC-DNA than ct-DNA, indicating that it prefers the regular alternating purine and pyrimidine structure given by the synthetic DNAs.
Figure 5.
ICD on addition of (a) M and (b) P (20 μM) to AT-DNA (solid line), GC-DNA (dashed line), and ct-DNA (dotted line) (10:1 base/metal complex mixing ratio).
Flow LD Spectroscopy.
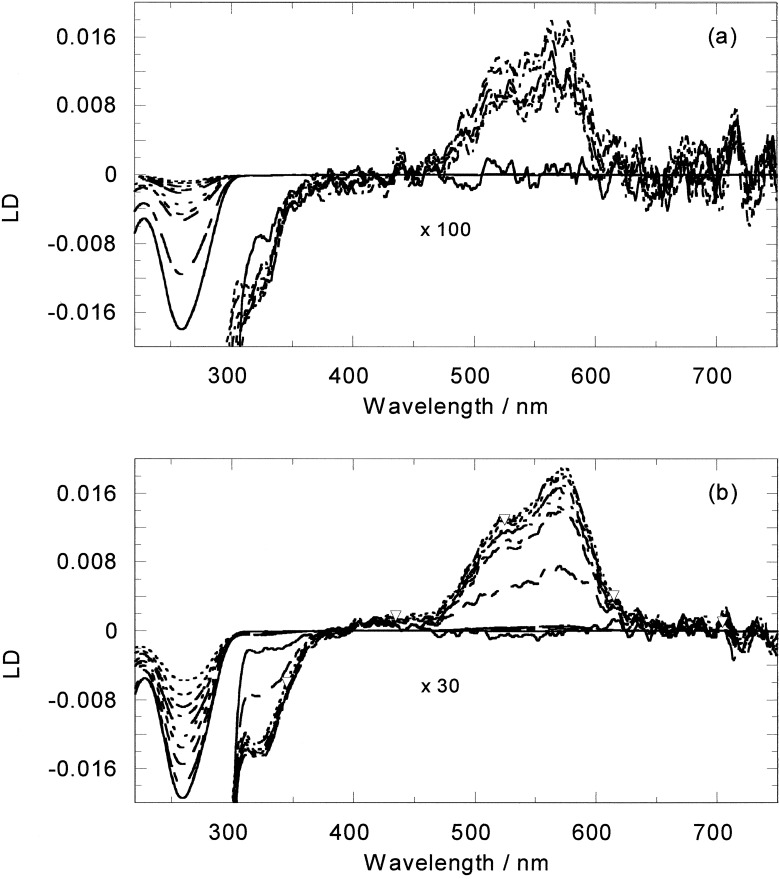
LD is the difference in absorption of light polarized parallel and perpendicular to an orientation direction. In the case of DNA-ligand systems it is usually a good indicator of the orientation of the ligand on the DNA. However, for the racemic metallo helicate the changes in the DNA LD spectrum upon addition of the ligand were dominated by the loss of DNA orientation associated with the bending or coiling of the DNA by the ligand (20). Flow LD data for the two enantiomers with ct-DNA therefore were collected to determine the average bending effect of the enantiomers compared with the racemate. The LD signal of ct-DNA in the presence of the M enantiomer is very small in the MLCT band and the DNA signal itself decreases dramatically even at very small mixing ratios (Fig. 6). The DNA solution was also observed to become significantly more viscous on the addition of M. At 100:1 DNA base/M, a 40% orientation loss was observed, at 50:1 two-thirds of the LD DNA intensity signal is lost, and at 20:1 ratio less than 5% of the signal remains (compared with 30% and 60% losses at these ratios for the racemate; ref. 20). Once again, P behaves differently, although still giving an increase in solution viscosity. The changes in the DNA LD region induced by Δ are less. For a 100:1 ratio there is only 8% loss in the LD signal intensity and for a 15:1 ratio a 43% loss. A change in the DNA LD signal shape was observed for ratios below 20:1, with the signal becoming broader, suggesting the existence of two different DNA-binding modes.
Figure 6.
LD spectra of free and (a) M or (b) P-bound ct-DNA (200 μM). Solid line is DNA alone, decreasing mixing ratios correspond to decreases in the 260-nm LD signal. Ratios are (a) 100:1, 50:1, 40:1, 35:1; 30:1; 27:1, 25:1, 22:1, 20:1; and (b) 100:1; 50:1, 40:1, 35:1; 30:1; 25:1, 20:1, 16:1, 13:1. Long wavelength region is shown to scale and ×100 (a) or ×30 (b) as indicated.
The MLCT LD for P is larger than that for M and increases in magnitude up to a 20:1 ratio (as would be expected for increasing M concentration in the absence of DNA bending) and then levels out, again suggesting a change in binding above this ratio.
High-Temperature Absorption.
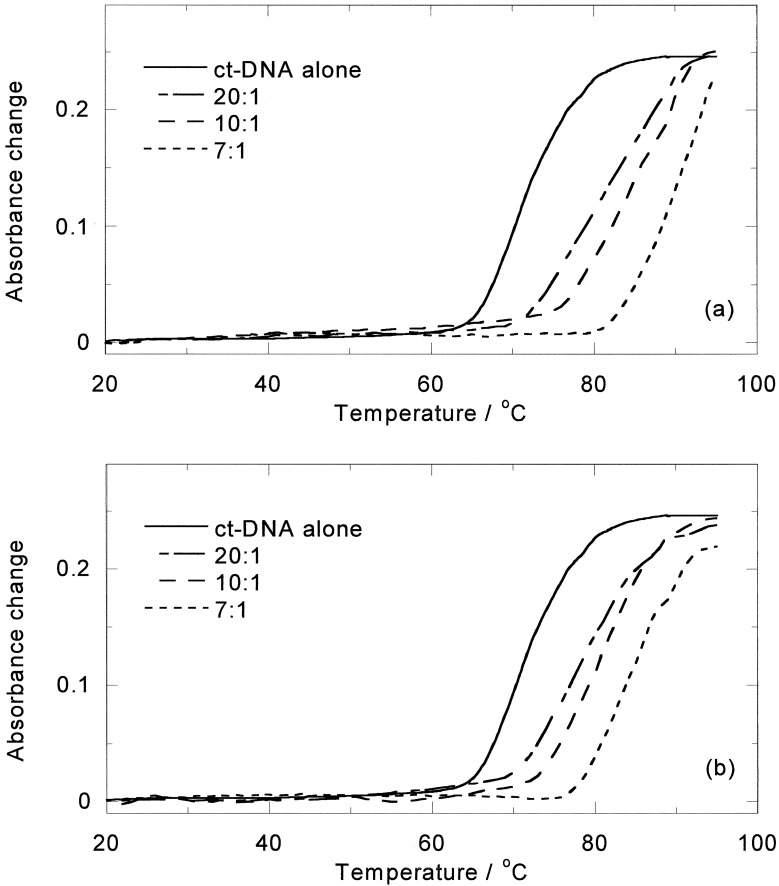
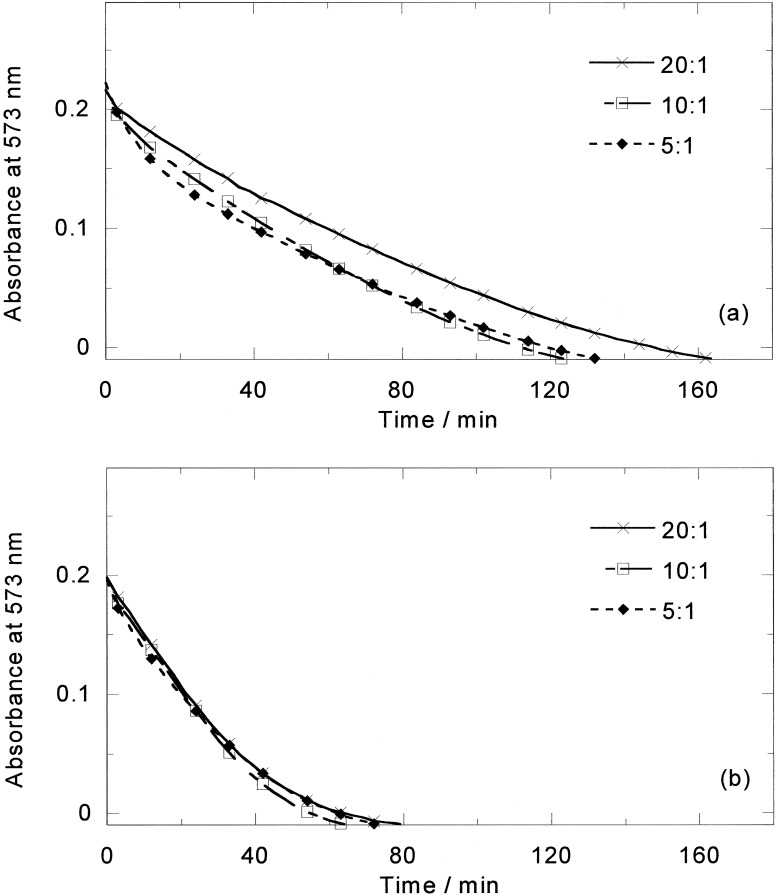
On heating aqueous solutions of either enantiomer or the racemic mixture at 85°C, in the absence of DNA, the purple color quickly disappears from solutions, indicating breakdown of the complex. However, in the presence of DNA the color fades but does not disappear on a time scale of hours. The high-temperature CD spectrum (24) of [Fe2L3]4+ with ct-DNA suggests that enrichment of one enantiomer (now known to be M) takes place, indicating either preferential binding or preferential stabilization of that enantiomer. The effect of temperature on the two enantiomers with ct-DNA therefore was investigated. On the addition of M to a ct-DNA solution, the DNA melting temperature was found to increase from 71°C to 82°C and 89°C for M and 78°C and 81°C for P at 20:1 and 10:1 mixing ratios (Fig. 7). Conversely the DNA was also found to stabilize M (half-life ≈3 h), although it had little effect on P (half-life ≈20 min), as shown in Fig. 8, which summarizes the ICDs at 85°C. For M, the rate of decomposition is slowest up to a 20:1 DNA base/M, suggesting that up to ≈20:1 M is bound in a mode where it is protected from degradation. At higher loadings other modes are occupied that, similar to the P modes, afford little protection. Thus the previous racemate CD signal increase was caused by the more rapid degradation of the P enantiomer (relative to the DNA-stabilized M enantiomer) rather than temperature-induced change of binding mode or Pfeiffer effect.
Figure 7.
Melting curves of ct-DNA (100 μM) in the presence of (a) M and (b) P. Mixing ratios are indicated.
Figure 8.
Absorbance at 573 nm vs. time at 85°C of (a) ct-DNA−M (20 μM) and (b) ct-DNA-P at different DNA base/metal complex ratios indicated on spectra.
AFM.
To get a molecular-level picture of the binding and DNA bending or coiling, we acquired AFM tapping mode images of the cylinder with a fragment of a linear DNA 200 bp (Fig. 9). The images are entirely consistent with both the LD data for the enantiomers and the racemate's AFM images collected previously. The M enantiomer kinks and coils the DNA significantly more than does the P enantiomer whose effect is still significant when compared with the free DNA. In the image corresponding to the racemic cylinders, approximately 50% of the fragments are kinked more than the rest, confirming the effect observed for the separate enantiomers.
Figure 9.
AFM images of a fragment of 200-bp linear DNA incubated at 37°C for 5 h with the iron cylinder. (a) Free DNA. (b) DNA incubated with enantiomer M at ratio 10:3. (c) DNA incubated with enantiomer P at ratio 10:3. (d) DNA incubated with rac-[Fe2(LL)3]4+ at ratio 10:3.
Discussion
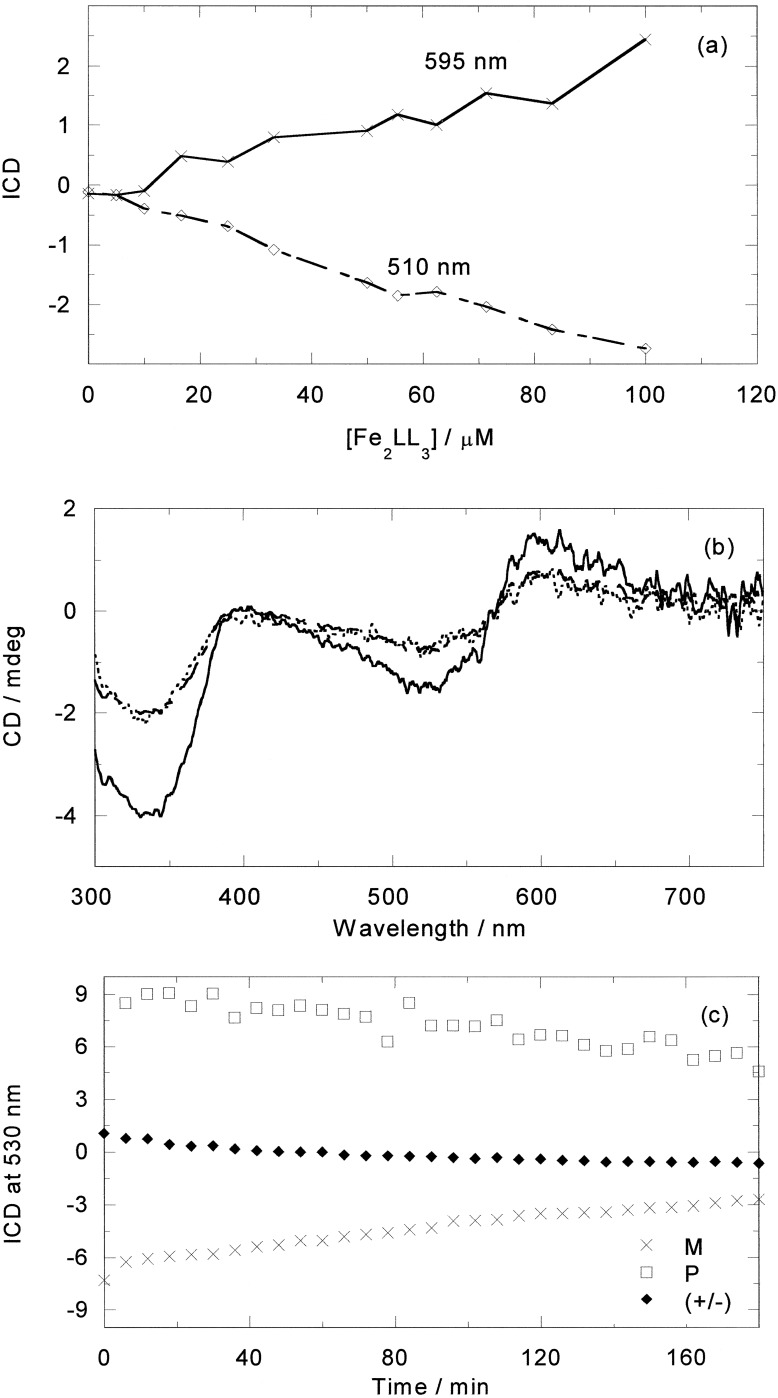
From the spectroscopic data, we can conclude that both enantiomers of [Fe2L3]4+ bind strongly to DNA, with binding modes that are clearly very different as are their structural effects on the DNA. Comparison of the previously obtained CD results for the racemate with ct-DNA reveal that its behavior is the “average” of the results reported here for the two enantiomers, suggesting that each enantiomer interacts independently from the other one. A plot of the ICD of the racemate with DNA versus metal complex concentration at wavelengths selected to probe only the M (≈595 nm) or only the P (≈510 nm) enantiomers (Fig. 10a) gives two symmetrical straight lines, suggesting that for each M binding a P also binds (because the binding constants are very high), thus indicating no quantitative enantiomeric binding preference at these concentrations. To confirm this, a racemate ICD spectrum was reconstructed from a 50:50 mix of the M and P results (Fig. 10b). The reconstruction overlays with the experimentally obtained racemate spectrum, indicating that the enantiomers interact with the DNA independently, suggesting that the enantiomers target different regions of the DNA. The racemate temperature-dependent behavior in the presence of ct-DNA can be similarly accounted for (Fig. 10c).
Figure 10.
(a) ICD vs. (±)-[Fe2L3]4+ concentration at 510 and 595 nm; [ct-DNA] = 500 μM. (b) ICD of (±)-[Fe2L3]4+ and M plus P (200 μM ct-DNA, 20 μM metal complex). (c) ICD at 530 nm at 85°C of (±)-[Fe2L3]4+ (20 μM) and each enantiomer (10 μM) in the presence of ct-DNA (200 μM) as a function of time.
The LD data in particular suggest that each enantiomer occupies one binding mode until the ligand loading is in excess of ≈20:1 DNA base/ligand (i.e., 10 bp/1 enantiomer). Given the independent action of the two enantiomers this finding is consistent with the previously observed ≈10:1 change point observed for the racemate. It is intriguing that this corresponds to one ligand per turn of the DNA helix, which may explain the dramatic bending effect of the M enantiomer—if it adopts a DNA-bending binding mode then this phasing would ensure that the effect reinforced rather than cancelled. The precise orientations of the enantiomers are difficult to determine from the LD experiments because no independent measure of the DNA orientation is possible (attempts to use e.g., methylene blue as a calibrant foundered on the fact that such ligands are displaced by the metallo helicate; data not shown). However, we can qualitatively conclude from our knowledge of the helicate transition moments (24) that both enantiomers, below a 20:1 ratio, bind with the molecule long axis more parallel than perpendicular to the DNA helix axis.
On the basis of our previous NMR work (20), we can conclude that the M enantiomer's first binding mode is along the major groove. The P enantiomer's preferred binding mode must be (i) less protected, in that the DNA does not significantly increase the resistance of the metal complex to thermal degradation (presumably reactivity of the imine bond) and (ii) is not competitive with the M binding mode. It does, however, have a definite CD signature and orientation on the DNA, indicating it is not a loose association with the backbone. The binding position of this enantiomer was not revealed by 1H NMR nuclear Overhauser effect spectroscopy experiments, indicating either that residence time in any one specific location is short and exchange is fast on the NMR time scale or that the P cylinder is distant from the DNA protons. The independent nature of the two enantiomers' binding to DNA leads us to conclude that P is unlikely also to bind in the major groove in the low-concentration binding mode. Its overall orientation along the helical axis leads us to speculate that P lies along the surface of the minor groove perhaps interacting with the two phosphate backbones. Steric considerations appear to preclude extensive insertion of the cylinder into this minor groove (the DNA region CD is not significantly changed at low cylinder concentrations).
The bimetallo triple helicate structure can be viewed as two tris-chelate transition metal complexes, such as [Ru(1,10-phenanthroline)3]2+, stacked together and binding to DNA as a unit. Our previous experimental and molecular modeling studies of Λ-[Ru(1,10-phenanthroline)3]2+ at high ligand load (modeled by two or three ligands stacked together) showed a preference for what we referred to as the major groove partial insertion binding mode (11). In this mode one of the phenanthroline chelates slots into the edge of a base pair but does not cause it to move significantly apart as in an intercalative mode. Inspection of the NMR structure (20) of M-[Fe2L3]4+ is consistent with this description of its binding.
The binding mode of Δ-[Ru(1,10-phenanthroline)3]2+ at high ligand load was found to be more complicated than for the Λ enantiomer (11). In particular, there was evidence of the binding of Δ-[Ru(1,10-phenanthroline)3]2+ in both grooves. Molecular modeling and LD orientations led us to conclude that Δ favored two minor groove binding modes, one where a chelate is slotted into the groove and one where two chelates span the groove, rather than the partially inserted major groove binding mode. With the bi-metallo triple helicate a binding mode analogous to this could be adopted. An M enantiomer major groove binding mode (as determined from NMR) and a possible minor groove P enantiomer binding mode are illustrated in Figs. 11 and 12, which are published as supporting information on the PNAS web site, www.pnas.org.
Conclusion
The two enantiomers of [Fe2L3]4+ thus may be concluded to bind in different binding modes. The M enantiomer preferentially adopts a major groove mode with partial insertion of one of its chelates between DNA bases and the P enantiomer may sit along the minor groove spanning the two phosphate backbones.
To pursue our goal of DNA major groove sequence selectivity by synthetic supramolecular assemblies we are currently derivatizing the M enantiomer backbone to achieve additional hydrogen bonding and steric effects with the major groove. Our preliminary efforts in this direction indicate that at least an element of sequence selectivity can be achieved in such a fashion.
The other intriguing feature of both enantiomers' interaction with DNA is its significant effect on the DNA's extent of coiling while having little effect on the very local structure of base-base interactions that affect the CD spectra. In this way they resemble the molecules involved in packaging DNA into chromosomes. The effect is particularly dramatic with the major groove binding M enantiomer. It is anticipated that tuning sequence selectivity into the backbone of this enantiomer will enable us to target sequences to be coiled.
Supplementary Material
Acknowledgments
We thank the Leverhulme Trust (Grant F/125/BC to C.J.I.), the University of Warwick (to I.M.), and the Engineering and Physical Sciences Research Council Life Sciences Initiative (Grant GR/M91105) for support.
Abbreviations
- ct
calf thymus
- LD
linear dichroism
- EB
ethidium bromide
- AFM
atomic force microscopy
- MLCT
metal-to-ligand charge-transfer
- ICD
induced CD
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sinden R R. DNA Structure and Function. London: Academic; 1994. [Google Scholar]
- 2.Dickerson R E. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branden C, Tooze J. Introduction to Protein Structure. 2nd Ed. New York: Garland; 1999. [Google Scholar]
- 4.Thuong N T, Helene C. Angew Chem Int Ed Engl. 1993;32:666–690. [Google Scholar]
- 5.Nielsen P E, Haaima G. Chem Soc Rev. 1997;26:73–78. [Google Scholar]
- 6.White S, Szewczyk J W, Turner J M, Baird E E, Dervan P B. Nature (London) 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 7.Baguley B C. Mol Cell Biochem. 1982;43:167–181. doi: 10.1007/BF00223008. [DOI] [PubMed] [Google Scholar]
- 8.Denny W A. Anti-Cancer Drug Des. 1989;4:241–263. [PubMed] [Google Scholar]
- 9.Lippard S J, Berg J M. Principles of Bioinorganic Chemistry. Mill Valley, CA: University Science Books; 1994. [Google Scholar]
- 10.Lippert B, editor. Cisplatin, Chemistry and Biochemistry of a Leading Anti-Cancer Drug. Weinheim, Germany: Wiley-VCH; 1999. [Google Scholar]
- 11.Coggan D Z, Haworth I S, Bates P J, Robinson A, Rodger A. Inorg Chem. 1999;38:4486–4497. doi: 10.1021/ic990654c. [DOI] [PubMed] [Google Scholar]
- 12.Onfelt B, Lincoln P, Norden B. J Am Chem Soc. 1999;121:10846–10847. [Google Scholar]
- 13.Erkkila K E, Odom D T, Barton J K. Chem Rev. 1999;99:2777–2795. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
- 14.Palmer C R, Sloan L S, Adrian J C, Cuenoud B, Paolella D N, Schepartz A. J Am Chem Soc. 1995;117:8899–8907. [Google Scholar]
- 15.Lasey R C, Banerji S S, Ogawa M Y. Inorg Chim Acta. 2000;300–302:822–828. [Google Scholar]
- 16.Hastings C A, Barton J K. Biochemistry. 1999;38:10042–10051. doi: 10.1021/bi982039a. [DOI] [PubMed] [Google Scholar]
- 17.Lehn J-M. Supramolecular Chemistry: Concepts and Perspectives. Weinheim, Germany: VCH; 1995. [Google Scholar]
- 18.Philp D, Stoddart J F. Angew Chem Int Ed Engl. 1996;35:1155–1196. [Google Scholar]
- 19.Schoentjes B, Lehn J-M. Helv Chim Acta. 1995;79:1–12. [Google Scholar]
- 20.Hannon M J, Moreno V, Prieto M J, Moldrheim E, Sletten E, Meistermann I, Isaac C J, Sanders K J, Rodger A. Angew Chem Int Ed Engl. 2001;40:879–884. doi: 10.1002/1521-3773(20010302)40:5<879::AID-ANIE879>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 21. Hannon, M. J., Painting, C. L., Hamblin, J., Jackson, A. & Errington, W. (1997) Chem. Commun., 1807–1808.
- 22. Hannon, M. J., Meistermann, I., Isaac, C. J., Blomme, C., Aldrich-Wright, J. & Rodger, A. (2001) Chem. Commun. 1078–1079.
- 23.Rodger A. Methods Enzymol. 1993;226:232–258. doi: 10.1016/0076-6879(93)26012-x. [DOI] [PubMed] [Google Scholar]
- 24.Rodger A, Sanders K J, Hannon M J, Meistermann I, Parkinson A, Vidler D S, Haworth I S. Chirality. 2000;12:221–236. doi: 10.1002/(SICI)1520-636X(2000)12:4<221::AID-CHIR9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.