Abstract
Evidence is emerging that mechanical stretching can alter the functional states of proteins. Fibronectin (Fn) is a large, extracellular matrix protein that is assembled by cells into elastic fibrils and subjected to contractile forces. Assembly into fibrils coincides with expression of biological recognition sites that are buried in Fn's soluble state. To investigate how supramolecular assembly of Fn into fibrillar matrix enables cells to mechanically regulate its structure, we used fluorescence resonance energy transfer (FRET) as an indicator of Fn conformation in the fibrillar matrix of NIH 3T3 fibroblasts. Fn was randomly labeled on amine residues with donor fluorophores and site-specifically labeled on cysteine residues in modules FnIII7 and FnIII15 with acceptor fluorophores. Intramolecular FRET was correlated with known structural changes of Fn in denaturing solution, then applied in cell culture as an indicator of Fn conformation within the matrix fibrils of NIH 3T3 fibroblasts. Based on the level of FRET, Fn in many fibrils was stretched by cells so that its dimer arms were extended and at least one FnIII module unfolded. When cytoskeletal tension was disrupted using cytochalasin D, FRET increased, indicating refolding of Fn within fibrils. These results suggest that cell-generated force is required to maintain Fn in partially unfolded conformations. The results support a model of Fn fibril elasticity based on unraveling and refolding of FnIII modules. We also observed variation of FRET between and along single fibrils, indicating variation in the degree of unfolding of Fn in fibrils. Molecular mechanisms by which mechanical force can alter the structure of Fn, converting tensile forces into biochemical cues, are discussed.
Extracellular matrices (ECM) are complex supramolecular assemblies that control cell signaling and behavior. While many ECM proteins have been characterized, little is known about how matrix assembly alters their structure and confers functions not present in individual proteins. Less is known about mechanisms by which cell contractile forces applied to protein assemblies regulate protein function. Many ECM proteins, such as fibronectin (Fn), laminin, and thrombospondin, are large (>100 kDa), multifunctional proteins composed of repeating, structurally defined modules often less than 100 aa each. The multimodular structure may serve as a convenient way to integrate multiple functions in one molecule. For example, some modules carry cell adhesion sites, whereas others carry sites for binding other proteins and for self-assembly. Exposure of functional sites may be controlled by the organization of such sites in extracellular matrices, and by the application of force by cells (1–3). However, the molecular mechanisms regulating ECM protein assembly and the exposure of binding sites remain unclear.
Fn is an ECM protein that undergoes cell-mediated assembly into insoluble, elastic fibrils. In blood and when secreted by cells such as fibroblasts, Fn exists as a soluble dimer. The two strands of the dimer are composed of three types of repeating globular modules (FnI, FnII, and FnIII) linearly connected by short chains of variable flexibility, and are linked at their C termini by a pair of disulfide bonds (4, 5). While the soluble form of Fn is relatively inactive, assembly into fibrils results in expression of most of Fn's biological activities. These activities include mediation of cell adhesion, proliferation, and differentiation, as well as embryogenesis and wound healing (6). Fibrillar networks of Fn mechanically couple cells to their environment. It has been shown that cell contractility is required for assembly of Fn into fibrils (7–11), and that stretching exposes cryptic sites embedded in the protein (12, 13). Furthermore, Fn fibrils are highly elastic and subject to cell-generated tension (14). Thus, Fn is an excellent model protein for investigating how protein function is controlled by matrix assembly and stretching.
Experimental techniques are needed to establish how cells alter the structure of proteins in fibrillar matrices and how structural changes relate to changes in function. We have recently demonstrated the application of fluorescence resonance energy transfer (FRET) between multiple donor and acceptor fluorophores attached to Fn to detect different Fn conformations in cell culture (15). As Fn unfolds, nanometer-scale increases in the mean distance between donors and acceptors cause decreases in FRET, observed as a decrease in donor emission and an increase in acceptor emission. Fn molecules unfolded to different degrees in cell culture are distinguished visibly by the color of emission in fluorescence microscopy, and spectrally by using a microscope-attached spectrometer. Before cell experiments, we correlated FRET with known Fn unfolding behavior in solution by denaturing Fn with guanidium chloride. We then added labeled Fn to the culture medium of NIH 3T3 fibroblasts, and allowed the cells to incorporate it into matrix fibrils. Based on intramolecular FRET, we have previously shown that Fn in cell fibrils is highly extended relative to Fn diffusely bound to the cell membrane. Here we demonstrate that cytoskeletal tension generated by surface adherent fibroblasts stretches Fn molecules within fibrils so that FnIII modules are unfolded, and that disruption of cytoskeletal tension using cytochalasin D allows refolding. These results give insight into the molecular basis of Fn matrix structure and elasticity.
Materials and Methods
Fn Labeling.
Fn labeling, correlation of FRET to Fn unfolding in solution, and microscopy and spectroscopy have been described (15). Briefly, human plasma Fn (>95% purity, Life Technologies) was randomly labeled on cysteine residues within modules FnIII7 and FnIII15 with the acceptor fluorophore tetramethylrhodamine-5-maleimide (Molecular Probes). The conjugate was separated from unbound fluorophores by size exclusion chromatography, then site-specifically labeled with the amine-reactive donor fluorophore, Oregon Green 488 carboxylic acid-succinimidyl ester-6-isomer (Molecular Probes). Fn labeled with donors and acceptors (Fn-D/A) was purified as before, and labeling ratios were calculated. The two-step labeling process resulted in an average of 7.0 Oregon Green donors and 3.7 tetramethylrhodamine acceptors per Fn dimer for the batch used in all experiments presented in this paper. Sensitivity of FRET to Fn unfolding was evaluated by denaturing Fn-D/A in PBS with guanidine hydrochloride (Gdn⋅HCl), using concentrations ranging from 0 to 8 M. The resulting curve was used to correlate FRET to the degree of Fn-D/A unfolding and to evaluate Fn-D/A unfolding in cell culture. Dilution of Fn-D/A with PBS had no effect on the ratio of donor and acceptor emission, indicating an absence of energy transfer between adjacent proteins in solution. Measurement of FRET in solution was performed by using an epifluorescence microscope with an attached spectrometer.
Incorporation of Labeled Fn into Cell Matrix Fibrils.
Sterile glass coverslips were coated with unlabeled Fn by adsorption from 25 μg/ml Fn in PBS for 1 h at 25°C. NIH 3T3 fibroblasts (American Type Culture Collection) were plated at a density of 5 × 103 cells per cm2 in DMEM with 10% FBS. Unbound cells were removed by rinsing at 1 h, and a mixture of Fn-D/A with a 10-fold excess of unlabeled Fn was added to the culture medium to give a final Fn concentration of 100 μg/ml. The 10-fold excess of unlabeled Fn was used to prevent energy transfer between adjacent proteins. Samples were incubated for 24 h to allow cells to incorporate Fn-D/A into mature matrix fibrils. To remove newly adherent Fn-D/A from fibrils before fixing the samples, medium containing Fn-D/A was replaced with growth medium supplemented with 100 μg/ml unlabeled Fn, and cells were incubated for an additional 3 h. Cells were fixed with 4% paraformaldehyde in PBS for 30 min and mounted on glass slides with ProLong anti-photobleaching reagent (Molecular Probes).
Treatment with Cytochalasin D (cytoD) After Matrix Assembly.
To disrupt contractile forces, cells were treated with cytoD, a reagent that caps the fast-growing end of actin filaments, disrupting their polymerization in the cytoskeleton. CytoD was added to the growth medium at a final concentration of 10 μM, 1 h before fixing the cells.
Fluorescence Microscopy and Spectroscopy of Individual Fn Fibrils.
Imaging and spectroscopy of Fn-D/A in cell culture was performed by using an inverted epifluorescence microscope (Nikon TE 200) with an attached spectrometer (Acton 150, Acton Research Corporation, Acton, MA), and have been described (15). Spectra were collected only from fibrils that were visibly separate from cells to avoid superposition of spectra from Fn-D/A bound to the cell surface and in fibrils. A fibril was centered on the charge-coupled device camera, and a rectangular slit was inserted in the emitted light path to isolate the fibril of interest, blocking fluorescence from surrounding cells and fibrils. A spectroscopy grating (300 grooves per mm, 500-nm blaze angle) was inserted in place of the imaging mirror. A spectral image of the emission from an entire fibril was analyzed by performing a line scan, which integrated emission from a 0.5-μm segment along the fibril. Spectra were collected roughly every 1 μm along a fibril, and spectra with weak, background-level intensities were not used for data analysis. Spectra were normalized to the donor peak, and acceptor peak intensity was calculated as the average intensity 2.25 nm to either side of the 570-nm acceptor peak. Eleven fibrils from untreated samples and eight fibrils from samples treated with cytoD were analyzed.
Results
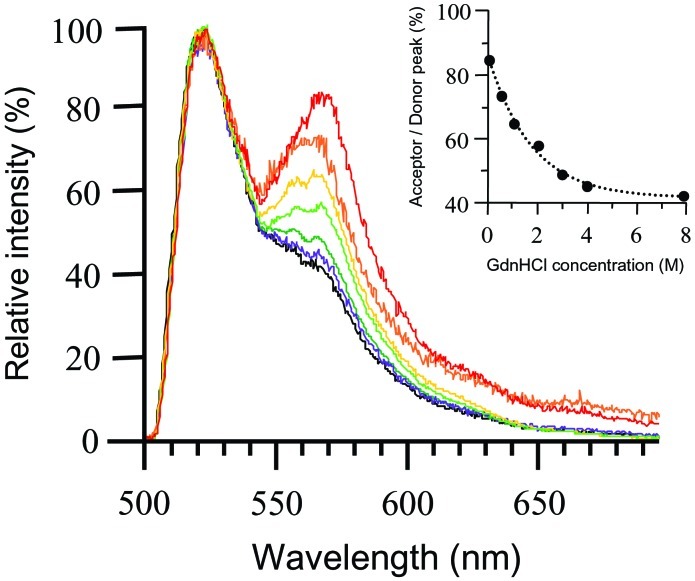
Fn was labeled with donor and acceptor fluorophores using two reactions: amine residues were randomly labeled with Oregon Green, the donor fluorophore, and the free cysteines in modules FnIII7 and FnIII15 were site-specifically labeled with tetramethylrhodamine, the acceptor. We tested the sensitivity of FRET to conformational changes by unfolding Fn-D/A in solution using Gdn⋅HCl. Fig. 1 shows the decrease in FRET as Fn-D/A is exposed to progressively higher concentrations of Gdn⋅HCl in PBS (15). Energy transfer involves an increase in acceptor emission accompanied by a decrease in donor emission; however, we have normalized spectra to the donor peak so that changes in energy transfer are reflected only by changes in the acceptor peak. The decrease in FRET from 0 to 2 M Gdn⋅HCl (Fig. 1) indicated Fn unfolding, and was likely caused by separation of Fn's two strands because of disruption of ionic interactions that stabilize the globular state (16). Additional decreases in FRET at >2 M Gdn⋅HCl indicated further Fn unfolding. In these stronger denaturing conditions, it has been shown that Fn modules progressively unfold (17). Thus, decreases in FRET at >2 M Gdn⋅HCl were likely the result of donor–acceptor separation caused by module unfolding. Fn-D/A in 8 M Gdn⋅HCl exhibited a spectrum indistinguishable from that of a mixture of free donor and acceptor fluorophores in solution, indicating complete loss of FRET. The fluorescence remaining at 570 nm after complete loss of FRET was contributed by the spectral tail of the donor emission and by direct excitation of the acceptor.
Figure 1.
Sensitivity of energy transfer to Fn-D/A unfolding in denaturing solution. Fn-D/A emission spectra were measured by using an epifluorescence microscope with an attached spectrometer. Spectra were normalized to the donor peak so that changes in energy transfer were indicated by changes in the acceptor peak. Fn-D/A was progressively denatured over a range of Gdn⋅HCl concentrations, from 0 M to 8 M. Fn-D/A in 0 M Gdn⋅HCl is shown in red, 0.5 M in orange, 1 M in yellow, 2 M in light green, 3 M in dark green, 4 M in purple, and 8 M in black. The emission spectrum of Fn-D/A in 8 M Gdn⋅HCl was indistinguishable from that of a mixture of free donor and acceptor fluorophores in PBS. The small shoulder at 570 nm at 8 M Gdn⋅HCl was not caused by energy transfer, but by superposition of the donor fluorescence tail and fluorescence from direct excitation of the acceptor. (Inset) FRET (acceptor peak divided by donor peak) as a function of Gdn⋅HCl concentration.
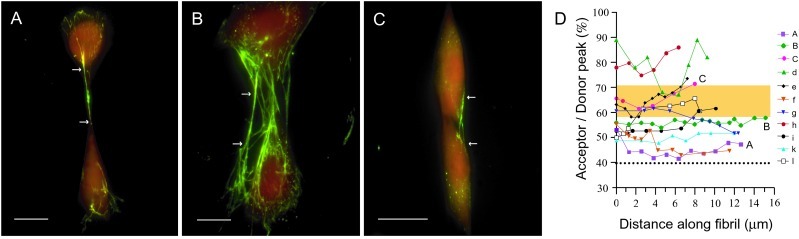
Fig. 2 A–C shows fluorescence images of Fn-D/A in matrix fibrils stretched between NIH 3T3 fibroblasts adherent to Fn-coated glass. Fibrils appear predominantly green and cell surfaces appear red, indicating a lower level of energy transfer in the fibrillar protein. This color difference, complemented by fluorescence spectra collected from individual fibrils (Fig. 2E), indicated that fibrillar Fn-D/A was more extended than Fn-D/A diffusely bound to the cell surface. FRET associated with cell-surface Fn-D/A was relatively uniform, and was comparable to that of Fn-D/A in PBS with no denaturant. FRET in fibrillar Fn was comparable to that of Fn-D/A in 0.5 M to 4 M Gdn⋅HCl solution. Further spectroscopic analysis of fibrils revealed significant variation in FRET both along individual fibrils and between fibrils. Some fibrils exhibited uniform FRET along the entire fibril (<5% variation), while others varied widely (up to 25%) (Fig. 2D).
Figure 2.
Fluorescence images and spectra of Fn-D/A in fibroblast matrix fibrils. (A–C) Fn-D/A was added to the culture medium of NIH 3T3 fibroblasts and incorporated into their fibrillar matrix. Excess unlabeled Fn was added to prevent intermolecular energy transfer. Fibrils visibly separated from other fibrils and from cells were analyzed. Arrows indicate beginning and end points of a series of spectra collected along a fibril. Spectra were integrated from 0.5-μm fibril segments spaced roughly 1 μm apart. (Scale bars, 10 μm.) (D) FRET varied between fibrils and along individual fibrils. Spectra labeled A, B, and C correspond to fibrils in images. Other fibrils that were analyzed are designated by lowercase letters, d–l. The shaded region indicates the range of FRET observed for Fn-D/A in fibrils of cells treated with cytochalasin D, a reagent that disrupts cytoskeletal tension (n = 78, 8 fibrils). The dashed line at the bottom indicates the level of FRET from Fn-D/A in 8 M Gdn⋅HCl in PBS.
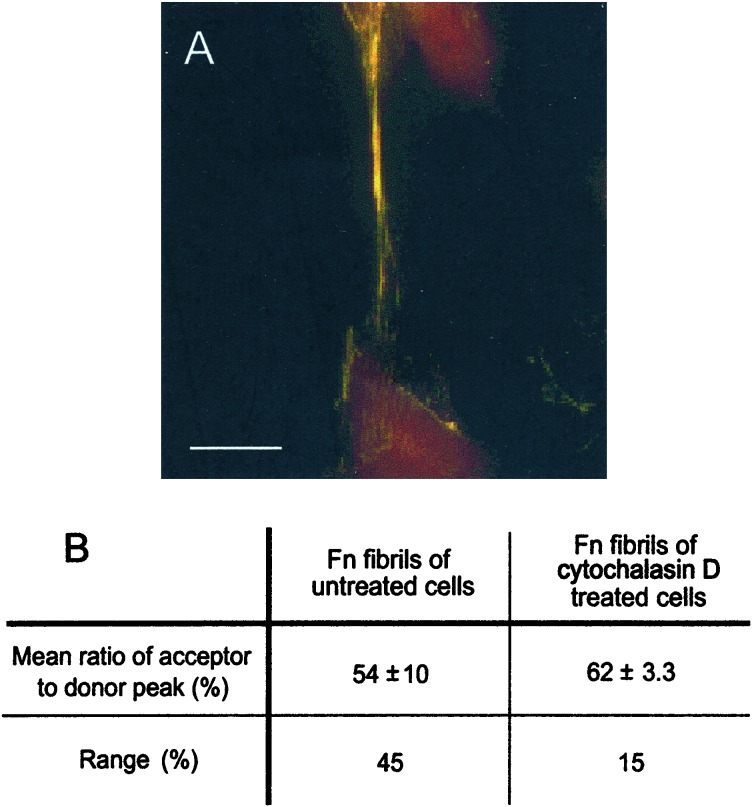
To disrupt cytoskeletal tension applied to fibrils, we treated cells with cytoD for 1 h after 24 h of incubation with Fn-D/A. In fluorescence images, we observed a shift in the predominant fibril color from green to yellow (Fig. 3A). This shift indicated an increase in FRET, suggesting that Fn-D/A in fibrils of cytoD-treated cells was less unfolded than in untreated samples. The increase in FRET was confirmed by using spectroscopy. The mean FRET associated with Fn-D/A in fibrils after treatment with cytoD was 62% ± 3.3% (n = 78, 8 fibrils), significantly higher than that of untreated samples, 54% ± 10% (n = 115, 11 fibrils) (P < 0.001, two-sample t test). The range of FRET in cytoD-treated samples (15%) was much narrower than that of untreated samples (45%).
Figure 3.
Effect of cytochalasin D on conformation of Fn-D/A in matrix fibrils. (A) Fluorescence image of fibril after treatment of cells with 10 μM cytoD for 1 h after incorporation of Fn-D/A into fibrils. Fibrils in cell samples treated with cytoD appeared predominantly yellow, indicating a higher level of FRET than in the predominantly green fibrils of untreated samples. The increase in FRET indicated that cytoD allowed refolding of Fn-D/A in fibrils relative to untreated samples. (Scale bar, 10 μm.) (B) The mean level of FRET (acceptor peak divided by donor peak) from fibrils in untreated samples, 54% ± 10% (n = 115, 11 fibrils), was significantly lower than in samples treated with cytoD, 62 ± 3.3% (n = 78,8 fibrils) (P < 0.001, two-sample t test). The range of FRET in fibrils of cytoD-treated cells (15%) was also narrower than that of untreated cells (45%).
Discussion
Proteins such as titan and tenascin have been unfolded by using atomic force microscopy and optical tweezers (18, 19), but the question of whether cells unfold proteins or protein modules under physiological conditions remains unanswered (14, 20). By measuring FRET between donor and acceptor fluorophores conjugated to Fn, we were able to investigate the effect of cell-generated tension on the structure of Fn in fibrillar matrices. Biochemical experiments using circular dichroism and fluorescence polarization have shown that denaturant concentrations of up to 2 M Gdn⋅HCl in PBS disrupt electrostatic interactions that hold Fn in a compact conformation (21). These conditions cause Fn to open into an extended conformation while leaving the secondary structure of individual modules intact (16). Exposure of Fn-D/A to 2 M Gdn⋅HCl resulted in a decrease in FRET (acceptor peak divided by donor peak) from 85% at 0 M to 60% (Fig. 1) because of a loss of energy transfer between Fn's two strands as they dissociate from each other. FRET remaining at 2 M Gdn⋅HCl would then occur mainly between donors and acceptors in close proximity along the strands. High-resolution structures of FnIII modules have shown that different modules are structurally homologous and measure 3.2 nm from N to C terminus (22–25). Because FRET is limited to donors and acceptors located within 10 nm of each other (26), only donors within modules FnIII5–9 and FnIII13–15 are sufficiently close to acceptors located within FnIII7 and FnIII15 to permit energy transfer. From 2 M to 4 M Gdn⋅HCl, FnIII modules unfold sequentially, beginning with the least stable modules (16, 27). The decrease in FRET from 60% at 2 M Gdn⋅HCl to 42% at 4 M is thus caused by unfolding of FnIII modules located within 10 nm of acceptor-labeled cysteines. The sensitivity of FRET to the range of unfolding using Gdn⋅HCl demonstrated that FRET was sensitive to a range of Fn conformations, from relatively compact to extended with FnIII modules unfolded.
The most immediate observation from fluorescence images of cells was that Fn-D/A in matrix fibrils was predominantly green, whereas the cell surface was red (Fig. 2 A–C). Spectra taken from 0.5-μm regions spaced 1 μm apart along the fibrils showed that FRET varied as little as 3% and as much as 25% along an entire fibril (Fig. 2D). FRET in most fibrils was comparable to that of Fn-D/A in 1 M to 4 M Gdn⋅HCl solutions. The range of FRET suggested that Fn-D/A in some but not all fibrils was extended, with unfolded FnIII modules near the acceptor sites on FnIII7 and FnIII15. We use the term “hyperextended” to describe these conformations of Fn: extended with at least one module unraveled. The second observation was that different Fn fibrils exhibited different average levels of FRET. Cells build up tension on the ECM by mechanically coupling the ECM to the contractile cytoskeleton with transmembrane integrins. To determine whether different levels of cytoskeletal tension were responsible for differences in unfolding of fibrillar Fn, we disrupted cell contractile forces with cytoD, which disrupts cytoskeletal tension by capping the fast-growing end of actin filaments. The mean level of FRET associated with fibrils in cytoD-treated samples, 62% ± 3.3%, was significantly higher than that of fibrils in untreated samples, 54% ± 10% (P < 0.001, two-sample t test) (Fig. 3). FRET in fibrils of cytoD-treated samples was equivalent to that of Fn-D/A in 0.5 M to 2 M Gdn⋅HCl solutions, corresponding to a conformation with FnIII modules intact. Thus, release of cell contractility appeared to allow Fn to relax by the refolding of FnIII modules.
These results provide insight into the molecular basis of Fn fibril elasticity. By using cells that express a Fn–green fluorescent protein chimera, Ohash et al. (14) showed that Fn fibrils contract up to fourfold after being surgically cut. Two models have been proposed to account for the elasticity of Fn fibrils. The first model proposes that Fn dimers within fibrils are structured such that their N-terminal regions are aligned with the fibril axis, and C-terminal regions fold on themselves normal to the fibril axis. Mechanical tension pulls the molecule into alignment with the fibril axis, causing the fibril to stretch (14, 17). According to this model, the level of FRET in fibrillar Fn-D/A under tension should be larger or equal to that of Fn-D/A extended with its FnIII modules intact—the level of FRET in 0.5 M to 2 M Gdn⋅HCl, 60% to 75% (Fig. 1). Because FRET in fibrils under tension was predominantly lower than these values, our results do not support the first model. The second model proposes that elasticity of Fn fibrils originates from unraveling and refolding of Fn modules (12–14). Because FRET in a number fibrils was nearly as low as Fn-D/A denatured with 8 M Gdn⋅HCl, our results suggest that some FnIII modules are unfolded in Fn within fibrils. The increase in energy transfer after treatment with cytoD suggests that FnIII modules refold when tension is released. Atomic force microscopy and steered molecular dynamics simulations have shown that forced unfolding of a single FnIII module results in an extended peptide about 10 times the length of the native module (13, 28). FnIII modules lack internal disulfide bonds, making them mechanically less stable than FnI or FnII modules. Force-induced unfolding of FnIII modules provides a basis for one proposed mechanism of Fn self-assembly into fibrils. As neighboring Fn molecules refold upon release of tension, they may noncovalently bind each other by undergoing β-strand or β-sheet exchange between FnIII modules (29).
The observation that Fn in fibroblast fibrils is stretched into extended and partially unraveled, or “hyperextended,” conformations has functional implications. There are at least three mechanisms by which hyperextension of Fn can alter its functional states: (i) by exposing cryptic sites that are buried between Fn modules in the native state; (ii) by changing the relative distance of two or more binding sites that bind the same receptor; and (iii) by straightening the hydrophilic loops that carry binding sites (30). Cryptic sites have been identified on FnIII1, FnIII7–8, and FnIII10 that must be exposed to initiate Fn matrix assembly (31–34). Steered molecular dynamics simulations predict that, before unraveling of FnIII modules, cryptic sites buried between adjacent modules can become exposed in an intermediate state of the force-induced unfolding pathway of FnIII modules (28). Stretching FnIII modules into this intermediate state requires less force than breaking apart the first β-strands. Steered molecular dynamics simulations also predict that stretching the FnIII10 module into this intermediate state reduces α5β1 integrin binding (35). This reduction in integrin affinity is caused by an increase in the distance between the arginine–glycine–aspartic acid (RGD) loop and its synergy site, from 3.2 nm to 5.6 nm. The synergy site is required for α5β1 to bind Fn, but is not required for other integrins. While α5β1-mediated signaling may dominate in cells bound to unstretched Fn, other integrin signaling pathways may compete as the synergy site is separated from RGD. If enough force is exerted to unravel FnIII modules, exposed loops containing sites such as the RGD sequence will be straightened, reducing integrin affinity (36). This scenario is one example of how variation in the unfolding of Fn within fibrils may change the biochemical cues along a fibril.
Experiments have shown that conformational changes of Fn have profound biological consequences. For example, substrate-induced conformational changes of adsorbed Fn can modulate integrin binding and cause a switch between proliferation and differentiation in myoblasts (37). Changes in Fn matrix architecture affect cell signaling patterns that control cell proliferation. Fn mutants missing FnIII1–7 form a structurally distinct matrix that inhibits progression from G0/G1 into S phase, whereas matrices assembled from native Fn stimulate cell growth (1).
Maintaining Fn fibrils under tension exacts an energetic price, which fibroblasts may pay to regulate the exposure of Fn's recognition sites. Mechanical tension could serve as the means by which cells alter the biochemical signals presented by their environment. Once they adhere to a surface and produce an ECM, cells may begin to regulate the chemical stimuli presented by their environment by applying force.
We observed differences in the degree of Fn unfolding within Fn fibrils of contractile cells. The force experienced by individual fibronectin molecules likely depends on whether the fibril is freely suspended, interconnected with other fibrils or EMC proteins, or attached to the substrate. Depending on the number of attachments and the force applied by neighboring cells, the presentation and activity of functional sites may vary along a fibril.
Fn is an ECM protein for which there is experimental evidence that cell tension causes protein unfolding. As part of a larger supramolecular assembly, Fn is mechanically coupled to other proteins so that force applied to Fn can be transferred to these proteins, potentially altering their structures as well. Several ECM proteins, including tenascin and several types of collagen, contain FnIII modules (38–40), and these may unfold in synchrony with those of Fn. The method of FRET to study Fn conformation can also be applied to other ECM proteins to better understand how they are integrated into the ECM and affected by cell contractile forces. Further research is needed to understand how protein functions such as cell binding, protein binding, and self-assembly are affected by cell-generated mechanical force. These are initial steps in establishing how integration of ECM proteins into supramolecular assemblies enables cells to mechanically regulate the biological activity of their environments.
Acknowledgments
We gratefully acknowledge contributions by Lichia Feng and William Little, and the use of the University of Washington Engineered Biomaterials Tissue Culture Laboratory, which is funded by the National Science Foundation. This work was supported by National Institutes of Health Research Grant 5R01GM49063 (to V.V.), and the Graduate Student Award from the Center for Nanotechnology University Initiative Fund, University of Washington (to L.B.).
Abbreviations
- ECM
extracellular matrices
- Fn
fibronectin
- FRET
fluorescence resonance energy transfer
- Fn-D/A
Fn labeled with donors and acceptors
- Gdn⋅HCl
guanidine hydrochloride
- cytoD
cytochalasin D
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sechler J L, Schwarzbauer J E. J Biol Chem. 1998;273:25533–25536. doi: 10.1074/jbc.273.40.25533. [DOI] [PubMed] [Google Scholar]
- 2.Sechler J L, Corbett S A, Wenk M B, Schwarzbauer J E. Ann NY Acad Sci. 1998;857:143–154. doi: 10.1111/j.1749-6632.1998.tb10114.x. [DOI] [PubMed] [Google Scholar]
- 3.Darribere T, Schwarzbauer J E. Mech Dev. 2000;92:239–250. doi: 10.1016/s0925-4773(00)00245-8. [DOI] [PubMed] [Google Scholar]
- 4.Leahy D J, Aukhil I, Erickson H P. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 5.Spitzfaden C, Grant R P, Mardon H J, Campbell I D. J Mol Biol. 1997;265:565–579. doi: 10.1006/jmbi.1996.0736. [DOI] [PubMed] [Google Scholar]
- 6.Hynes R O. Fibronectins. New York: Springer; 1990. [Google Scholar]
- 7.Baneyx G, Vogel V. Proc Natl Acad Sci USA. 1999;96:12518–12523. doi: 10.1073/pnas.96.22.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Magnusson M K, Mosher D F. Mol Biol Cell. 1997;8:1415–1425. doi: 10.1091/mbc.8.8.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hocking D C, Smith R K, McKeown-Longo P L. J Cell Biol. 1996;133:431–444. doi: 10.1083/jcb.133.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zamir E, Katz B Z, Aota S, Yamada K M, Geiger B, Kam Z. J Cell Sci. 1999;112:1655–1669. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- 11.Zamir E, Katz M, Posen Y, Erez N, Yamada K M, Katz B Z, Lin S, Lin D C, Bershadsky A, Kam Z, Geiger B. Nat Cell Biol. 2000;2:191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- 12.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin A M, Burridge K. J Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson H P. Proc Natl Acad Sci USA. 1994;91:10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohashi T, Kiehart D P, Erickson H P. Proc Natl Acad Sci USA. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baneyx G, Baugh L, Vogel V. Proc Nat Acad Sci USA. 2001;98:14464–14468. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan M Y, Medow M S, Newman S A. Biochem J. 1990;270:33–38. doi: 10.1042/bj2700033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson K J, Sage H, Briscoe G, Erickson H P. J Biol Chem. 1999;274:15473–15479. doi: 10.1074/jbc.274.22.15473. [DOI] [PubMed] [Google Scholar]
- 18.Fisher T E, Carrion-Vazquez M, Oberhauser A F, Li H, Marszalek P E, Fernandez J M. Neuron. 2000;27:435–446. doi: 10.1016/s0896-6273(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 19.Oberhauser A F, Marszalek P E, Erickson H P, Fernandez J M. Nature (London) 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 20.Erickson H P, Carrell N, McDonagh J. J Cell Biol. 1981;91:267–269. doi: 10.1083/jcb.91.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sjoberg B, Eriksson M, Osterlund E, Pap S, Osterlund K. Eur Biophys J. 1989;17:5–11. doi: 10.1007/BF00257140. [DOI] [PubMed] [Google Scholar]
- 22.Huber A H, Wang Y E, Bieber A J, Bjorkman P J. Neuron. 1994;12:717–731. doi: 10.1016/0896-6273(94)90326-3. [DOI] [PubMed] [Google Scholar]
- 23.Main A L, Harvey T S, Baron M, Boyd J, Campbell I D. Cell. 1992;31:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- 24.Dickinson C D, Veerapandian B, Dai X P, Hamlin R C, Xuong N H, Ruoslahti E, Ely K R. J Mol Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 25.Leahy D J, Hendrickson W A, Aukhil I, Erickson H P. Science. 1992;258:987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 26.Lakowicz J R. Principles of Fluorescence Spectroscopy. New York: Plenum; 1986. [Google Scholar]
- 27.Markovic Z, Engel J. Hoppe-Seylers Z Physiol Chem. 1983;364:551–561. doi: 10.1515/bchm2.1983.364.1.551. [DOI] [PubMed] [Google Scholar]
- 28.Craig D, Krammer A, Schulten K, Vogel V. Proc Natl Acad Sci USA. 2001;98:5590–5595. doi: 10.1073/pnas.101582198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Litvinovich S V, Brew S A, Aota S, Akiyama S K, Haudenschild C, Ingham K C. J Mol Biol. 1998;280:245–258. doi: 10.1006/jmbi.1998.1863. [DOI] [PubMed] [Google Scholar]
- 30.Vogel V, Thomas W E, Craig D W, Krammer A, Baneyx G. Trends Biotechnol. 2001;19:416–423. doi: 10.1016/S0167-7799(01)01737-1. [DOI] [PubMed] [Google Scholar]
- 31.Litvinonich S V, Ingham K C. J Mol Biol. 1995;248:611–626. doi: 10.1006/jmbi.1995.0246. [DOI] [PubMed] [Google Scholar]
- 32.Ingham K C, Brew S A, Huff S, Litvinovich S V. J Biol Chem. 1997;272:1718–1724. doi: 10.1074/jbc.272.3.1718. [DOI] [PubMed] [Google Scholar]
- 33.Hocking D C, Sottile J, McKeown-Longo P J. J Biol Chem. 1994;269:19183–19187. [PubMed] [Google Scholar]
- 34.Langenbach K J, Sottile J. J Biol Chem. 1999;274:7032–7038. doi: 10.1074/jbc.274.11.7032. [DOI] [PubMed] [Google Scholar]
- 35.Krammer A, Craig D, Thomas W E, Schulten K, Vogel V. Matrix Biol. 2001;21:139–147. doi: 10.1016/s0945-053x(01)00197-4. [DOI] [PubMed] [Google Scholar]
- 36.Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Proc Natl Acad Sci USA. 1999;96:1351–1356. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia A J, Vega M D, Boettiger D. Mol Biol Cell. 1999;10:785–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engel J. Matrix Biol. 1996;15:295–299. doi: 10.1016/s0945-053x(96)90130-4. [DOI] [PubMed] [Google Scholar]
- 39.Ricard-Blum S, Dublet B, van der Rest M. Unconventional Collagens Types VI, VII, VIII, IX, X, XIV, XVI and XIX. Oxford: Oxford Univ. Press; 2000. 155. [Google Scholar]
- 40.Joester A, Faissner A. Matrix Biol. 2001;20:13–22. doi: 10.1016/s0945-053x(00)00136-0. [DOI] [PubMed] [Google Scholar]