Abstract
Diiron and dimanganese proteins catalyze a wide range of hydrolytic and oxygen-dependent reactions. To probe the mechanisms by which individual members of this class of proteins are able to catalyze such a wide range of reactions, we have prepared a model four-helix bundle with a diiron site located near the center of the bundle. The four-helix bundle is constructed by the noncovalent self-assembly of three different chains (Aa, Ab, and B) that self-assemble into the desired heterotetramer when mixed in a 1:1:2 molar ratio. On addition of ferrous ions and oxygen, the protein forms a complex with a UV-visible spectrum closely resembling that of peroxo-bridged diferric species in natural proteins and model compounds. By combining a small collection of n variants of these peptides, it should now be possible to prepare an n3 member library, which will allow systematic exploration of the features giving rise to the catalytic properties of this class of proteins.
Dinuclear metalloproteins comprise a large number of functionally diverse enzymes that catalyze a variety of hydrolytic and oxygen-dependent reactions. How the protein matrix is able to tune the activity of these cofactors to effect such a wide range of reactions is an intriguing question. One subset of diiron/dimanganese proteins includes the Mn catalases, radical-forming R2 subunit of ribonucleotide reductase, hydroxylases such as methane monooxygenase and toluene oxygenase, hydrocarbon desaturases, and ferroxidases (1–6). Although the overall folds of these proteins differ, their diiron and dimanganese sites are each ensconced within a four-helix bundle with one or two copies of a Glu-Xxx-Xxx-His sequence motif.
To help test and further elucidate the features that define the reactivity of individual members of this family, we have designed the Dueferri (DF1 and DF2) family of proteins as paradigms for diiron proteins (7–9). DF1 idealizes the approximate C2 symmetry of the natural proteins; it comprises two noncovalently associated helix–loop–helix motifs that bind the diiron cofactor near the center of the structure. The structure of diZn (II) (7) and diMn (II) (9) variants of DF1 have been determined by x-ray crystallography, and the metal-binding sites of these proteins are very similar to those of diMn (II) bacterioferritin (10), diMn (II) (11), and diFe (II) ribonucleotide reductase (12), and diFe (II) ACP Δ9-desaturase (13). In DF1, two Glu sidechains bridge both metal ions, whereas the other two carboxylates interact with a single metal ion in a bidentate chelating interaction. The two His residues form additional monodentate ligands.
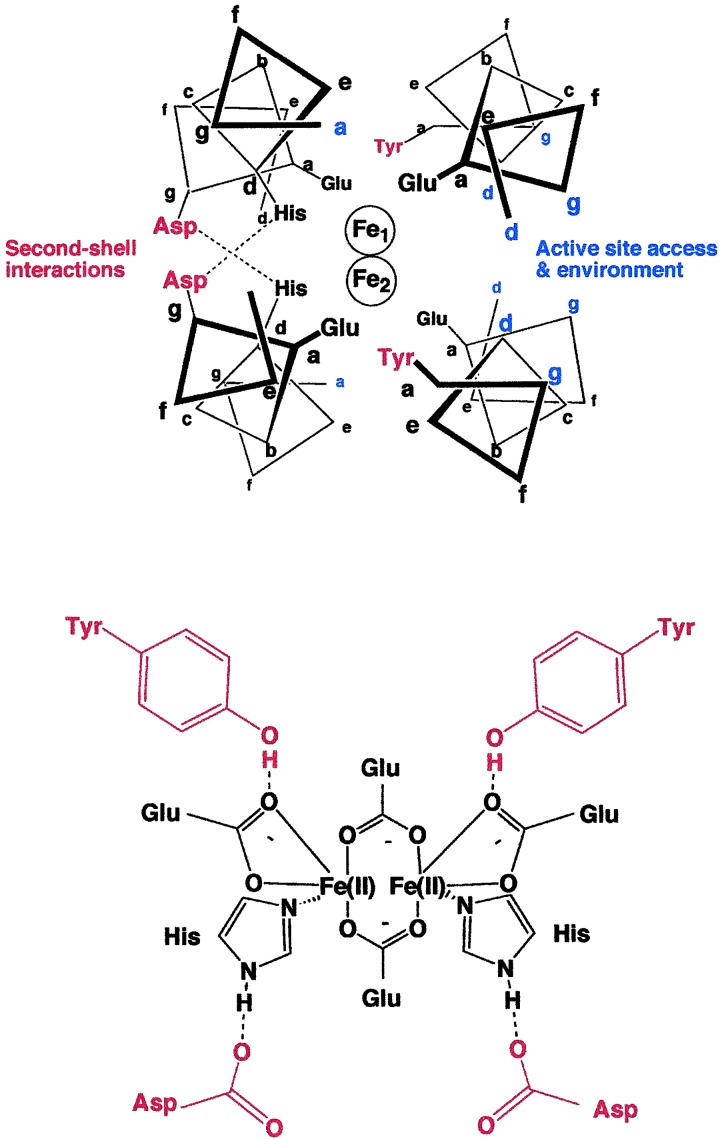
The electrostatic environment, polarity, and solvent accessibility of the metal-binding site are likely to influence its electrochemical midpoint potential and reactivity. The minimalist design of DF1 renders it an excellent candidate for determining how these features influence the properties of diiron proteins. Fig. 1 illustrates the active site of DF1 and highlights the features that directly affect the immediate environment of the active site. The protein forms an antiparallel four-stranded coiled coil in which the four Glu ligands occupy “a” positions of the center heptad repeat, and the His sidechains occupy “d” positions on two of the helices. The absence of His residues at the equivalent “d” positions of the other two helices (Fig. 1) results in an open coordination site on each of the metal ions. Small residues (Ala or Gly) at these “d” positions facilitate the entry of metal ions, O2 and small molecule ligands into the active site, whereas a Leu in this position occludes the active site and inhibits their entry (8).
Figure 1.
(Upper) Schematic view of a 12-residue slice through the active site of the crystal structure of active site of DF1. The diagram is based on the crystallographic structures of di-Mn(II) (9) and di-Zn(II) (7) derivatives of DF1. Second-shell hydrogen bonds are shown in red, and some of the positions that help define the environment of the active site are in blue. (Lower) Schematic of the first- and second-shell ligands defining the metal-binding site (this diagram is meant to describe the atomic connectivities but not the precise geometry of the site).
Second-shell interactions are important for maintaining the structures of metal-binding sites (14) and also for tuning their electrochemical midpoint potentials (15, 16). In DF1, the nonbridging Glu carboxylates receive a hydrogen bond from a Tyr sidechain at an “a” position of a neighboring helix (Fig. 1). Similarly, an Asp residue forms a hydrogen bond with the imidazole ɛN of a His ligand. In natural diiron proteins, Asp, Asn, Ser, Glu, or Gln form analogous second-shell hydrogen bonds with the His ligands. Although these second-shell and primary ligands shape the immediate environment of the metal ions, other more subtle but nevertheless important features might include distant electrostatic interactions, distortions of the helices, and the presence of aromatic sidechains that can form radical intermediates.
We would like to evaluate how systematic changes to the structure of DF1 affect its metal-binding, substrate-binding, and catalytic properties. In general, it would be useful to examine both natural as well as unnatural amino acids at a large number of positions. However, even if only five or six positions within the protein are substituted with a restricted number of sidechains, a very large number of combinations are generated. Furthermore, we would like to examine thoroughly the reaction of each one of these variants with a number of small molecule ligands and substrates. Such an extensive study would require the generation and purification of significant quantities of hundreds to thousands of variants.
To advance this goal, we intend to design a tetramer consisting of four disconnected helices, which could be separately synthesized, purified, and then combinatorially assembled to create an array of the desired helical bundles. For an ABCD heterotetramer (in which each of the monomers was different), 104 combinations could be prepared by combining only 10 variants of each of the individual chains. Previously, strategies have been developed for the design of two-stranded coiled coils that specifically self-assemble to form AB heterodimers, with minimal formation of homodimeric species (17); similarly, ABC heterotrimers (18–20) and A2B2 heterotetrameric (21) coiled coils have been designed. Recently, we described the design of an A2B2 heterotetrameric version of DF1 (22).
Here, we use a similar strategy to create a three-component A⋅A′⋅B2 heterotetrameric complex; extension of this strategy should allow the design of fully asymmetric complexes. Furthermore, we demonstrate the utility of this approach by designing a variant that interacts with Fe(II) in the presence of O2 with kinetics that are markedly different from DF1. Although DF1 reacts relatively slowly with Fe(II) and O2 to form an oxo-bridged diferric species, the variant reacts rapidly to form a stable species with a UV-visible spectrum similar to those of previously characterized peroxo-diferric species.
Materials and Methods
Materials.
The B peptide was synthesized and purified as described (22), and the Aa and Ab peptides were prepared by analogous procedures. All buffers, metal ions, and other chemicals were of the highest quality commercially available.
General Procedures.
Circular dichroism (CD) spectroscopy was carried out by using methods and equipment described (22). The Co(II) titration was also conducted as described (8).
Size-Exclusion Chromatography.
Peptides were mixed in the appropriate molar ratios at approximately 20 μM (in each peptide) and chromatographed through a Superdex 75 FPLC (Amersham Pharmacia) column equilibrated in 10 mM 4-morpholinepropanesulfonic acid buffer, pH 7.0, containing 100 mM NaCl at a flow rate of 1.0 ml/min. As a standard for the tetrameric state, we used DFtet, which cleanly forms a tetramer as assessed by analytical ultracentrifugation. The column was also calibrated by using aprotinin, cytochrome C, and carbonic anhydrase, which confirmed the tetrameric molecular weight of DFtet and AaAbB2. The DFtet A peptide, which forms monomers in the absence of metal ions, was used as a standard for the monomeric molecular weight of Aa and Ab.
Fe(II)-Binding/Oxidation.
Kinetic experiments were performed under aerobic conditions at pH 6.5 (0.15 M Mes/0.15 M NaCl; 40 μM AaAbB2). The reaction was initiated by addition of 2.0 equivalents (per tetramer) Fe(II) from a stock solution (10 mM ferrous ammonium sulfate/0.01% sulfuric acid), and spectra were measured on a Hewlett–Packard diode array spectrometer. For experiments performed under anaerobic conditions, a sample of AaAbB2 was introduced into a cuvette fitted with a septum and made anaerobic by repeated evacuation and flushing with argon. The Fe(II) solution was similarly made anaerobic by flushing with argon and introduced into the cuvette by using a gas-tight syringe. In the absence of oxygen, no significant change in the UV-visible spectrum of the sample occurred on addition of iron. When the sample was opened to the air, a spectrum characteristic of the Fe–peroxo complex gradually developed.
Results
Design.
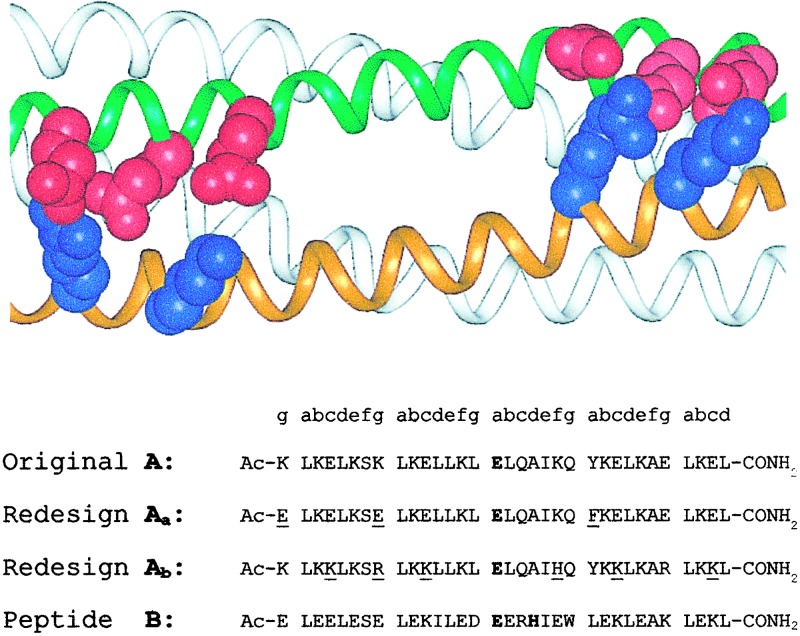
Previously, we described an A2B2 system, designated DFtet, that interacts with Co(II), Fe(II), and Fe(III) in a manner analogous to DF1. The individual peptides in this series are 33 residues in length and contain charged sidechains at interfacial (“c,” “g,” “b,” and “e”) positions in an arrangement intended to stabilize the heterotetramer relative to the homomeric species. The A peptides contained a single metal-chelating Glu ligand and the Tyr second-shell ligand. The B peptides contain a Glu-Xxx-Xxx-His sequence that contributes the bridging Glu and the His ligand, as well as the Asp second-shell ligand (Fig. 2). To prepare less symmetrical tetramers, we prepared two electrostatically complementary versions of the A peptide, which should specifically assemble with two equivalents of B to form the desired A,A′,B heterotetramer (Fig. 2). Along one face of the helix of the original A, negatively and positively charged groups had been placed near the N and C termini, respectively, to stabilize an antiparallel arrangement of the helices. In the current work, the sequences of these two helices were varied such that one contains a strip of Glu along the entire length of the interface, and the other contains Lys and Arg at equivalent positions (Fig. 2). We designate the peptide with the acidic replacements helix Aa and the other Ab.
Figure 2.
Redesign of DFtet to create an AaAbB2 heterotetramer. The sidechains introduced to encourage pairing of the Aa and the Ab helices are shown in red (Glu) and blue (Lys and Arg) (Upper). Changes between the DFtet A peptide and Aa or Ab are underlined in the sequences below.
We also made two additional substitutions to probe specific mechanistic questions. DF1 has two Tyr sidechains that form second-shell interactions with the nonbridging Glu residues, whereas in natural proteins only one of the structurally equivalent Glu residues is generally constrained by a second-shell interaction. Thus, DF1 might be too constrained to allow rapid catalysis. We therefore changed Tyr-23 to Phe in the Aa peptide. We also introduced a surface His residue in Ab to allow the attachment of probes in future experiments designed to investigate the photochemical generation of radicals at the remaining Tyr residue.
Characterization of Solution Properties.
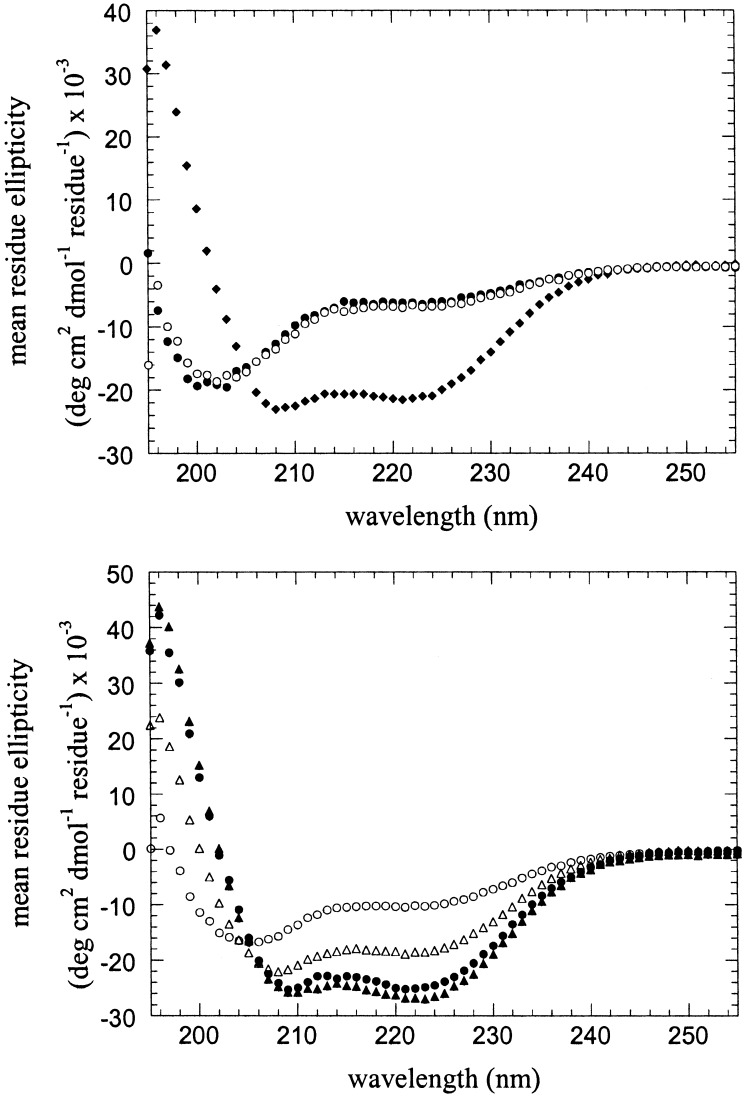
Far UV CD and size-exclusion chromatography were used to demonstrate that the peptides assemble into the desired tetramer, in the presence as well as the absence of metal ions. At pH 7.0, the individual Aa and B peptides (30 μM in 10 mM 4-morpholinepropanesulfonic acid/100 mM NaCl, pH 7.0) appear to be unfolded (Fig. 3 Upper), as assessed by the lack of a strong n–π* transition at 222 nm or an exciton-split π–π* transition at 208 and 195 nm that would be indicative of a helical conformation. This behavior of B is consistent with its previously characterized behavior. Size-exclusion chromatography of Aa indicated that it behaved similar to the parent A peptide, which is known to be monomeric under these conditions. By contrast, the Ab peptide appeared to be helical by CD spectroscopy (θ222 = −21,000 deg cm2⋅dmol−1). Size-exclusion chromatography indicated that it eluted at the volume expected for the dimer.
Figure 3.
CD spectra of isolated peptides and the Aa, Ab, and B peptides in combination. (Upper) CD spectra of the individual Aa (filled circles), Ab (diamonds), and B (open circles) peptides; only the Ab peptide shows any appreciable helicity. (Lower) CD spectra of a 1:1:2 mixture of the Aa, Ab, and B peptides in the absence of divalent metal ions (filled circles) and in the presence of 50 μM zinc acetate (filled triangles). For comparison, the CD spectra calculated by summation of the spectra of the individual peptides are also shown for a 1:1:2 mixture of Aa, Ab, and B in the absence of metal ions (open circles) and in the presence of 50 μM zinc acetate (open triangles). In all experiments, the buffer was 10 mM 4-morpholinepropanesulfonic acid, pH 7.0/100 mM NaCl; the total concentration of peptide(s) was 30 μM.
When Aa, Ab, and B are mixed in a 1:1:2 molar ratio, an α-helical CD spectrum is observed, indicating that all three components are folded. The shape and intensity of the spectrum are nearly identical to that observed for DFtet and DF1. Fig. 3 Lower compares the helical spectrum observed for a 1:1:2 mixture of the Aa, Ab, and B peptides versus the random coil-like theoretical curve expected if the peptides did not interact (generated by summing the experimental spectra of the individual peptides). The magnitude of [θ222] and [θ208] for the ternary mixture is also greater than that observed for the Aa:B, Aa:Ab, and Ab:B binary mixtures, which show an intermediate degree of structure formation on mixing (data not shown).
We also examined the spectra of the peptides in the presence of Zn(II), which binds tightly to DF1 and DFtet. As described previously, Zn(II) binds to B and induces a helical conformation (θ222 = −23,000 deg cm2⋅dmol−1) in this peptide. This metal ion failed to induce a helical spectrum in either Aa [θ222 = −5,300 deg cm2⋅dmol−1) after addition of one equivalent of Zn(II)], or a further increase in the magnitude of the ellipticity for Ab. However, when Aa, Ab, and B are mixed in a 1:1:2 molar ratio with two equivalents of Zn(II), a spectrum is observed, which is nearly identical to that observed in the absence of Zn(II), indicating a similar secondary structure for the AaAbB2 tetramer in the presence and absence of metal ions. However, thermal-unfolding studies indicated that the helical conformation was greatly stabilized in the presence of metal ions. In the absence of Zn(II), a cooperative unfolding transition is observed with a midpoint near 70°C (at 7.5 μM tetramer concentration); by contrast, the protein remained folded up to 95°C in the presence of 50 μM Zn(II). These results, together with size-exclusion chromatography, indicate that the peptides specifically self-assemble to form a tetramer, whose conformation is stabilized by binding Zn(II).
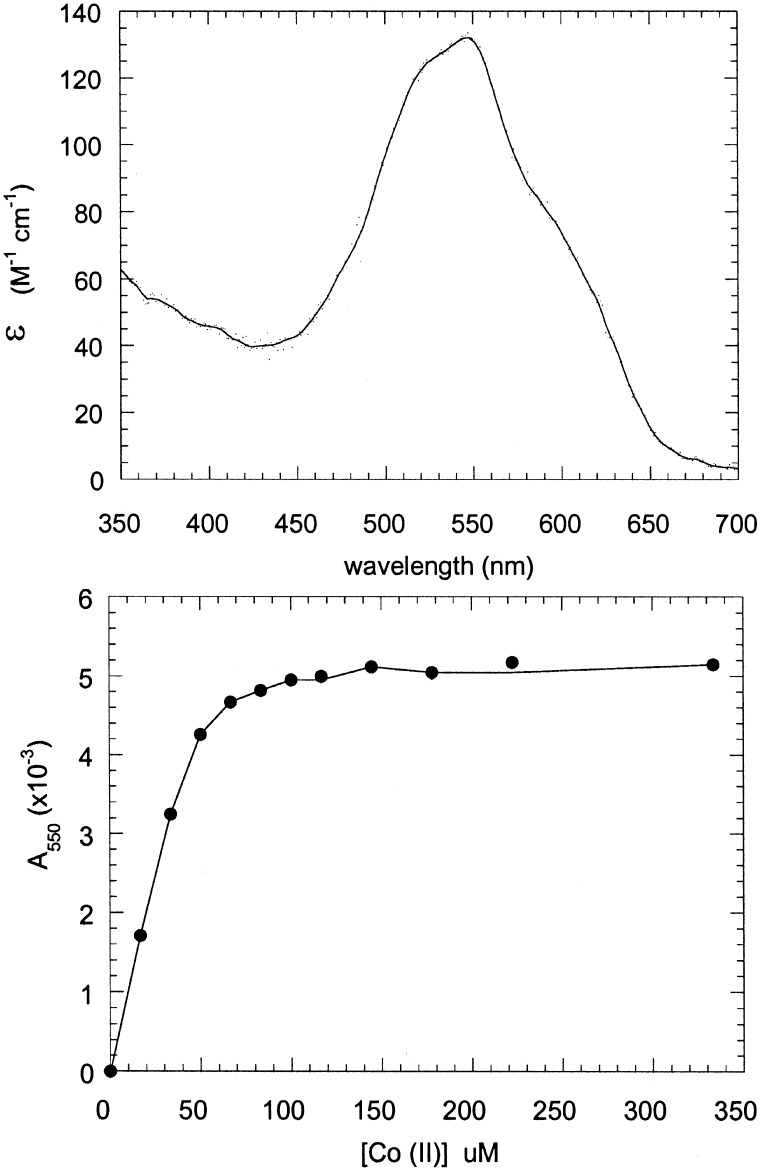
Binding of Co(II).
The absorbance spectrum of Co(II) is highly sensitive to environment, and its extinction coefficient increases from approximately 10 to 100 to 500 M−1 cm as the coordination number increases from 4 to 5 to 6 (23). Fig. 4 illustrates the UV-visible spectrum of Co(II) in the presence of the AaAbB2 heterotetramer; the spectrum is nearly identical to that of the Co(II) complex of derivatives of DF2 (8) and DFtet (22). Further, a titration of 40 μM tetramer with Co(II) indicated that the metal ion bound tightly with the expected stoichiometry of 2 Co(II) per tetramer (Fig. 4).
Figure 4.
Cobalt binding to the Aa,Ab,B2 tetramer. (Upper) Spectrum of Co(II) bound to the Aa,Ab,B2 tetramer. The concentration of the peptides was 40 μM in tetramer (80 μM in metal-binding sites). The extinction coefficient at 550 nm was 140 M−1⋅cm−1 (per tetramer). (Lower) Titration of the Aa,Ab,B2 tetramer with CoCl2. The concentration of peptides was 40 μM in tetramer (80 μM in metal-binding sites). The curve saturates at between 80 and 100 μM, indicating that two cobalt ions are bound per tetramer.
Interactions with Fe(II) and O2.
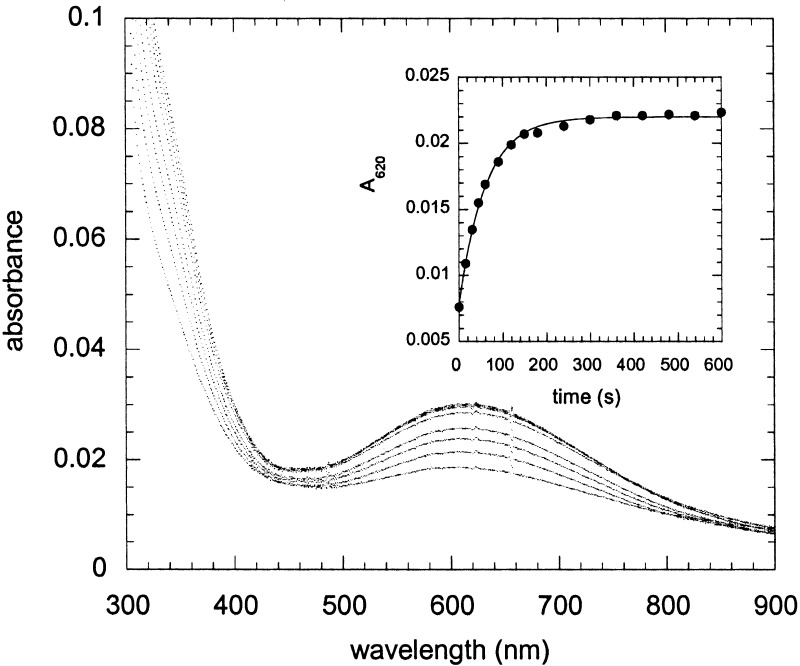
Although the AaAbB2 peptides behave similarly to DF2 and DFtet in their binding to Co(II) and Zn(II), this protein shows marked differences in its interaction with Fe(II) under aerobic conditions. The ferroxidase activity of DF2 and DFtet has been characterized; these proteins display a modest rate enhancement for the air oxidation of Fe(II) to Fe(III) (8, 22). Under single turnover conditions [50 μM protein, 2.0 Fe(II) per active site], an oxo-bridged diferric species is formed over the course of ≈1 h. By contrast, addition of Fe(II) to AaAbB2 leads to the formation of a novel spectroscopic species (Fig. 5) in an apparent first-order process (the time course follows first-order kinetics for over four half-lives); at 25 μM tetramer concentration, the apparent rate constant reaches a limiting value of 2 min−1 at initial concentrations of Fe(II) greater than 100 μM.
Figure 5.
Reaction of Aa,Ab,B2 tetramer with Fe(II) in the presence of oxygen. Spectra of Aa,Ab,B2 (25 μM in tetramer) measured at various times after the addition of ferrous ammonium sulfate, final concentration 50 μM, in air-saturated buffer (150 mM Mes, pH 6.5/150 mM NaCl). The spectra shown were measured at 15, 30, 45, 60, 90,120, 150, and 180 s after mixing. (Inset) The kinetics of peroxide formation followed by the increase in absorbance at 620 nm; the data are fit to a first-order reaction with kapparent = 0.98 ± 0.03 min−1.
Although the oxo-bridged species formed by DF2 and DFtet show strong metal-to-ligand charge transfer bands near 305 nm and only very weak d–d transitions at longer wavelengths, AaAbB2 shows an additional strong band centered near 625 nm, with an extinction coefficient of approximately 1,000 M−1⋅cm−1 (based on the initial tetramer concentration). This band was not observed when Fe(II) was added anaerobically but was formed on addition of O2. The spectrum is very similar to that observed in the cis-μ-1,2-peroxo-bridged diferric complex observed in ferritin (λmax = 650 nm, ɛ = 1,000 M−1⋅cm−1) (24, 25) but at somewhat shorter wavelength than the corresponding species in the enzymes ACP 9Δ-desaturase (λmax = 700 nm, ɛ = 1,200 M−1⋅cm−1) (26), methane monooxygenase (λmax = 725 nm ɛ = 1,800 M−1⋅cm−1) (for a review, see ref. 27), and a mutant of the R2 subunit of ribonucleotide reductase (λmax = 700 nm ɛ = 1,800 M−1⋅cm−1) (28). Hemerythrins also form peroxide complexes in which peroxide is bound in an end-on manner to a single site, but they are characterized by an absorption maximum that is shifted to approximately 500 nm (29, 30). A further but less likely possibility is that the absorption represents a tyrosinate-Fe(III) ligand-to-metal charge transfer band, although again these have absorption maxima around 500 nm (31).
To confirm that this species was associated with the tetrameric complex, the protein was separated by size-exclusion chromatography. A single major peak comprising approximately 90% of the UV-absorbing material eluted at the same position as that of the tetrameric form of DFtet. The remaining material eluted at somewhat greater apparent molecular weight, suggesting that some higher-order aggregate is also formed. The tetrameric material was analyzed by mass spectrometry and reverse-phase HPLC; each monomer was present in the appropriate stoichiometry and had not been covalently modified.
Conclusion
Here we designed a system that allows the combinatorial exploration of hypotheses concerning the structural bases for the function of diiron proteins. To rigorously evaluate mechanistic aspects of the reactivity of this cofactor, we wish to generate a large number of variants, which can be spectroscopically interrogated in a relatively high-throughput manner. Thus, it is necessary to prepare an array of purified proteins at defined concentrations to allow direct spectroscopic and catalytic measurements of individual library members (e.g., in 96-well format). The production of such variants has been facilitated by the use of noncovalent self-assembly to prepare the proteins.
The AaAbB2 variant of the DFtet tetramer appears to satisfy many of the requirements for such a protein. The assembly appears to be relatively specific and results in the formation of the desired tetramer, even in the absence of transition metal ions. CD studies showed that a 1:1:2 mixture of the Aa, Ab, and B peptides had much greater helicity than the individual components or the various binary mixtures. Further size-exclusion chromatography showed the formation of a tetramer only when the three components were present in the appropriate ratios. Together, these studies indicate that the assembly process is specific for the desired heterotetramer relative to other possible species. Thus, electrostatic interactions between interfacially located sidechains appear to be sufficient to direct preferential assembly, even in the presence of 0.10 M NaCl. Although more discriminating disulfide-crosslinking studies might be helpful to fully quantify the extent of the hetero-tetramer formation, the observed degree of specificity is sufficient for the purpose of constructing combinatorial libraries.
Interestingly, the kinetics of the reaction of AaAbB2 with Fe(II) and oxygen and the products formed are entirely different from those formed in the original DF1 protein and DFtet. Although additional spectroscopic studies are necessary to confirm the identity of this species, its UV-visible spectrum is similar to that found in the peroxo-bridged diferric intermediates found in a variety of diiron proteins (1–4, 25). In natural proteins, the bridged diferric peroxo complex is formed only transiently, whereas this putative species appears to be stable for hours at room temperature in AaAbB2, and only minor decomposition occurs after one day at 4°C. We note that it has been difficult to design proteins or small molecule complexes that stably form these peroxo-diferric species in water at room temperature (27). Thus, if this species can be demonstrated to belong to an oxo-bridged diferric species, it will be interesting to determine the precise structural features that give rise to its stability.
Although additional experiments are required to place the identity of this inorganic species on a firm footing, these data nevertheless demonstrate the promise of the combinatorial approach for identifying potential candidates for more detailed analysis. Having demonstrated the potential of the system, we are now preparing a small focused library of variants. The initial screening of this library has already identified individual members that show a number of interesting variations in their spectroscopic and chemical properties.
Abbreviation
- CD
circular dichroism
References
- 1.Nordlund P, Eklund H. Curr Opin Struct Biol. 1995;5:758–766. doi: 10.1016/0959-440x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 2.Lange S J, Que L., Jr Curr Opin Chem Biol. 1998;2:159–172. doi: 10.1016/s1367-5931(98)80057-4. [DOI] [PubMed] [Google Scholar]
- 3.Waller B J, Lipscomb J D. Chem Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 4.Feig A L, Lippard S J. Chem Rev. 1994;94:759–805. [Google Scholar]
- 5.Yocum C F, Pecoraro V L. Curr Opin Chem Biol. 1999;3:182–187. doi: 10.1016/S1367-5931(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 6.Dismukes G C. Chem Rev. 1996;96:2909–2926. doi: 10.1021/cr950053c. [DOI] [PubMed] [Google Scholar]
- 7.Lombardi A, Summa C, DeGrado W F. Proc Natl Acad Sci USA. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasternak A, Kaplan J, Lear J D, DeGrado W F. Protein Sci. 2001;10:958–969. doi: 10.1110/ps.52101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Costanzo L, Wade H, Geremia S, Randaccio L, Pavone V, DeGrado W F, Lombardi A. J Am Chem Soc. 2002;123:12749–12757. doi: 10.1021/ja010506x. [DOI] [PubMed] [Google Scholar]
- 10.Frolow F, Kalb A J, Yariv J. Nat Struct Biol. 1994;1:453–460. doi: 10.1038/nsb0794-453. [DOI] [PubMed] [Google Scholar]
- 11.Atta M, Nordlund P, Aberg A, Eklund H, Fontecave M. J Biol Chem. 1992;267:20682–20688. [PubMed] [Google Scholar]
- 12.Andersson M E, Högbom M, Rinaldo-Matthis A, Andersson K K, Sjöberg B-M, Nordlund P. J Am Chem Soc. 1999;121:2346–2352. [Google Scholar]
- 13.Lindqvist Y, Huang W, Schneider G, Shanklin J. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 14.Christianson D W, Fierke C A. Acc Chem Res. 1996;29:331–339. [Google Scholar]
- 15.Gooden D B, McRee D E. Biochemistry. 1993;32:3313–3324. [PubMed] [Google Scholar]
- 16.Vance C K, Miller A-F. J Am Chem Soc. 1998;120:461–467. [Google Scholar]
- 17.Lumb K J, Kim P S. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 18.Nautiyal S, Alber T. Protein Sci. 1999;8:84–90. doi: 10.1110/ps.8.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardi A, Bryson J W, DeGrado W F. Biopolymers. 1996;40:495–504. doi: 10.1002/(SICI)1097-0282(1996)40:5%3C495::AID-BIP7%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 20.Nautiyal S, Woolfson D N, King D S, Alber T. Biochemistry. 1995;34:11645–11651. doi: 10.1021/bi00037a001. [DOI] [PubMed] [Google Scholar]
- 21.Fairman R, Chao H G, Lavoie T B, Villafranca J J, Matsueda G R, Novotny J. Biochemistry. 1996;35:2824–2829. doi: 10.1021/bi952784c. [DOI] [PubMed] [Google Scholar]
- 22. Summa, C. M., Rosenblatt, M., Hong, J.-K., Lear, J. D. & DeGrado, W. F. (2001) J. Mol. Biol., in press. [DOI] [PubMed]
- 23. Bertini, I. & Luchinat, C. in Advances in Inorganic Biochemistry, eds. Eichhorn, G. L. & Marzilli, L. G. (Elsevier, New York), Vol. 6, pp. 71–111. [PubMed]
- 24.Pereira A S, Small W, Krebs C, Tavares P, Edmondson D E, Theil E C, Huynh B H. Biochemistry. 1998;37:9871–9876. doi: 10.1021/bi980847w. [DOI] [PubMed] [Google Scholar]
- 25.Hwang J, Krebs C, Huynh B H, Edmondson D E, Theil E C, Penner-Hahn J E. Science. 2001;287:122–125. doi: 10.1126/science.287.5450.122. [DOI] [PubMed] [Google Scholar]
- 26.Broadwater J A, Ai J, Loehr T M, Sanders-Loehr J, Fox B G. Biochemistry. 1998;37:14664–14671. doi: 10.1021/bi981839i. [DOI] [PubMed] [Google Scholar]
- 27.Merkx M, Kopp D A, Sazinsky M H, Blazyk J L, Müller J, Lippard S J. Angew Chem Int Ed. 2001;2001:2782–2807. [PubMed] [Google Scholar]
- 28.Moenne-Loccoz P, Baldwin J, Ley B A, Loehr T M, Bollinger J M., Jr Biochemistry. 1998;37:14659–14663. doi: 10.1021/bi981838q. [DOI] [PubMed] [Google Scholar]
- 29.Dunn J B, Addison A W, Bruce R E, Loehr J S, Loehr T M. Biochemistry. 1977;16:1743–1749. doi: 10.1021/bi00627a035. [DOI] [PubMed] [Google Scholar]
- 30.Mizoguchi T J, Kuzelka J, Spingler B, DuBois J L, Davydov R M, Hedman B, Hodgson K O, Lippard S L. Inorg Chem. 2001;40:4662–4673. doi: 10.1021/ic010076b. [DOI] [PubMed] [Google Scholar]
- 31.Logan D T, deMare F, Persson B O, Slaby A, Sjoberg B M, Nordlund P. Biochemistry. 1998;37:10798–107807. doi: 10.1021/bi9806403. [DOI] [PubMed] [Google Scholar]