Abstract
Fundamental to most genetic analysis is availability of genomic DNA of adequate quality and quantity. Because DNA yield from human samples is frequently limiting, much effort has been invested in developing methods for whole genome amplification (WGA) by random or degenerate oligonucleotide-primed PCR. However, existing WGA methods like degenerate oligonucleotide-primed PCR suffer from incomplete coverage and inadequate average DNA size. We describe a method, termed multiple displacement amplification (MDA), which provides a highly uniform representation across the genome. Amplification bias among eight chromosomal loci was less than 3-fold in contrast to 4–6 orders of magnitude for PCR-based WGA methods. Average product length was >10 kb. MDA is an isothermal, strand-displacing amplification yielding about 20–30 μg product from as few as 1–10 copies of human genomic DNA. Amplification can be carried out directly from biological samples including crude whole blood and tissue culture cells. MDA-amplified human DNA is useful for several common methods of genetic analysis, including genotyping of single nucleotide polymorphisms, chromosome painting, Southern blotting and restriction fragment length polymorphism analysis, subcloning, and DNA sequencing. MDA-based WGA is a simple and reliable method that could have significant implications for genetic studies, forensics, diagnostics, and long-term sample storage.
For genomic studies, the quality and quantity of DNA samples is critical. High-throughput genetic analysis requires large amounts of template for testing, yet typically the yield of DNA from individual patient samples is limited. Forensic and paleoarcheology work also can be severely limited by DNA sample size. An important goal is to supply a sufficient amount of genomic sequence for a variety of procedures as well as long-term storage for future work and archiving of patient samples. Methods include the time-consuming process of creating of Epstein–Barr virus-transformed cell lines and whole genome amplification (WGA) by random or degenerate oligonucleotide-primed PCR (DOP-PCR) (1–3). However, PCR-based WGA methods may generate nonspecific amplification artifacts (2), give incomplete coverage of loci (4), and generate DNA less than 1 kb long (1–3) that cannot be used in many applications.
Recently, a rolling circle amplification (5) method was developed for amplifying large circular DNA templates such as plasmid and bacteriophage DNA (6). Using φ29 DNA polymerase and random exonuclease-resistant primers, DNA was amplified in a 30°C reaction not requiring thermal cycling. This is made possible in part by the great processivity of φ29 DNA polymerase, which synthesizes DNA strands 70 kb in length (7). Here we extend the use of exonuclease-resistant primers and φ29 DNA polymerase to WGA. The amplification is surprisingly uniform across the genomic target, with the relative representation of different loci differing by less than 3-fold. In contrast, PCR-based WGA methods exhibited strong amplification bias ranging from 4 to 6 orders of magnitude. Multiple displacement amplification (MDA)-generated DNA product is >10 kb, and its performance is demonstrated for a variety of applications, including single nucleotide polymorphism (SNP) analysis, restriction fragment length polymorphism (RFLP), and comparative genome hybridization. MDA was capable of accurate WGA from <10 human cells. This simple and robust method also uniformly amplified the human genome directly from whole blood without a requirement for DNA purification.
Materials and Methods
DNA and Enzymes.
A panel of human genomic DNA samples, the Human Variation Panel-Caucasian Panel of 100 (reference number HD100CAU) was obtained from Coriell Cell Repositories, Camden, NJ. Human genomic DNA also was obtained from Promega. Thiophosphate-modified random hexamer (5′-NpNpNpNpsNpsN-3′) was synthesized at Molecular Staging, φ29 DNA polymerase was from Amersham Pharmacia Biosciences, and yeast pyrophosphatase was from Roche Molecular Biochemicals. Restriction endonucleases were from New England Biolabs. DNA size markers (100-bp DNA ladder, 1-kb DNA ladder) were from GIBCO/BRL.
DNA Preparation from Blood and Cell Lines.
Human blood samples were from Grove Hill Medical Center Laboratory, New Britain, CT. U266 myeloma cell line (American Type Culture Collection) was passaged according to the accompanying protocol. Cells were lysed in alkaline lysis solution by a modification of Zhang et al. (3). Briefly, blood was diluted 3-fold, and tissue culture cells were diluted to 30,000 cells/ml in PBS. Blood or cells (35 μl) were lysed by dilution with 35 μl of alkaline lysis solution (400 mM KOH, 100 mM DTT, 10 mM EDTA) and incubated 10 min on ice. The lysed cells were neutralized with 35 μl of neutralization solution (prepared by mixing 4 ml of 1 M HCl and 6 ml of 1 M Tris⋅HCl buffer, pH 7.5, final pH of the solution is 0.6). The lysed blood or cells (1 μl) were used directly as template in MDA reactions as described.
Amplification of Human Genomic DNA by MDA.
Human genomic DNA (300 ng to 0.03 ng, as indicated) was placed into 0.2-ml tubes in a total volume of 100 μl containing 37 mM Tris⋅HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 5 mM (NH4)2SO4, 1 mM dNTPs, 50 μM exonuclease-resistant hexamer, 1 unit/ml of yeast pyrophosphatase, and 800 units/ml φ29 DNA polymerase. Radioactively labeled α-[32P] dCTP, approximately 240 cpm/pmol total dCTP, was added as indicated. Reactions were incubated for 18 h at 30°C and terminated by heating to 65°C for 3 min. Acid-precipitable radioactive deoxyribonucleotide was determined with glass fiber filters to quantitate product yield. A template heat denaturation step was included or omitted, as indicated. For heat treatment the DNA template was incubated in 50 μl at 95°C for 3 min and chilled to 4°C in a PCR System Thermocycler (Applied Biosystems), and then brought to a final volume of 100 μl with the composition described above.
Analysis of Amplification Products.
Three-microliter reaction aliquots were cleaved with restriction endonuclease AluI and analyzed by electrophoresis through a 1.0% agarose gel in TBE buffer (90 mM Tris-borate, pH 8.0/2 mM EDTA), stained with GelStar or SYBR green (Molecular Probes), and imaged with a Storm 860 PhosphorImager (Amersham Biosciences). Denaturing gel analysis was carried out by electrophoresis through a 1.0% agarose gel in 30 mM NaOH, 1 mM EDTA. The radioactive products in the dried gel were visualized with the PhosphorImager.
Southern Analysis.
Ten micrograms of whole genome-amplified DNA or human genomic DNA controls was digested with EcoRI restriction endonuclease and separated through a 1% agarose gel in TBE buffer. Southern analysis (8) was performed by using a Hybond-N+ membrane (Amersham Biosciences). An exon fragment probe of parathyroid hormone (p20.36) and RFLP marker probes for the D13S12 (p9D11) and thyroglobulin (pCHT.16/8) loci were obtained from American Type Culture Collection. Probes were radiolabeled by using the NEBlot random primer labeling method (New England Biolabs). The membrane was prehybridized for 1 h and hybridized to the radiolabeled probe overnight in a membrane hybridization buffer (Amersham Biosciences). The hybridized membrane was washed in 2× SSC and 0.1% SDS twice for 5 min at room temperature, 1× SSC and 0.1% SDS for 15 min at 42°C, and 0.1× SSC twice for 15 min at 65°C. The membrane was then exposed overnight and analyzed by using the PhosphorImager.
Quantitative PCR Analysis.
TaqMan analysis was performed by using the Applied Biosystems 7700 according to the manufacturer's specifications with 1 μg of amplified DNA as template. TaqMan assay reagents for the eight loci tested were obtained from Applied Biosystems. Three replicate TaqMan assays were averaged for each of the eight loci for each DNA amplification reaction. Error bars represent 1 SD from the mean. The eight loci and their chromosome assignments were: acidic ribosomal protein (1p36.13), connexin 40 (1q21.1), chemokine (C-C motif) receptor 1 (3p21), chemokine (C-C motif) receptor 6 (6q27), chemokine (C-C) receptor 7 (17q21), CXCR5 Burkitt lymphoma receptor 1 (chromosome 11), c-Jun (1p32-p31), and MKP1 dual specificity phosphatase 1 (5q34). Connexin 40 is located near the centromere, and chemokine (C-C motif) receptor 6 is located near the telomere. A standard curve for input template was generated to determine the loci copy number in amplified DNA relative to that of genomic DNA. The standard curve was generated from 0, 0.001, 0.01, 0.1, 0.5, and 1 μg of unamplified genomic DNA. This standard curve was used for the determination of locus copy number in the WGA product by determining the threshold cycle number by real-time PCR (9). The locus representation in an amplified DNA sample is expressed as a percent, relative to input genomic DNA. It is defined as the locus copy number in 1 μg of amplified DNA divided by the locus copy number in 1 μg of genomic DNA control. Amplification bias between two loci is the ratio between the two locus representation values. The maximal amplification bias for a group of loci is the ratio between the locus representations for the two loci with the highest and the lowest locus representations.
Amplification of Human Genomic DNA by DOP-PCR.
Human genomic DNA (ranging from 300 ng to 0.03 ng) was amplified as described (1). Radioactively labeled α-[32P] dCTP was added to the reaction for the quantitation of PCR product yield. TaqDNA polymerase was from Invitrogen. DOP-PCR amplifications were carried out by using a GeneAmp 9700 PCR System thermocycler (Applied Biosystems). Locus representation in the DOP-PCR product was quantitatively analyzed by using the TaqMan assay (Applied Biosystems).
Amplification of Human Genomic DNA by Primer Extension Preamplification (PEP).
Human genomic DNA (ranging from 300 ng to 0.03 ng) was placed into 0.2-ml tubes in a total volume of 60 μl, yielding final concentrations of 33 μM PEP random primer (5′-NNN NNN NNN NNN NNN-3′) as described (3). Radioactively labeled α-[32P] dCTP was added to the reaction for the quantitation of PCR product. PEP reactions were carried out with a GeneAmp 9700 PCR System thermocycler (Applied Biosystems). Locus representation in the PEP product was quantitatively analyzed with the TaqMan assay (Applied Biosystems).
Genotyping of SNPs.
The SNPs analyzed here had the following chromosomal locations: 1822, 251, and 221, 13q32; 465, 458, and 474, 19q13; VCAM, 1p31; IL-8, 4q13; and PDK2–2, 17p. Assays were carried out by using Coriell Human Variation Panel samples (nos. 12, 70, 75, and 86) as described (10). The 24 RFLP genotypes of Table 1 were obtained from MDA carried out with genomic DNA from Coriell Human Variation Panel sample catalog 73–96. They were genotyped at SNP1822 by PCR amplification (1822 forward primer-5′-GGA TAA TAC TTC TGA ATG ACT AAA G-3′ and 1822 backward primer- 5′-TAG TAT GAA TAT ATT TAA GAT TCA GTG-3′) followed by restriction endonuclease digestion with enzyme TaqI. The size of the PCR-amplified fragment is 350 bp. TaqI enzyme digested the 1822 (C) allele into two fragments (250 bp and 100 bp) whereas the 1822 (T) allele was undigested. The genotypes obtained were compared with the known sequences of SNP1822 determined for these Coriell DNA samples by DNA sequencing (10).
Table 1.
DNA amplification yield after MDA, DOP-PCR, and PEP
| Process | Template DNA, ng | Genome copies | DNA yield, ng | Yield, fold |
|---|---|---|---|---|
| MDA | 300 | 90,000 | 32,000 | 100 |
| 30 | 9,000 | 34,000 | 1,000 | |
| 3 | 900 | 30,000 | 10,000 | |
| 0.3 | 90 | 33,000 | 100,000 | |
| DOP | 100 | 30,000 | 8,400 | 84 |
| 10 | 3,000 | 5,900 | 590 | |
| 1 | 300 | 6,800 | 6,800 | |
| 0.1 | 30 | 6,100 | 61,000 | |
| PEP | 100 | 30,000 | 820 | 8 |
| 10 | 3,000 | 780 | 78 | |
| 1 | 300 | 360 | 360 | |
| 0.1 | 30 | 340 | 3,400 |
Comparative Genome Hybridization.
Amplified and nonamplified DNA preparations were compared by modifications of published procedures (11). Genomic DNA preparations were nick-translated to incorporate nucleotides modified with biotin for amplified samples or digoxigenin for unamplified control samples. One microgram of both amplified and unamplified DNA was precipitated in the presence or absence of 30 μg of CotI DNA and resuspended in 10 μl of 20% dextran sulfate/4× SSC, with the later addition of 5 μl of hybridization-grade formamide. After denaturation of normal metaphase chromosome spreads in 70% formamide/2× SSC, the hybridization mix was added and the slides were incubated for 3 days. After washing off excess probe, specific hybridization signals were detected by avidin-FITC and antidigoxigenin rhodamine. Captured images of metaphase chromosomes were analyzed by using the Applied Imaging (Santa Clara, CA) cgh software program and fluorescence profiles were generated. As controls, differentially labeled amplified or unamplified DNAs were mixed, hybridized, detected, and subjected to ratio analysis as outlined above.
Results
Isothermal rolling circle amplification (5) with random hexamer primers and φ29 DNA polymerase was recently described for circular DNA (6) (Fig. 1A). Surprisingly, these reagents will also readily amplify linear, human genomic DNA in a cascading, strand displacement reaction termed MDA (12) (Fig. 1B). Amplification of genomic DNA by MDA at 30°C proceeds for 4–6 h (Fig. 1C), after which a plateau is reached, resulting in approximately the same yield of DNA product in all reactions. Average product length exceeds 10 kb as determined by alkaline gel electrophoresis (Fig. 1D). The yield of amplified DNA from a 100-μl reaction was consistently in the range of 20 to 30 μg, regardless of the amount of starting material over a 5-log range (100 fg-10 ng). This consistent yield is important for some applications, allowing use of samples of different concentration or quality and providing reproducible yields for subsequent genetic analysis without the need to measure or adjust DNA concentration.
Figure 1.
MDA strategy and product characterization. (A) Random-primed rolling circle amplification of circular DNA templates. DNA synthesis is initiated with random oligonucleotide primers. 3′ Ends are indicated by arrowheads. Thickened regions indicate primers. Secondary priming events occur on the displaced product DNA strands. (B) Scheme for MDA of genomic DNA. Secondary priming events are initiated from primary products. (C) Effect of template concentration on amplification yield. A total of 100 fg to 100 ng human genomic DNA was amplified by MDA at 30°C as described. Aliquots were taken from single reactions at the times indicated to quantitate DNA synthesis. ●, 10 ng genomic DNA template; ○, 1 ng; ▴, 100 pg; ▵, 10 pg; ■, 1 pg; □, 100 fg; ⧫, primers omitted. (D) Denaturing gel analysis of amplification product size. Radioactively labeled amplification products shown in C were electrophoresed through an alkaline agarose gel (1%), and the dried gel was exposed to a phosphor screen and imaged as described. Reaction products were loaded in order of increasing DNA template amount, as indicated above the gel.
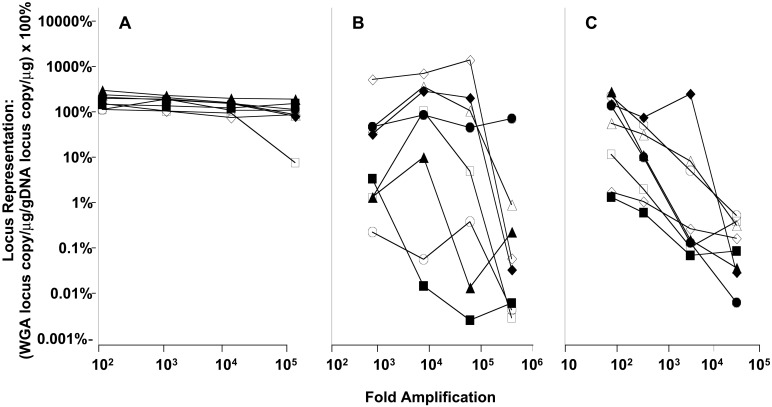
For many applications, WGA must provide complete coverage of the genome with minimal amplification bias. A certain amount of sequence bias may be anticipated with any amplification method and may result from factors such as priming efficiency, template accessibility, GC content, and proximity to telomeres and centromeres. Amplification bias of MDA was examined by TaqMan quantitative PCR for eight genes, including one near the centromere of chromosome 1 (connexin 40) and one adjacent to the telomere of chromosome 6 [chemokine (C-C motif) receptor 6]. MDA reactions contained from 300 down to 0.3 ng of human genomic template DNA, or between 90,000 and 90 copies of the genome, and amplification yield was calculated on the basis of total DNA synthesis as determined by the incorporation of radioactive deoxyribonucleotide (Table 1). For 100-, 1,000-, and 10,000-fold amplifications from genomic DNA target, the maximum ranges in locus representation were 120% to 324%, 109% to 249%, and 78% to 218%, respectively. These data yield maximum amplification biases for MDA of only 2.70, 2.28, and 2.79, respectively, expressed as the ratio between the loci representations of the most highly and the least highly represented genes (Fig. 2A). In contrast, DOP-PCR, a WGA method based on PCR (1, 2), had an amplification bias that varied from a low of 103 to a high of 106 (Fig. 2B). PEP, another PCR-based WGA method (3), exhibited an amplification bias that varied from a low of 102 to a high of 104 (Fig. 2C). PEP has been reported to generate an amplification bias of up to 50-fold even between two alleles of the same gene (4). Significantly, the ≈3-fold amplification bias of MDA remained almost constant between 100- and 100,000-fold amplification (Fig. 2A).
Figure 2.
Effect of amplification on gene representation bias. Amplification reactions omitted a heat denaturation step and were carried out as described. Reactions contained 300 ng, 30 ng, 3 ng, or 0.3 ng of template DNA. The relative representation of eight loci was determined by using TaqMan quantitative PCR. The x axis represents the fold amplification in the amplified DNA used as template for quantitative PCR; the y axis is the locus representation in the amplified DNA relative to the input genomic DNA, expressed as a percent, and is calculated as the yield of quantitative PCR product from 1 μg of amplified DNA divided by the yield from 1 μg of genomic DNA control. The results for eight loci are indicated as follows: CXCR5, ◊; connexin40, ▵; MKP1, □; CCR6, ○; acidic ribosomal protein, ⧫; CCR1, ▴; cJUN, ■; CCR7, ●. (A) Percent representation for eight loci derived from MDA-amplified DNA. (B) Percent representation for eight loci present in DNA amplified by using DOP-PCR. (C) Percent representation for eight loci present in PEP-amplified DNA.
Surprisingly, TaqMan quantification indicated that the average gene locus was enriched by MDA in amplified DNA; locus representation was >100% of the representation in genomic DNA (Fig. 2A). Whereas representation of mitochondrial DNA was the same between starting genomic DNA and amplified DNA (data not shown), Southern blots (data not shown) and chromosome painting experiments (see Fig. 5) indicate that repetitive sequences are under-represented in the MDA product, thus conferring an effective enrichment of genes. Overall, the amplified DNA is enriched between 80% and 225% for the eight genes amplified 10,000-fold (Fig. 2A). Additional studies will be necessary to identify the extent and type of repetitive element under-representation.
Figure 5.

Representation bias assessed by competitive genome hybridization to metaphase chromosomes. Amplification reactions included a heat denaturation step, and amplification was carried out as described. Amplified (100,000-fold) and unamplified DNA samples were nick-translated to incorporate biotin nucleotide and digoxigenin nucleotide, respectively. The probes were mixed in equimolar amounts of nucleotide and hybridized without CotI suppression. Specific signals were detected by avidin FITC (amplified DNA, green) and antidigoxigenin rhodamine (unamplified DNA, red).
In early experiments, the double-stranded genomic DNA target was denatured at 95°C for 5 min before the MDA reaction to facilitate annealing of random primers. However, heating to 95°C tends to damage the DNA target. Under the conditions of an MDA reaction, genomic DNA was substantially degraded at 95°C, as determined by alkaline agarose gel electrophoresis (not shown). As expected, the rate and yield of MDA reactions decreased when the DNA target had been subjected to longer 95°C denaturation steps, as determined by radioactive deoxyribonucleotide incorporation (not shown). Surprisingly, the denaturation step could be completely omitted, giving the most efficient amplification. Apparently, the high concentration of random primers (50 μM) is sufficient to allow a slow, initial priming step to occur. However, once the reaction is initiated, the strand-displacing mechanism of MDA will release single-stranded template for ongoing priming and amplification (Fig. 1B). Random priming in the absence of the target denaturation step does not result in large amplification bias (Fig. 2A). Even if priming of the double-stranded target is initially favored at certain sequences, the highly processive φ29 DNA polymerase will rapidly generate long single-stranded product, giving even coverage of the genome.
The use of a 95°C denaturation step also reduces the specificity of MDA. With denaturation, locus representation ranged from only about 2% to 80% relative to genomic DNA control (not shown). Presumably, much of the DNA product is amplification artifact that does not contain genomic sequence as observed for PCR-based WGA (2). Possibly, DNA fragments generated at 95°C contribute to nonspecific priming of amplification artifacts. With denaturation omitted, locus representation improved to 80–225% for DNA amplified 10,000-fold, relative to the control (Fig. 2A). Amplification bias was also reduced to <3-fold when the target was not denatured (Fig. 2A) compared with about 40-fold with denaturation (not shown).
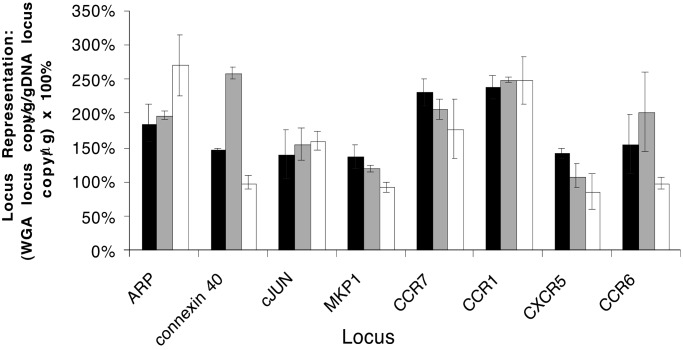
For many diagnostic and research applications genomic DNA is obtained from crude biological sources including blood or cells grown in tissue culture. MDA was successfully carried out directly from whole blood after a brief alkaline treatment. Amplification bias for MDA using blood was indistinguishable from the purified genomic DNA control (Fig. 3). In this experiment the maximum ranges in locus representation for blood and pure DNA were 137% to 238% and 108% to 259%, respectively. These corresponded to amplification biases of 1.74 and 2.40, respectively, demonstrating the consistency of MDA regardless of the purity of the starting material. MDA was also successful starting with approximately 10 cells grown in tissue culture. The maximum range in locus representation for cells was 86% to 271%, which corresponded to an amplification bias of 3.15. Although the DNA template released by the alkaline lysis procedure likely became denatured during this process, it did not exhibit the reduced functionality shown by heat-denatured DNA, presumably because the alkaline treatment does not damage the DNA in the way that heat treatment does.
Figure 3.
Gene coverage comparison of MDA-amplified DNA from blood, tissue culture cells, and pure DNA. Quantitative PCR was used to measure the yield of specific DNA fragments in amplified DNA preparations relative to genomic DNA. Reactions omitted a heat denaturation step, and alkaline lysis of blood and cells was carried out as described. The representation of eight loci in the amplified samples relative to pure DNA was determined by using TaqMan quantitative PCR. Black bars depict the locus representation of DNA amplified directly from blood. Gray bars depict the locus representation of DNA amplified from purified genomic DNA (30 ng, representing 9,000 gene copies). White bars depict the locus representation of DNA amplified from tissue culture cells (10 cell equivalents of DNA).
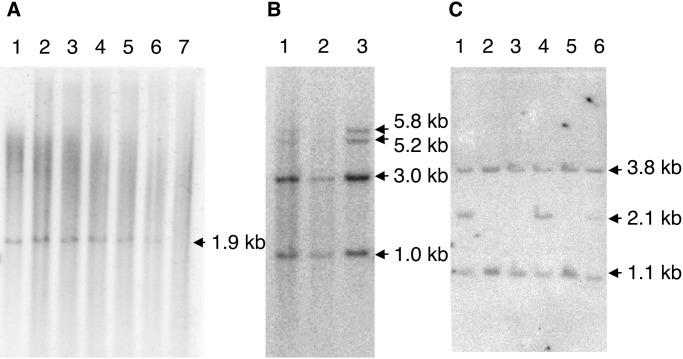
For general utility, WGA product must perform similarly to unamplified genomic DNA during subsequent genetic analysis. To examine the integrity of amplified DNA, a restriction fragment within the human parathyroid hormone gene (chromosome 11p15.2–15.1) was analyzed on Southern blots. A 1.9-kb restriction enzyme fragment was observed for MDA-based WGA products amplified from as few as 10 genomic copies (or 106-fold amplification; Fig. 4A). Whereas PCR-based WGA methods typically generate products of only several hundred nucleotides in length (1–3), MDA-based WGA products were of sufficient length and integrity for RFLP-based genotyping: 16 random individuals were correctly genotyped by the presence of a 2.1-kb and 1.1-kb PstI fragment (five representative samples are shown in Fig. 4C). Omission of heat denaturation before MDA was important for detection of restriction fragments >5 kb in length (Fig. 4B), consistent with degradation of the DNA target at 95°C. RFLP-based testing of forensic material may represent a useful application for MDA-based WGA of small samples.
Figure 4.
Southern blot analysis of the human parathyroid hormone (PTH) and thyroglobulin genes and RFLP marker D13S2 for MDA-generated DNA target. (A) MDA reactions included a heat denaturation step and amplification was carried out as described. EcoRI DNA digests were probed by using a radioactively labeled genomic fragment of the PTH gene (p20.36) that hybridized to an ≈1.9-kb DNA fragment. The EcoRI-cleaved DNA preparations were genomic DNA, DNA amplified by MDA from varying amounts of input genomic DNA, as indicated, or an MDA reaction that lacked input genomic DNA template. The position of the 1.9-kb genomic DNA fragment is indicated (lane 1). Genomic DNA is shown: (lane 2) ×100, (lane 3) ×1,000, (lane 4) ×10,000, (lane 5) ×100,000, (lane 6) ×1,000,000, and (lane 7) 0 template. (B) MDA reactions included or omitted a heat denaturation step, as indicated, of genomic target DNA heterozygous for two thyroglobulin alleles, and amplification was carried out as described. TaqI DNA digests were probed by using a radioactively labeled genomic fragment of the thyroglobulin gene (pCHT.16/8) that hybridized to invariant 1-kb and 3-kb DNA fragments and a polymorphic 5.8-kb (allele A) or 5.2-kb (allele B) DNA fragment. The TaqI-cleaved DNA preparations were (lane 1) genomic DNA, (lane 2) DNA amplified by MDA reaction (×10,000) with a 95°C preheating step, and (lane 3) an MDA reaction (×10,000) without the preheating step. (C) Reactions lacked a heat denaturation step, but otherwise amplification was carried out as described. PstI DNA digests were probed by using a radioactively labeled genomic fragment of the RFLP marker D13S12 locus (p9D11) that hybridized to an invariant 3.8-kb DNA fragment and a polymorphic 2.1-kb (allele A) or 1.1-kb (allele B) DNA fragment. The PstI-cleaved DNA preparations were genomic DNA and five different patient DNAs amplified by MDA (×10,000 amplification): (lane 1) patient 5 (AB), (lane 2) patient 4 (BB), (lane 3) patient 3 (BB), (lane 4) patient 2 (AB), (lane 5) patient 1 (BB), and (lane 6) genomic DNA (AB). AB and BB represent the diploid genotypes of the D13512 locus.
MDA product was tested for reproducibility and coverage in assays of SNPs. Samples of both amplified and unamplified genomic DNA from 24 individuals were genotyped by using RFLP for a SNP located on chromosome 13q32 (10). In all 24 cases the amplified DNA gave the correct genotype (Table 2). A 10,000-fold amplified MDA product was also tested at another 10 loci by using a high-throughput, ligation-based SNP assay (Table 3) (10). Results were indistinguishable from unamplified genomic DNA genotyped by the same method (Table 2). MDA provided complete coverage for all 10 loci for all genomic DNA preparations obtained from Coriell. The amplified DNA performed normally as substrate in this real-time kinetic assay. MDA-based WGA offers a useful alternative to multiple locus-specific PCR preamplifications for large SNP scoring studies and, in particular, where sensitive kinetic assays are used and where sample availability is limited.
Table 2.
Assay of genomic samples from 24 individuals for SNP1822
| Individual | Genotype | Individual | Genotype |
|---|---|---|---|
| 1 | C/T | 13 | C/T |
| 2 | C/C | 14 | C/T |
| 3 | C/T | 15 | C/C |
| 4 | C/T | 16 | C/T |
| 5 | C/C | 17 | C/C |
| 6 | T/T | 18 | C/T |
| 7 | C/C | 19 | C/T |
| 8 | T/T | 20 | C/T |
| 9 | T/T | 21 | C/C |
| 10 | C/T | 22 | C/T |
| 11 | C/T | 23 | C/T |
| 12 | C/T | 24 | C/T |
Table 3.
Assay of 10 different SNPs
| SNP | Genotype | Coriell catalog designation no. |
|---|---|---|
| 1822 | C/C | 70 |
| 251 | A/A | 75 |
| 221 | T/T | 75 |
| 465 | T/T | 70 |
| 458 | G/G | 12 |
| 474 | G/G | 86 |
| VCAM | C/C | 70 |
| IL-8 | T/T | 12 |
| SNP21 | G/G | 75 |
| PDK2 | G/G | 85 |
Whole genome MDA was also tested for uniformity of chromosome coverage by comparative genomic hybridization. Equal amounts of biotin-labeled amplified and digoxigenin-labeled unamplified DNA were cohybridized in the presence or absence of CotI DNA to suppress repetitive DNA cross-hybridization. With suppression of signals from repetitive sequences using CotI DNA, the hybridization patterns of MDA probes and unamplified probes were indistinguishable even after 100,000-fold amplification (not shown). Without CotI suppression, however, the MDA-amplified probe gave reduced centromeric signals, indicating some loss of the repetitive centromeric sequences (Fig. 5). The uniformity of signal along the length of the chromosome arms was further evidence that MDA-based WGA does not induce significant amplification bias. These results indicated that MDA compared favorably with DOP-PCR for preparation of chromosome painting probes (13–15), but that unlike the latter, suppression hybridization may be unnecessary for detection of single copy sequences. This method should be invaluable for DNA probe preparation for comparative genome hybridization, karyotyping, and chip-based genetic analysis from limited patient DNA sources such as needle biopsy material or amniocentesis samples.
Discussion
Genomic studies often require large quantities of high-quality DNA for analysis and long-term storage. A simple, isothermal reaction has been developed that provides amplification of limiting samples of genome DNA with less than a 3-fold bias between loci tested. A 100-μl reaction generates 20–30 μg of DNA product regardless of the amount of starting template. Therefore, MDA may be amenable to automated, high-throughput formats such as DNA sequencing and SNP analysis where consistent DNA concentrations are required from numerous samples that may be of uncertain concentration or quality. The products of MDA are consistently >10 kb in length, allowing accurate RFLP analysis.
MDA can be carried out directly from a variety of biological samples commonly used for diagnostic purposes. DNA from blood and tissue culture cells amplified with the same low sequence bias as purified genomic DNA. An absence of significant bias observed in MDA-based WGA may be explained by the extraordinary processivity of φ29 DNA polymerase (7); tight binding of the polymerase to the template assures replication through obstacles caused by DNA primary or secondary structure.
MDA does not depend on high-temperature denaturation of genomic DNA to provide single-stranded template for the random priming. Indeed, omission of denaturation reduces template degradation and improves the specificity of the amplification for genomic sequence. Elevated temperature is known to increase the rate of depurination in DNA and hydrolysis at the resulting apurinic sites. This process may account for the DNA breakage observed at high temperature. Whereas PCR-based WGA products contain up to 70% amplification artifacts (2), MDA-based WGA products appear to be almost entirely derived from genomic and mitochondrial DNA (data not shown and Trevor Hawkins, personal communication). Locus representation was 80–225% of levels in the starting genomic DNA target. However, when template was denatured at 95°C, locus representation dropped to between about 2% and 80% of the control, suggesting that much of the DNA product was amplification-generated artifact. Bias was also reduced from 40-fold to less than 3-fold by eliminating the denaturation step.
We have compared the amplification bias of MDA to that of two PCR-based WGA methods, DOP and PEP. In our hands, bias for these methods ranged between 102- and 106-fold compared with less than 3-fold for MDA. It is problematic to compare our data to the published reports on DOP and PEP, which were carried out on different targets and through application of different methods of analysis. PEP was initially described in terms of DNA amplification from a single cell, and the loss of some loci in the amplified DNA was noted (3). The amplification of loci was estimated to be at least 30-fold. We observe a 102-fold bias in this range (Fig. 2C), consistent with the loss of some loci that was reported. Other studies also have used PEP for the amplification of DNA from single cells (4, 16, 17) and the preferential amplification of one allele at heterozygous loci was noted (4). Subsequent studies have expanded the use of PEP to the amplification of DNA from 100 to 1,000 cells, tissues, mouth swab samples, or microdissected tumors (18–20). However, amplification bias was not determined in these studies. For DOP, loss of loci was reported for amplifications of about 20,000-fold (2), and it is noted that a significant percent of the amplified DNA is some nonspecific product not containing genomic sequence.
Apparently, random priming on the double-stranded DNA target is efficient enough for an initial priming step followed by a strand-displacement mechanism generating single-stranded template for the exponential amplification. With locus representations of 80–225%, MDA product contained an average of about 150% of the locus copy number in starting DNA. Chromosome painting experiments indicated that repetitive sequences were under-represented in MDA product, conferring an effective enrichment of genes. One hypothesis for this observation is that primers corresponding to highly repetitive elements became depleted during MDA.
For genome subcloning and sequencing, MDA appears to have several intrinsic advantages over PCR-based methods. The product size of >10 kb is compatible with genome subcloning. Because no in vivo propagation of amplified material is necessary, MDA may represent an efficient method for amplifying “poisonous” genomic sequences. In addition, φ29 DNA polymerase used for MDA has an error rate of 1 in 106–107 (21) in contrast to ≈3 in 104 for PCR with TaqDNA polymerase (22). Therefore, PCR accumulates about one mutation per 900 bases (23) after 20 cycles. Sequence error rates of cloned MDA-based WGA products appear to be similar to those for cloned genomic DNA (Trevor Hawkins, personal communication).
In summary, the utility of MDA-based WGA was demonstrated for a variety of uses including quantitative PCR, SNP genotyping, Southern blot analysis of restriction fragments, and chromosome painting. Several situations may be contemplated where MDA may represent the method of choice for WGA. First, applications where faithful replication during WGA is necessary, such as molecular cloning, or single cell analysis. Second, applications where adequate genome representation during WGA is critical, such as genomewide SNP genotyping studies, and third, where minimization of bias during WGA is important, particularly cytogenetic testing such as prenatal diagnosis (24), comparative genome hybridization (13–15), and assessment of loss of heterozygosity (18). Additional situations where there is a significant need for WGA include amplification of DNA from microdissected tissues (14), buccal smears (19), and archival, anthropological samples (25). Finally, DNA amplified from sorted, individual chromosomes may be used for the generation of whole chromosome-specific painting probes (26).
Acknowledgments
We thank Dr. Paul Lizardi, who did pioneering work on multiple displacement amplification in the laboratory of Dr. David Ward at Yale University. We gratefully acknowledge the help of John Nelson, Wenzhuo Li, Lin Huang, Carl W. Fuller, and others at Amersham Pharmacia Biotech who provided highly purified φ29 DNA polymerase and unpublished results of WGA experiments. We also thank Rajanikanta Bandaru and Debra Itzkowitz for synthesis of oligonucleotides.
Abbreviations
- WGA
whole genome amplification
- DOP-PCR
degenerate oligonucleotide-primed PCR
- MDA
multiple displacement amplification
- SNP
single nucleotide polymorphism
- RFLP
restriction fragment length polymorphism
- PEP
primer extension preamplification
References
- 1.Telenius H, Carter N P, Bebb C E, Nordenskjold M, Ponder B A, Tunnacliffe A. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 2.Cheung V G, Nelson S F. Proc Natl Acad Sci USA. 1996;93:14676–14679. doi: 10.1073/pnas.93.25.14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paunio T, Reima I, Syvanen A C. Clin Chem. 1996;42:1382–1390. [PubMed] [Google Scholar]
- 5.Lizardi P M, Huang X, Zhu Z, Bray-Ward P, Thomas D C, Ward D C. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 6.Dean F B, Nelson J R, Giesler T L, Lasken R S. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco L, Bernad A, Lazaro J M, Martin G, Garmendia C, Salas M. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 8.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 9.Applied Biosystems. User Bulletin 2: ABI PRISM 7700 Sequence Detection System. Foster City, CA: Applied Biosystems; 1997. , part number 4303859B, Dec. 11. [Google Scholar]
- 10. Faruqi, A. F., Hosono, S., Driscoll, M. D., Dean, F. B., Alsmadi, O., Bandaru, R., Kumar, G., Grimwade, B., Zong, Q., Sun, Z., et al. (2001) BMC Genomics2, http://www.biomedcentral.com/1471-2164/2/4. [DOI] [PMC free article] [PubMed]
- 11.Kallioniemi A, Kallioniemi O P, Sudar D, Rutovitz D, Gray J W, Waldman F, Pinkel D. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 12. Lizardi, P. M. (2000) U.S. Patent 6,124,120.
- 13.Wells D, Sherlock J K, Handyside A H, Delhanty J D. Nucleic Acids Res. 1999;27:1214–1218. doi: 10.1093/nar/27.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S H, Godfrey T, Jensen R H. J Urol. 1999;162:1512–1518. [PubMed] [Google Scholar]
- 15.Klein C A, Schmidt-Kittler O, Schardt J A, Pantel K, Speicher M R, Riethmuller G. Proc Natl Acad Sci USA. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt K, Lazzeroni L C, Foote S, Vollrath D, Fisher E M, Goradia T M, Lange K, Page D C, Arnheim N. Am J Hum Genet. 1994;55:423–430. [PMC free article] [PubMed] [Google Scholar]
- 17.Snabes M C, Chong S S, Subramanian S B, Kristjansson K, DiSepio D, Hughes M R. Proc Natl Acad Sci USA. 1994;91:6181–6185. doi: 10.1073/pnas.91.13.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulson T G, Galipeau P C, Reid B J. Genome Res. 1999;9:482–491. [PMC free article] [PubMed] [Google Scholar]
- 19.Gillespie K M, Valovin S J, Saunby J, Hunter K M, Savage D A, Middleton D, Todd J A, Bingley P J, Gale E A. Tissue Antigens. 2000;56:530–538. doi: 10.1034/j.1399-0039.2000.560607.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang V W, Bell D A, Berkowitz R S, Mok S C. Cancer Res. 2001;61:4169–4174. [PubMed] [Google Scholar]
- 21.Esteban J A, Salas M, Blanco L. J Biol Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 22.Eckert K A, Kunkel T A. PCR Methods Appl. 1991;1:17–24. doi: 10.1101/gr.1.1.17. [DOI] [PubMed] [Google Scholar]
- 23.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 24.Harper J C, Wells D. Prenatal Diagn. 1999;19:1193–1199. doi: 10.1002/(sici)1097-0223(199912)19:13<1193::aid-pd728>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 25.Buchanan A V, Risch G M, Robichaux M, Sherry S T, Batzer M A, Weiss K M. Hum Biol. 2000;72:911–925. [PubMed] [Google Scholar]
- 26.Guillier-Gencik Z, Bernheim A, Coullin P. Cytogenet Cell Genet. 1999;87:282–285. doi: 10.1159/000015450. [DOI] [PubMed] [Google Scholar]