Abstract
Sialic acids are widely expressed as terminal carbohydrates on glycoconjugates of eukaryotic cells. Sialylation is crucial for a variety of cellular functions, such as cell adhesion or signal recognition, and regulates the biological stability of glycoproteins. The key enzyme of sialic acid biosynthesis is the bifunctional UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase (UDP-GlcNAc 2-epimerase), which catalyzes the first two steps of sialic acid biosynthesis in the cytosol. We report that inactivation of the UDP-GlcNAc 2-epimerase by gene targeting causes early embryonic lethality in mice, thereby emphasizing the fundamental role of this bifunctional enzyme and sialylation during development. The need of UDP-GlcNAc 2-epimerase for a defined sialylation process is exemplified with the polysialylation of the neural cell adhesion molecule in embryonic stem cells.
Sialic acid is the most abundant terminal monosaccharide of glycoconjugates of the eukaryotic cell surface (1, 2). It is involved in a variety of cellular functions, such as cell–cell interaction including metastasis formation and progression of a variety of tumors, virus infection, and the biological stability of glycoproteins (3, 4). Of particular interest is the unique polysialylation of the neural cell adhesion molecule (NCAM), which is down-regulated during development. The expression of the polysialylated form of NCAM is known to be involved in learning and memory in the adult hippocampus. In addition, polysialylation of NCAM often correlates with tumorigenicity (5–9).
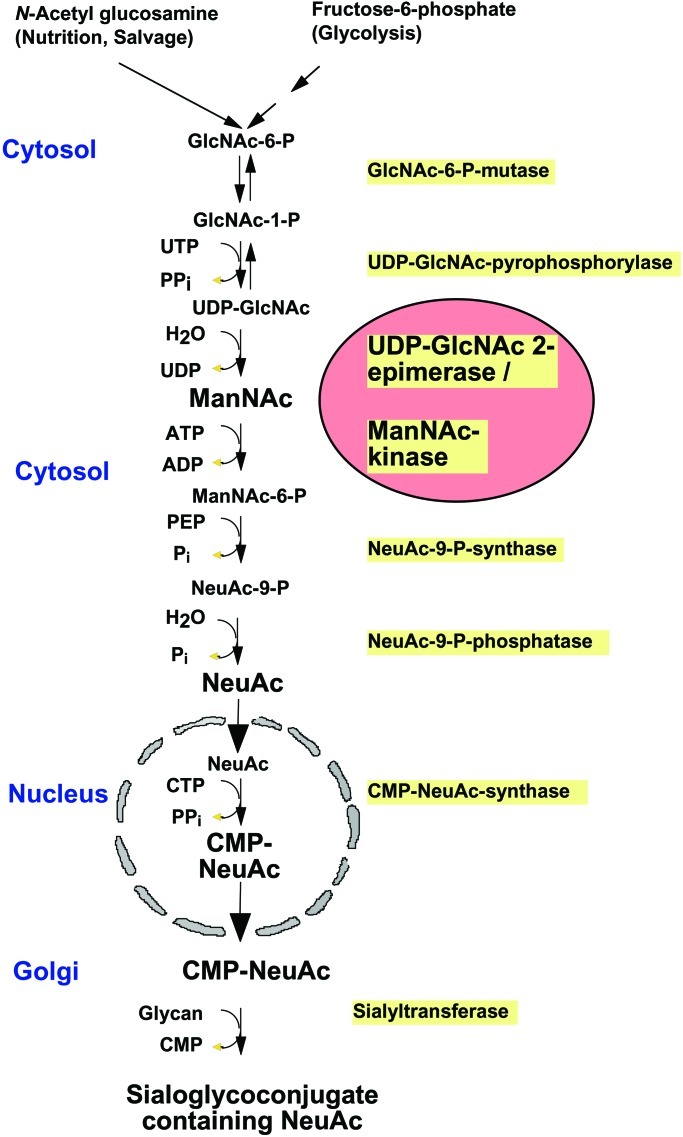
Sialic acids are synthesized in the cytosol from UDP-N-acetylglucosamine by four consecutive reactions. The first two steps in sialic acid biosynthesis (Fig. 1) are catalyzed by the bifunctional enzyme, UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase (UDP-GlcNAc 2-epimerase), which has been cloned and characterized in rodents and humans (10–13). It catalyzes both enzymatic reactions as a hexamer only (14). Recently, protein kinase C has been associated with UDP-GlcNAc 2-epimerase, regulating its enzymatic activity (15). Northern-blot analysis and in situ hybridization revealed the highest expression in liver. To a lesser extent the enzyme is also expressed in all other organs investigated. UDP-GlcNAc 2-epimerase is fully expressed at all stages during mouse development that have been investigated so far (11).
Figure 1.
Scheme representing the biosynthesis of sialic acid
The clinical relevance of the UDP-GlcNAc 2-epimerase was demonstrated by the detection of a binding defect of the feedback inhibitor CMP-sialic acid (16, 17) leading to sialuria (13, 18, 19). In this sialic acid storage disease, free sialic acid accumulates in the cytoplasm, which results in severe mental retardation of the surviving patients. The significance of the enzyme is further illustrated by the observation that, in a variant of HL60 cells, the low expression of sialic acids is correlated with dramatically reduced enzyme activity (20). Recently, it was proposed that mutations in the human UDP-GlcNAc 2-epimerase gene are responsible for hereditary inclusion body myopathy, a unique group of neuromuscular disorders characterized by adult onset, slowly progressive weakness, and a typical muscle pathology (21).
Methods
Gene Targeting.
The genomic clone containing the 5′ part of the UDP-GlcNAc 2-epimerase gene was isolated from a mouse 129Sv genomic library by hybridization with a rat 529-bp PCR fragment corresponding the UDP-GlcNAc 2-epimerase-coding region from nucleotide 430–959. A 5-kb fragment obtained by BamHI digestion containing the second exon of the UDP-GlcNAc 2-epimerase representing the coding region from nucleotide 164–619 was isolated and used for the target construct. A neomycin-resistance gene under the control of a phosphoglycerate promotor and a pgk poly(A) addition site were inserted in reverse orientation at the NcoI site of exon 2. A herpes simplex virus thymidine kinase gene, also under the control of a pgk promotor and a pgk poly(A) addition site, was inserted at the NcoI site of the 3.3-kb fragment. The sequenced targeting vector was linearized by using NotI and purified. Fifty micrograms of the DNA were then electroporated into E14.1 cells.
Genotyping.
Transfected embryonic stem cells or mouse tails tissue were lysed, and DNA was isolated as described elsewhere (31). DNA of individual embryonic stem cell clones was digested with BamHI, subjected to Southern blotting, then analyzed by hybridization with the probe mentioned above (probe 1, labeled to 108 cpm/μg by random priming).
Sequencing.
Sequencing was performed by using a LI-COR 4200 automatic sequencer (MWG Biotec, Ebersberg, Germany). Both strands of all cDNA molecules were sequenced at least twice and the results analyzed with use of the MacMolly Tetra software.
Generation of Embryonic Stem Cells.
Blastocysts from heterozygous matings were isolated at day 3.5 postcoitum and cultivated for another 4 days in single wells on embryonic fibroblasts. The outgrowth of the inner cell mass was picked, trypsinated, and replated. Colonies of embryonic stem (ES) cells were cultivated and expanded as described by Evans and Kaufmann (22). Resulting ES-cell lines were genotyped by Southern blotting.
Analytical Procedures.
UDP-GlcNAc 2-epimerase activity was measured as described (14).
Immunoprecipitations and Western Blotting.
Cells were lysed, and lysates were cleared by centrifugation (100,000 × g, 30 min). Five milligrams of protein A-agarose (Sigma) were precoated with 12 μg of UDP-GlcNAc 2-epimerase-specific polyclonal antibodies in PBS for 2 h at room temperature and washed twice with lysis buffer. Lysates were incubated for 2 h at 4°C with precoated protein A-agarose and pelleted by centrifugation. Before SDS/PAGE, pellets were washed three times with lysis buffer and once with PBS.
Samples were separated on SDS-polyacrylamide gels (Bio-Rad) and transferred to nitrocellulose filters. The blots were analyzed by using UDP-GlcNAc 2-epimerase-specific antibodies (11) and peroxidase-conjugated secondary antibodies (Dianova, Hamburg, Germany). Bands were visualized by enhanced chemiluminescence (Amersham Pharmacia) according to the manufacturer's instructions and exposing Kodak XR-5 films for times between 10 and 120 s.
Reverse Transcription–PCR.
The PCR reaction was performed under standard conditions. Total ES-cell RNA was transcribed to cDNA by using superscript reverse transcriptase (Life Technologies, Karlsruhe, Germany) according to the manufacturer's instructions. UDP-GlcNAc 2-epimerase-specific primers were annealed to cDNA and incubated with Taq polymerase (Perkin–Elmer) for 25 cycles (60 s, 94°C; 60 s, 58°C; 60 s, 72°C). As a control, glyceraldehyde-3-phosphate dehydrogenase was also analyzed.
Fluorescence-Activated Cell Sorter Analysis.
Cells (1 × 106) of each culture were then harvested and washed in PBS, centrifuged at 300 × g for 3 min, and resuspended in 500 μl of PBS containing 20 μg/ml Limax flavus agglutinin (LFA). Cells were then incubated on ice for 45 min and washed two times in 2 ml of PBS, before resuspending in PBS and analyzing on a Becton Dickinson FACScan instrument.
Results and Discussion
Here, we report the consequences of the targeted mutagenesis of the UDP-GlcNAc 2-epimerase gene. The strategy of its inactivation is demonstrated in Fig. 2. Heterozygous mice deficient in UDP-GlcNAc 2-epimerase have no obvious abnormalities. They were intercrossed and 134 offspring were genotyped; of these, 47 were wild type (+/+) and 87 were heterozygous (+/−). No homozygous mice could be identified. The fraction of wild-type mice was within the Mendelian expectations, assuming that inactivation of the UDP-GlcNAc 2-epimerase gene is lethal. Also by genotyping embryos between E10.5 and E17 of heterozygous parental mice, no −/− embryos were identified. Because the fraction of wild-type (+/+) and heterozygous (+/−) embryos was within the Mendelian expectations (1:2), we conclude that inactivation of the UDP-GlcNAc 2-epimerase gene is lethal before E 10.5. However, by genotyping embryos between E8.5 and E9.5, we could identify 10% homozygous UDP-GlcNAc 2-epimerase-deficient (−/−) embryos at E8.5 and 6% (−/−) embryos at E9.5, indicating that the inactivation is lethal between E8.5 and E9.5.
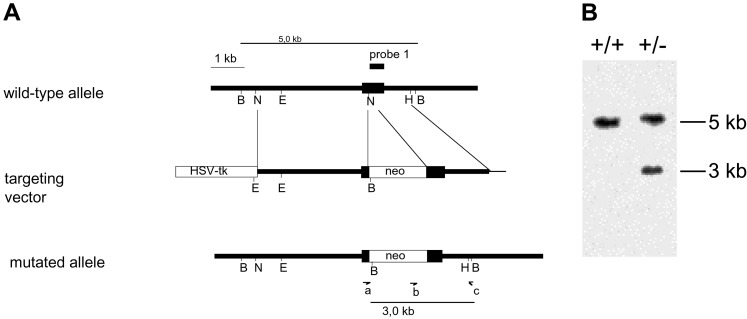
Figure 2.
(A) Strategy of constructing the UDP-GlcNAc 2-epimerase-targeting vector. A gene-targeting vector was used to generate murine 129 ES cells deficient in one copy of UDP-GlcNAc 2-epimerase by homologous recombination. The murine UDP-GlcNAc 2-epimerase gene was inactivated by replacing the second exon by a neo cassette. Homologous recombinant clones were used to create 129/Ola-C57BL/6J chimeric mice that transmitted the mutated UDP-GlcNAc 2-epimerase gene through the germ line (B = BamHI, E = EcoRI, H = HincII, n = NcoII). (B) Identification of homologous recombination events. Ten days after transfection, genomic DNA was isolated from individual cell clones. Homologous recombination was identified by Southern blot analysis as an additional 3-kb fragment in heterozygous cells (+/−). In contrast, in wild-type cells (+/+) only the 5-kb fragment is detectable.
The consequences of UDP-GlcNAc 2-epimerase inactivation were investigated in stem cell lines deficient for this enzyme. We generated 11 ES cell lines of day 3.5 postcoitum. Four of these had the wild-type (+/+) genotype, six were heterozygous (+/−), and one was homozygous (−/−) UDP-GlcNAc 2-epimerase-deficient. As viewed by light microscopy, no significant differences existed in the growth and morphology of all these cell lines.
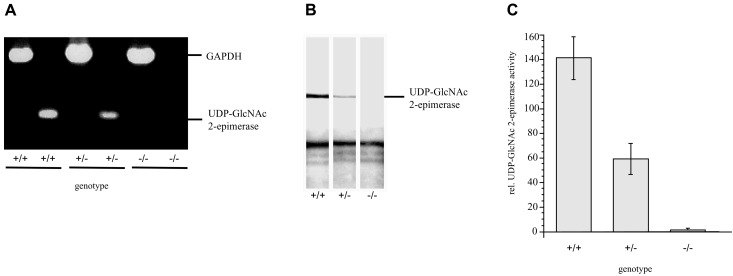
As expected, the homozygous deficient cells expressed no UDP-GlcNAc 2-epimerase-mRNA (Fig. 3A), and consequently no UDP-GlcNAc 2-epimerase could be detected by Western blot analysis (Fig. 3B). Furthermore, no enzyme activity was measured in the deficient ES cells (Fig. 3C). In all experiments, heterozygous ES cells expressed approximately half of the UDP-GlcNAc 2-epimerase mRNA, protein, or enzyme activity compared with wild-type cells (Fig. 3 A–C).
Figure 3.
(A) Reverse transcription–PCR of +/+, +/−, and −/− ES cell clones. Total RNA of wild-type (+/+), heterozygous (+/−), and homozygous deficient (−/−) ES cells were transcribed to cDNA and amplified by using specific primers to UDP-GlcNAc 2-epimerase or glyceraldehyde-3-phosphate dehydrogenase. (B) Western blot analysis of UDP-GlcNAc 2-epimerase-deficient ES cells. UDP-GlcNAc 2-epimerase of wild-type (+/+), heterozygous (+/−), and homozygous deficient (−/−) ES cells was immunoprecipitated and detected by Western blot analysis by using specific polyclonal antibodies to UDP-GlcNAc 2-epimerase. (C) Determination of the enzyme activity in UDP-GlcNAc 2-epimerase-deficient ES-cells. Cytosolic fractions of wild-type (+/+), heterozygous (+/−), and homozygous deficient (−/−) ES cells were prepared by ultracentrifugation, and UDP-GlcNAc 2-epimerase activity was determined. Bars represent mean values of three independent experiments.
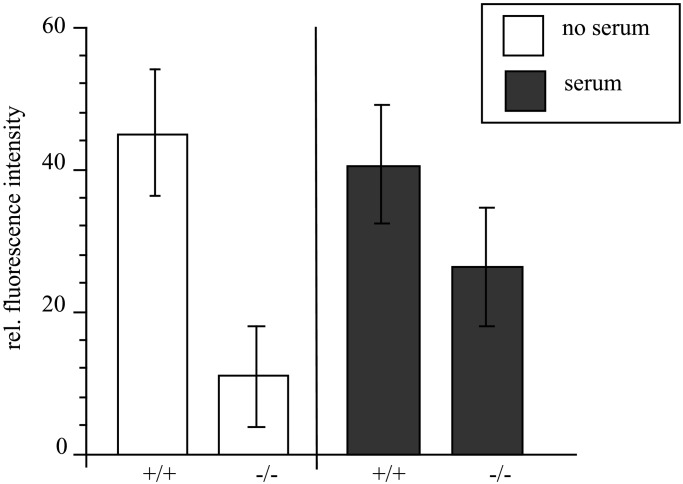
Because UDP-GlcNAc 2-epimerase is the key enzyme of sialic acid biosynthesis, deficient ES cells should not express sialic acids on their cell surface. By using the sialic acid-specific lectin L. flavus agglutinin in fluorescence-activated cell sorter analysis, we could demonstrate that the cell surface sialylation in the deficient ES cells is reduced by more than 75% compared with wild-type ES cells when cultured in medium containing serum-replacement factor (Fig. 4, open bars), which contains only little amount of sialylated glycoproteins. In contrast, by using serum-containing medium, which contains a lot of highly sialylated glycoproteins, the surface sialylation of deficient ES cells is reduced by only 28% compared with wild-type cells (Fig. 4, filled bars), indicating that sialylated glycoproteins are taken up from the medium and compensate the deficient biosynthesis of sialic acid. We then investigated the polysialylation of NCAM by Western blot analysis. Wild-type ES cells express a large amount of polysialic acid (PSA) on NCAM. Heterozygous ES cells express less PSA, and ES cells deficient in UDP-GlcNAc 2-epimerase do not express any PSA (Fig. 5). Because all these cells were cultured in the presence of serum, polysialylation of deficient ES cells cannot be restored by reutilization of sialylated glycoproteins from the medium. In contrast, polysialylation can be restored by feeding deficient ES cells with the natural precursor of sialic acid, N-acetylmannosamine (ManNAc), but not with N-acetylglucosamine (Fig. 5), which cannot be epimerized to ManNAc (Fig. 1).
Figure 4.
Cell surface sialylation of UDP-GlcNAc 2-epimerase-deficient ES cells. Wild-type (+/+) and homozygous deficient ES cell lines (−/−) were grown for 5 days in serum-replacement medium without serum (no serum, open bars), or in serum-containing medium (serum, filled bars). Cell surface expression of sialic acids was analyzed by fluorescence-activated cell sorter analysis with L. flavus agglutinin. Bars represent mean values of three independent experiments.
Figure 5.
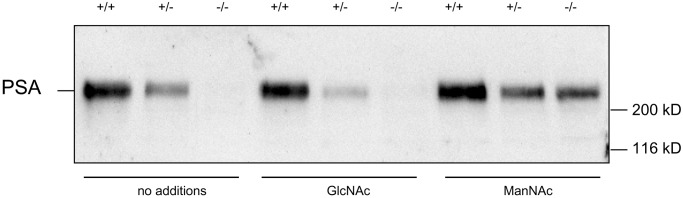
PSA expression of UDP-GlcNAc 2-epimerase-deficient ES cells. Solubilisates of wild-type (+/+), heterozygous (+/−), or UDP-GlcNAc 2-epimerase-deficient ES cells (−/−) cultured in FCS (no additions), FCS plus 10 mM GlcNAc (GlcNAc) or FCS plus 10 mM ManNAc (ManNAc) were separated on SDS-polyacrylamide gels and transferred to nitrocellulose filters. PSA was detected with a PSA-specific monoclonal antibody.
These data indicate that sialic acid can be synthesized from ManNAc without the UDP-GlcNAc 2-epimerase activity, but that the epimerization of UDP-GlcNAc to ManNAc is the rate-limiting step of sialic acid biosynthesis in vivo. However, the phosphorylation of ManNAc to ManNAc-6-phosphate can be catalyzed by sugar kinases other than that of the UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase, such as the GlcNAc-kinase (23). The total hexokinase activity is ≈1.9 mU/mg in all, UDP-GlcNAc 2-epimerase-deficient, heterozygous and wild-type ES cells (data not shown).
Our experiments demonstrate that endocytosed sialylated glycoproteins are a source of sialic acid enabling the sialylation of cell surface glycoproteins. In contrast, NCAM-specific polysialylation needs an active UDP-GlcNAc 2-epimerase to provide a sufficient amount of sialic acid. The observation that sialylation is not completely abolished in deficient ES cells is because medium containing serum-replacement factor is not completely free of sialic acids (63 nmol/ml). Experiments in the absence of serum could not be performed, because ES cells are not viable in the absence of serum or serum-replacement factor for longer culture periods (data not shown).
Therefore, in vivo a strong need exists for the biosynthetic machinery for sialic acid biosynthesis, but in vitro several UDP-N-GlcNAc 2-epimerase-deficient cell lines survive because of the presence of sialylated glycoproteins in the medium (20). Moreover, it should be noted that in vivo maternal, sialylated glycoproteins cannot be used by the embryo, because UDP-GlcNAc 2-epimerase inactivation is lethal during embryogenesis. The early lethality of the deficient mice might be explained by the loss of protection from proteolytic processes or by disturbed cell–cell adhesion and cell migration, because most of the cell adhesion molecules involved are sialylated glycoproteins (6, 24–26). In contrast to other nonlethal knockout models, such as the sialylated neural cell adhesion molecule (27), lack of sialylation can be compensated only by ManNAc and not by other, closely related amino sugars.
In agreement with our study, it has been demonstrated that inactivation of the N-acetylglucosaminyltransferase I, an enzyme that is necessary to build complex oligosaccharides of glycoproteins, is lethal between E9.5 and E10.5 (28, 29). Furthermore, mice expressing only the monosialylated ganglioside GM3 suffer from epileptical attacks and die early (30).
Our experiments demonstrate that in vivo the UDP-GlcNAc 2-epimerase is the key enzyme of sialic acid biosynthesis. No bypass pathway exists. The embryonic lethality of UDP-GlcNAc 2-epimerase-deficient mice proves that sialylation plays a crucial role during development showing that sialoglycans display essential functions during embryogenesis.
Acknowledgments
We thank Dr. R. Gerardy-Schahn for providing monoclonal antibody 735 to polysialic acid. These studies were supported by the Deutsche Forschungsgemeinschaft (Ho 1959/1-1,1-2, SFB 366), the Fonds der Chemischen Industrie, and the Sonnenfeld Stiftung.
Abbreviations
- NCAM
neural cell adhesion molecule
- UDP-GlcNAc 2-epimerase
UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase
- ES
embryonic stem
- PSA
polysialic acid
- ManNAc
N-acetylmannosamine
References
- 1.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Traving C, Schauer R. Cell Mol Life Sci. 1998;54:1330–1349. doi: 10.1007/s000180050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kageshita T, Hirai S, Kimura T, Hanai N, Ohta S, Ono T. Cancer Res. 1995;55:1748–1751. [PubMed] [Google Scholar]
- 4.Varki A. FASEB J. 1997;11:248–255. doi: 10.1096/fasebj.11.4.9068613. [DOI] [PubMed] [Google Scholar]
- 5.Finne J, Finne U, Deagostini-Bazin H, Goridis C. Biochem Biophys Res Commun. 1983;112:482–487. doi: 10.1016/0006-291x(83)91490-0. [DOI] [PubMed] [Google Scholar]
- 6.Sadoul R, Hirn M, Deagostini-Bazin H, Rougon G, Goridis C. Nature (London) 1983;304:347–349. doi: 10.1038/304347a0. [DOI] [PubMed] [Google Scholar]
- 7.van Zandwijk N, Mooi W J, Rodenhuis S. Lung Cancer. 1995;12, Suppl. 1:S27–S33. doi: 10.1016/0169-5002(95)00418-z. [DOI] [PubMed] [Google Scholar]
- 8.Rutishauser U. Curr Opin Cell Biol. 1996;8:679–684. doi: 10.1016/s0955-0674(96)80109-8. [DOI] [PubMed] [Google Scholar]
- 9.Perl A K, Dahl U, Wilgenbus P, Cremer H, Semb H, Christofori G. Nat Med. 1999;5:286–291. doi: 10.1038/6502. [DOI] [PubMed] [Google Scholar]
- 10.Stäsche R, Hinderlich S, Weise C, Effertz K, Lucka L, Moormann P, Reutter W. J Biol Chem. 1997;272:24319–24324. doi: 10.1074/jbc.272.39.24319. [DOI] [PubMed] [Google Scholar]
- 11.Horstkorte R, Nöhring S, Wiechens N, Schwarzkopf M, Danker K, Reutter W, Lucka L. Eur J Biochem. 1999;260:923–927. doi: 10.1046/j.1432-1327.1999.00253.x. [DOI] [PubMed] [Google Scholar]
- 12.Lucka L, Krause M, Danker K, Reutter W, Horstkorte R. FEBS Lett. 1999;454:341–344. doi: 10.1016/s0014-5793(99)00837-6. [DOI] [PubMed] [Google Scholar]
- 13.Seppala R, Lehto V P, Gahl W A. Am J Hum Genet. 1999;64:1563–1569. doi: 10.1086/302411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinderlich S, Stäsche R, Zeitler R, Reutter W. J Biol Chem. 1997;272:24313–24318. doi: 10.1074/jbc.272.39.24313. [DOI] [PubMed] [Google Scholar]
- 15.Horstkorte R, Nohring S, Danker K, Effertz K, Reutter W, Lucka L. FEBS Lett. 2000;470:315–318. doi: 10.1016/s0014-5793(00)01331-4. [DOI] [PubMed] [Google Scholar]
- 16.Warren L, Felsenfeld H. J Biol Chem. 1962;237:14121–14131. [PubMed] [Google Scholar]
- 17.Kornfeld S, Kornfeld R, Neufeld E, O‘Brian J. Biochemistry. 1964;52:371–379. doi: 10.1073/pnas.52.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montreuil J, Biserte G, Strecker G, Spik G, Fontaine G, Farriaux J P. Clin Chim Acta. 1968;21:61–69. doi: 10.1016/0009-8981(68)90011-9. [DOI] [PubMed] [Google Scholar]
- 19.Wilcken B, Don N, Greenaway R, Hammond J, Sosula L. J Inherited Metab Dis. 1987;10:97–102. doi: 10.1007/BF01800030. [DOI] [PubMed] [Google Scholar]
- 20.Keppler O T, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, Pawlita M. Science. 1999;284:1372–1376. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 21.Eisenberg I, Avidan N, Potikha T, Hochner H, Chen M, Olender T, Barash M, Shemesh M, Sadeh M, Grabov-Nardini G, et al. Nat Genet. 2001;29:83–87. doi: 10.1038/ng718. [DOI] [PubMed] [Google Scholar]
- 22.Hinderlich S, Berger M, Keppler O T, Pawlita M, Reutter W. Biol Chem. 2001;382:291–297. doi: 10.1515/BC.2001.036. [DOI] [PubMed] [Google Scholar]
- 23.Kawano T, Takasaki S, Tao T W, Kobata A. Int J Cancer. 1993;53:91–96. doi: 10.1002/ijc.2910530118. [DOI] [PubMed] [Google Scholar]
- 24.Barnes Y C, Skelton T P, Stamenkovic I, Sgroi D C. Blood. 1999;93:1245–1252. [PubMed] [Google Scholar]
- 25.Katoh S, Miyagi T, Taniguchi H, Matsubara Y, Kadota J, Tominaga A, Kincade P W, Matsukura S, Kohno S. J Immunol. 1999;162:5058–5061. [PubMed] [Google Scholar]
- 26.Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, et al. Nature (London) 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- 27.Ioffe E, Stanley P. Proc Natl Acad Sci USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ioffe E, Liu Y, Stanley P. Glycobiology. 1997;7:913–919. doi: 10.1093/glycob/7.7.913. [DOI] [PubMed] [Google Scholar]
- 29.Kawai H, Allende M L, Wada R, Kono M, Sango K, Deng C, Miyakawa T, Crawley J N, Werth N, Bierfreund U, et al. J Biol Chem. 2001;276:6885–6888. doi: 10.1074/jbc.C000847200. [DOI] [PubMed] [Google Scholar]
- 30.Evans M J, Kaufman M H. Nature (London) 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 31.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the Mouse Embryo. 2nd Ed. Oxford: Oxford Univ. Press; 1994. [Google Scholar]