Abstract
The cellular response to low tissue oxygen concentrations is mediated by the hypoxia-inducible transcription factor HIF-1. Under hypoxic conditions, HIF-1 activates transcription of critical adaptive genes by recruitment of the general coactivators CBP/p300 through interactions with its α-subunit (Hif-1α). Disruption of the Hif-1α/p300 interaction has been linked to attenuation of tumor growth. To delineate the structural basis for this interaction, we have determined the solution structure of the complex between the carboxy-terminal activation domain (CAD) of Hif-1α and the zinc-binding TAZ1 (CH1) motif of cyclic-AMP response element binding protein (CREB) binding protein (CBP). Despite the overall similarity of the TAZ1 structure to that of the TAZ2 (part of the CH3) domain of CBP, differences occur in the packing of helices that can account for differences in specificity. The unbound CAD is intrinsically disordered and remains relatively extended upon binding, wrapping almost entirely around the TAZ1 domain in a groove through much of its surface. Three short helices are formed upon binding, stabilized by intermolecular interactions. The Asn-803 side chain, which functions as a hypoxic switch, is located on the second of these helices and is buried in the molecular interface. The third helix of the Hif-1α CAD docks in a deep hydrophobic groove in TAZ1, providing extensive intermolecular hydrophobic interactions that contribute to the stability of the complex. The structure of this complex provides new insights into the mechanism through which Hif-1α recruits CBP/p300 in response to hypoxia.
Hypoxia-inducible factor 1 (HIF-1) activates genes that are crucial for cell survival under hypoxic conditions. HIF-1 is a basic helix-loop-helix–periodic-aryl hydrocarbon receptor-single-minded (Per–Arnt–Sim) domain transcription factor composed of two subunits, Hif-1α and the aryl hydrocarbon receptor nuclear translocator (1). It induces expression of proteins controlling glucose metabolism, cell proliferation, and vascularization, and thus plays a key role in the pathology of cancer, heart disease, and stroke (2, 3). The oxygen-sensing and transactivating functions of HIF-1 are contained within the Hif-1α subunit, which is overexpressed in most human cancers (4). In mice, deletion of Hif-1α results in neural and cardiovascular developmental arrest and embryonic death (5), underscoring the vital role of Hif-1α, and the maintenance of cellular oxygen homeostasis, in the correct functioning of the mammalian cell.
Several steps in the signal transduction mechanism by which low cellular oxygen concentration is converted to Hif-1α-mediated transcription of hypoxic response genes are now known (reviewed in refs. 6 and 7). Under normoxic conditions, Hif-1α becomes hydroxylated at a proline in the oxygen-dependent degradation domain, associates with the von Hippel–Lindau tumor suppressor protein, and is rapidly degraded in the cytoplasm via the ubiquitin-proteasome pathway (8–10). Under hypoxic conditions, proline hydroxylation no longer takes place and Hif-1α accumulates in the nucleus, where it forms a DNA-binding heterodimer with the aryl hydrocarbon receptor nuclear translocator and recruits the general coactivator CBP/p300 to initiate transcription at hypoxia-responsive elements.
CBP and its ortholog p300 are modular transcriptional coactivators that integrate multiple signal transduction pathways by serving as scaffolds for the assembly of transcriptional activation complexes and by modifying chromatin and transcription factors through their intrinsic acetyltransferase activity (reviewed in refs. 11–13). They are essential for such basic cellular functions as growth, differentiation, apoptosis, and DNA repair, and they play critical roles in embryonic development. CBP/p300 contain two transcriptional adapter zinc-binding (TAZ) motifs (14), which function as the sites of interaction with numerous transcription factors and viral oncogenes (15). The TAZ1 motif corresponds to cysteine/histidine-rich domain-1 (the CH1 domain); the TAZ2 domain and the zz zinc-binding domain together make up cysteine/histidine-rich domain-3 (the CH3 domain). The TAZ1 and TAZ2 domains each bind three Zn2+ ions through HCCC motifs that are completely conserved across all species sequenced to date (16). The solution structure of the isolated CBP TAZ2 domain (16) shows a novel, zinc-stabilized helical domain with an extensive, highly conserved hydrophobic core and pronounced surface grooves that are likely sites for protein–protein interactions.
Transcriptional regulation by Hif-1α is entirely dependent on the interaction between the Hif-1α C-terminal activation domain (CAD) and the TAZ1 domain of CBP/p300 (17, 18). In addition to the proline hydroxylation switch in the oxygen-dependent degradation domain, which governs the degradation of Hif-1α under normoxic conditions, a second hypoxic switch operates in the CAD: hydroxylation of an asparagine residue occurs under normoxic conditions and inhibits binding to CBP/p300. In hypoxia, asparagine hydroxylation is blocked and recruitment of the CBP/p300 coactivators is facilitated (19). To further our understanding of the mechanisms controlling the cellular response to hypoxia, we have undertaken a structural analysis of the interacting domains of the transcription factor Hif-1α and the TAZ1 (CH1) domain of CBP. Our structure reveals a highly intimate complex in which Hif-1α wraps almost completely around the TAZ1 domain. The Hif-1α activation domain, which is intrinsically unstructured in the free state, undergoes a folding transition in forming the complex with TAZ1. Although the structure of the TAZ1 domain in the complex is similar overall to that of the free TAZ2 domain (16), several key differences reveal a structural basis for the different specificity of CH1 and CH3 and their participation in separate transcriptional pathways.
Experimental Procedures
Protein Expression and Binding Assays.
Constructs of the Hif-1α C-terminal activation domain and CBP TAZ1 domain were designed on the basis of previous domain-mapping experiments (17, 20–22) and sequence conservation. The TAZ1 domain (residues 345–439 of mouse CBP) was cloned into pET21a and overexpressed in Escherichia coli BL21 DE3 [DNAY] in M9 minimal medium supplemented with 150 μM zinc sulfate. The protein was solubilized from inclusion bodies, purified, and reconstituted with zinc by using a procedure similar to that for TAZ2 (16). Binding of three zinc atoms per TAZ1 monomer was confirmed by electrospray mass spectrometry.
To avoid degradation in E. coli, the human Hif-1α CAD (residues 776–826) was coexpressed with TAZ1 (345–439) in a biscistronic expression vector (23). Upon low-temperature induction in medium supplemented with zinc sulfate, both TAZ1 and the HIF-1α CAD are expressed at high levels in the soluble fraction. The CAD (10 mg/liter) was isolated from the cleared cell lysate by anion-exchange chromatography and reversed-phase HPLC in acetonitrile/acetate, pH 6, and acetonitrile/trifluoroacetic acid buffers. Hif-1α peptides corresponding to residues 808–826 and 790–826 were prepared by automated synthesis (PerSeptive Biosystems, Framingham, MA). Folded TAZ1 and Hif-1α CAD were exchanged into NMR buffer (10 mM Tris-d11, pH 6.3, containing 10 mM DTT-d10, 0.5–0.7 mM ZnSO4, and 0.02% sodium azide in D2O or 90% H2O/10% D2O) and concentrated by using a Centriprep 3.
Binding was assayed by the yeast two-hybrid method, by fluorescence titrations, and by isothermal titration calorimetry (ITC). For yeast two-hybrid experiments, TAZ1 (345–439) was cloned into pGBT9 (DNA-binding domain fusion) and the Hif-1α (776−826), p53 (1–61), and E1A (15–77) activation domains were cloned into pGAD424 (activation domain fusion) (Matchmaker System, CLONTECH). Protein–protein interactions were assayed in yeast strain Y190 with selection on -HIS, -LEU, -TRP plates supplemented with 15 mM 3-aminotriazole. Positive transformants were further assayed for β-galactosidase activity toward the substrate ONPG. At least six clones per interacting pair of proteins were assayed. ITC data were collected at 25°C by using a Microcal MCS Titration calorimeter. Purified protein samples were dialyzed into ITC buffer (20 mM Tris, pH 6.8, containing 50 mM NaCl, 2 mM DTT) and centrifuged to remove aggregates. Protein concentrations were measured by standard bicinchoninic acid protein assay. Titration data were fit to a one-site model by using the Microcal data analysis software ORIGIN 2.3 (Microcal Software, Northampton, MA) and were not adjusted for buffer effects.
NMR Spectroscopy.
NMR spectra were acquired at 298 K on Bruker DRX600 and DRX800 spectrometers. Complexes were prepared from uniformly 13C,15N-labeled TAZ1 (345–439) and unlabeled Hif-1α (776–826) or 13C,15N-labeled Hif-1α (776–826) and unlabeled TAZ1 (345–439). The concentrations ranged between 1 and 1.5 mM labeled and 2.5 and 3.4 mM unlabeled protein. Data were processed by using NMRPipe (24) and analyzed by using NMRView (25). Backbone 13C, 15N, and 1H assignments were made by using three-dimensional HNCACB (26), CBCA(CO)NH (27), HBHA(CBCACO)NH (28), and HNCA (27) experiments. Assignment of side-chain resonances was accomplished by three-dimensional C(CO)NH-TOCSY, H(CCO)NH-TOCSY (29), and, CCH-correlated spectroscopy, CCH-TOCSY, and HCCH-TOCSY experiments (TOCSY, total correlation spectroscopy) (30). Stereospecific assignments and coupling constants were obtained from the analysis of three-dimensional HACAHB-correlated spectroscopy (31), HNHB, HNHA, and two-dimensional 13C-{13CO} and 13C-{15N} spin echo difference (32), and heteronuclear-edited nuclear Overhauser effect (NOE) spectroscopic data. Distance restraints were obtained by three-dimensional 13C- and 15N/13C-edited NOE spectroscopic experiments. three-dimensional 12C-filtered/13C-edited NOE spectroscopic experiments (33) were performed to obtain intermolecular distance restraints.
Structure Calculation and Analysis.
Distance restraints were generated with the program SANE (34) and classified according to NOE volumes, with upper bounds of 2.7 Å, 3.5 Å, 4.5 Å, 5.5 Å, and 1.8 Å lower bounds. Stereospecific assignments were included for 49 β-methylene proton pairs. Backbone φ and ψ angles in helical regions, as indicated by the chemical shift index (35), were restrained to −60° ± 30° and −40° ± 30°. On the basis of the analysis of coupling constant and NOE data, restraints for side chain χ1 angles were defined as (−60°, 60°, 180°) ± 30°. Helical hydrogen bond restraints were used in the early stages of the calculations to facilitate assignment of ambiguous NOEs but were omitted during refinement. The experimental restraints are summarized in Table 1.
Table 1.
NMR structure statistics
| NMR constraints | ||
|---|---|---|
| TAZ1 | Hif-1α | |
| Individual domains | ||
| Unambiguous distance restraints | ||
| Intraresidue | 448 | 145 |
| Sequential + medium range | 602 | 520 |
| Long range (|i − j| > 4) | 145 | 24 |
| Ambiguous distance restraints | 41 | 4 |
| Torsion angle restraints | 79 φ, 58 ψ, 45 χ1 | 29 φ, 16 ψ, 23 χ1 |
| TAZ1/Hif-1α complex | ||
| Intermolecular distance restraints | 92 | |
| Ambiguous restraints | 28 | |
| Structure statistics (20 structures) | |
|---|---|
| Violation statistics | |
| NOE violations >0.1 Å/structure | 8 ± 2 |
| Maximum NOE violation/structure, Å | 0.17 ± 0.05 |
| Dihedral constraint violations/structure | 0.7 ± 0.7 |
| Maximum torsion angle violation/structure, ° | 4 ± 4 |
| Energies | |
| Mean GB* constraint violation energy, kcal/mol | 8 ± 1 |
| Mean GB AMBER energy, kcal/mol | −8,496 ± 20 |
| Mean deviations from ideal covalent geometry | |
| Bond lengths, Å | 0.0061 ± 0.0002 |
| Bond angles, ° | 2.40 ± 0.03 |
| PROCHECK statistics | |
| Residues in most favored regions | 87.2 |
| Residues in allowed regions | 12.0 |
| Residues in generously allowed regions | 0.6 |
| Residues in disallowed regions | 0.1 |
| RMS† deviations from the average structure | |
| TAZ1/HIF-1α complex (residues 345–435; 792–825) | |
| Backbone atoms (N, Cα, C′), Å | 0.91 |
| All heavy atoms, Å | 1.39 |
| Helices only | |
| Backbone atoms (N, Cα, C′), Å | 0.45 |
| All heavy atoms, Å | 0.91 |
GB, generalized Born.
RMS, root-mean-square.
An initial set of 200 structures was calculated by torsion-angle dynamics with the program DYANA (36). Restraints on the zinc coordination geometry were applied as described (16). In the first cycle of structure calculation, some of the ambiguous NOEs for TAZ1 were assigned on the basis of a topological model generated from the CBP TAZ2 structure. Additional intra- and intermolecular NOE ambiguities were resolved during subsequent cycles of structure calculation by using the iterative SANE procedure (34). The 100 structures with the lowest target functions were refined in several cycles of molecular dynamics-simulated annealing by using the AMBER software package (37) and all-atom force field, modified to reduce charges to 20%. The final structures were refined through two cycles of vacuum-simulated annealing; in the first cycle, the system was heated to 1,000 K for the first 4 ps, followed by 4 ps of molecular dynamics, and final cooling to 0 K for the remaining 12 ps. Force constants were 20 kcal mol−1 Å−1 for NOE restraints and 150 kcal mol−1 rad−2 for dihedral angle restraints. A second cycle of vacuum-simulated annealing was performed at 500 K, followed by a final round of 1,000 steps of energy minimization. The 50 vacuum-refined structures with smallest restraint violations were subjected to a further cycle of simulated annealing by using a generalized Born potential (38, 39) to account for the effects of solvent. The first round of this calculation consisted of 1,000 steps of energy minimization, followed by a single 20-ps round of generalized Born-simulated annealing (heating to 1,000 K for 4 ps, annealing at 1,000 K for 4 ps, cooling for 12 ps), followed by a further 1,000 steps of energy minimization.
The 20 structures with lowest generalized Born AMBER energy and no distance constraint violations greater than 0.20 Å were chosen for structural analysis. Analysis with PROCHECK-NMR (40) shows that more than 99% of residues have backbone dihedral angles in allowed regions of the Ramachandran plot, with only 0.1% of residues in disallowed regions. Statistics are given in Table 1.
Results and Discussion
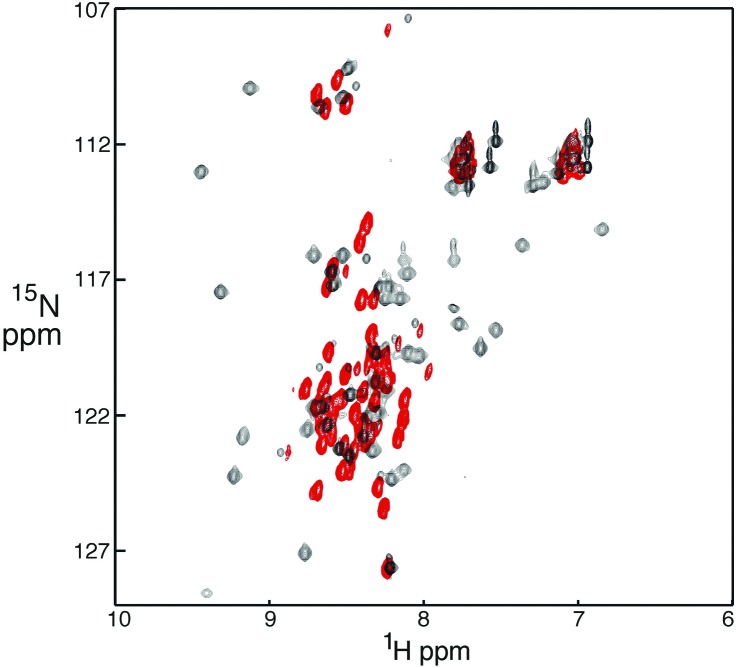
Most NMR experiments were performed with TAZ1 (345–439) isolated from inclusion bodies; the 1H-15N heteronuclear single quantum coherence spectrum of this construct was identical with that of TAZ1 obtained from the soluble cellular fraction and purified under native conditions, confirming that the refolded protein has the native fold. Hif-1α (776–826) in the unbound state is intrinsically disordered; its circular dichroism spectrum is characteristic of a random coil (data not shown) and the NMR spectrum lacks dispersion (Fig. 1, red cross peaks). The CAD becomes structured upon binding to TAZ1, as evidenced by the pronounced increase in dispersion of the amide proton resonances (Fig. 1, black cross peaks). Consistent with these observations, the Hif-1α construct is rapidly degraded in E. coli unless coexpressed with the TAZ1 domain.
Figure 1.
1H-15N heteronuclear single quantum coherence spectra (600 MHz) of Hif-1α (776–826) free (red) and bound to unlabeled TAZ1 (black).
Yeast two-hybrid assays confirmed that the constructs used for the NMR experiments, HIF-1α (776–826) and TAZ1 (345–439), interact specifically and with high affinity in vivo. No interactions between TAZ1 and activation domains derived from two other CBP-interacting proteins, p53 and E1A, were observed with this assay (data not shown). The dissociation constant of the TAZ1/HIF-1α CAD complex was determined to be 7 ± 1 nM by ITC. Because the high binding affinity made accurate determination of thermodynamic parameters difficult, ITC experiments were also performed with a construct truncated at the amino terminus, Hif-1α (790–826), which binds 5-fold more weakly (Kd = 34 ± 5 nM). Binding is enthalpy driven (ΔH0 = −21.8 ± 0.4 kcal⋅mol−1) and is entropically disfavored (TΔS0 = −11.6 ± 0.4 kcal⋅mol−1), consistent with the requirement for Hif-1α to fold upon binding. The shortest Hif-1α construct (residues 808–826) binds only weakly to TAZ1 (Kd > 500 μM). Although residues 813–826 of the CAD have been shown to be vital for binding to CBP/p300 and for Hif-1α-mediated transactivation (41, 42), residues 790–807 are clearly necessary for high-affinity binding.
Overall Structure of the Complex.
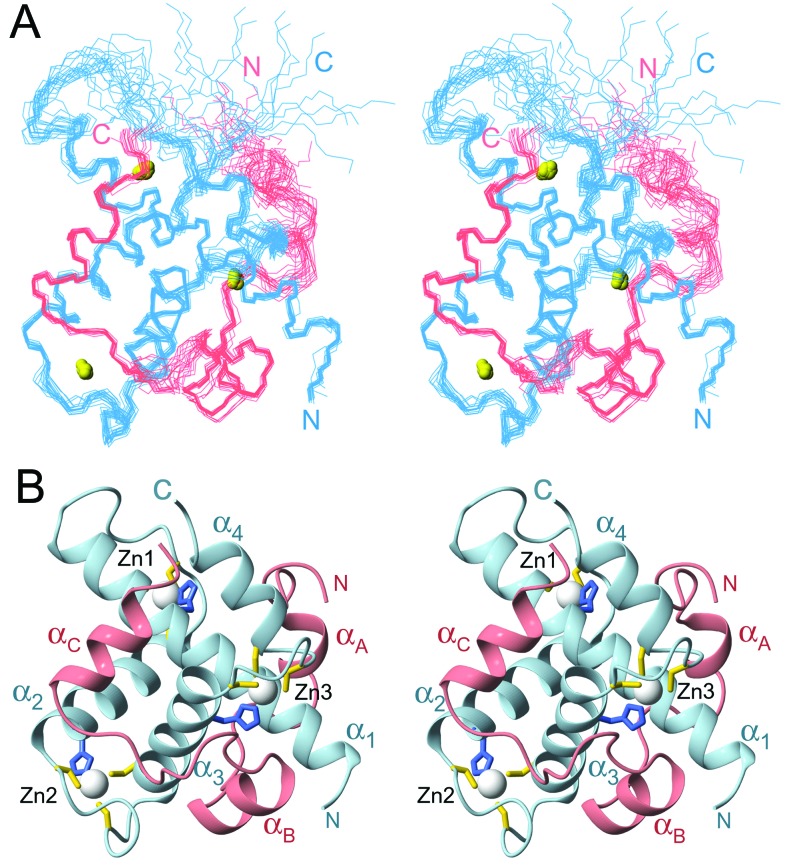
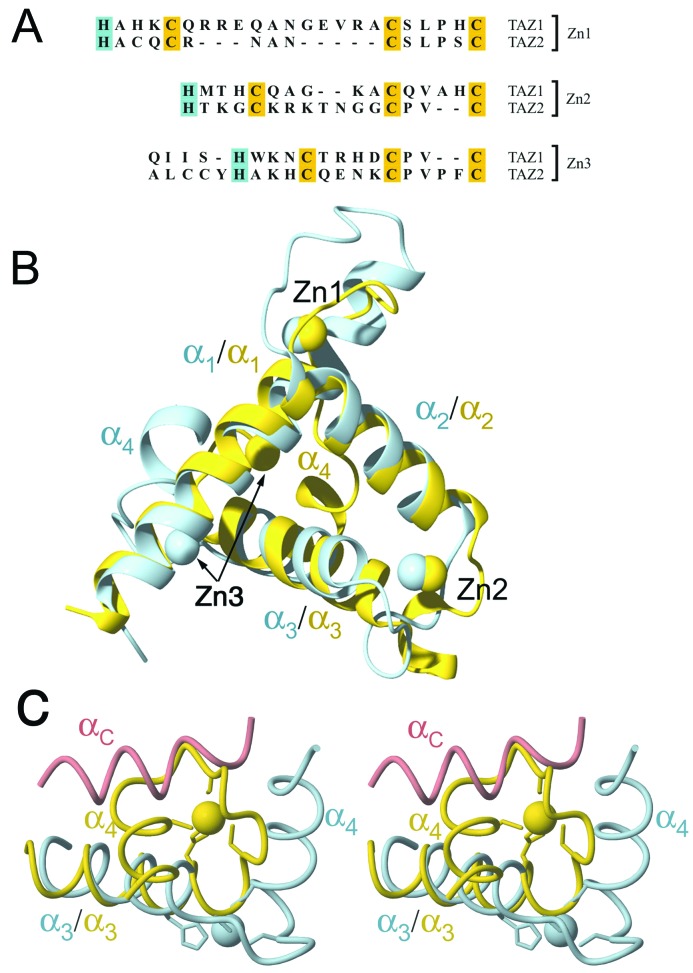
The family of NMR structures is generally well defined by the available constraints (Fig. 2A and Table 1). Residues near the N terminus of Hif-1α and residues 435–439 at the C terminus of TAZ1 are disordered, as are residues 372–380 in the first zinc-binding site of TAZ1. The TAZ1 structure consists of a bundle of four helices formed by residues Pro-347–Ala-372 (α1), His-383–His-396 (α2), Ala-406–Cys-421 (α3), and Cys-429–Asn-434 (α4) packed to enclose a hydrophobic core (Fig. 2B). The three zinc atoms are bound in similar coordination environments by His-362, Cys-366, Cys-379, Cys-384 (Zn1), His 393, Cys-397, Cys-403, Cys-408 (Zn2), and His-417, Cys-421, Cys-426, Cys-429 (Zn3). The histidine and the first cysteine ligand to each zinc are separated by one turn of helix, and the spacing between them (three residues, HX3C) is strictly conserved in both the TAZ1 and TAZ2 sequences (16) (Fig. 3A). The second Cys ligand is located in a loop of variable length, and the final Cys ligand is on the first turn of the following helix (Fig. 2B). The spacing between the second and third cysteine ligands is also variable (Fig. 3A).
Figure 2.
NMR structure of the Hif-1α:TAZ1 complex. (A) Stereo view of the best 20 structures superposed on backbone heavy atoms in ordered regions. The TAZ1 backbone is shown in blue, Hif-1α in pink, and the N and C termini of each chain are labeled in the corresponding colors. Bound zinc ions are shown as yellow spheres. (B) Ribbon representation of a single structure in a similar orientation to A. Helices α1–α4 of TAZ1 and αA–αC of the Hif-1α CAD are labeled. The zinc ions are represented as white spheres, and the side chains of the cysteine and histidine ligands are shown in yellow and blue, respectively.
Figure 3.
(A) Amino acid sequences of the zinc-binding sites (Zn1, Zn2, and Zn3) in the TAZ1 and TAZ2 domains of mouse CBP. The histidine (blue) and cysteine (yellow) ligands are highlighted. The sequence of part of helix α3 preceding the Zn3 site is shown; note the deletion in TAZ1 immediately preceding the histidine ligand. (B) Superposition of TAZ1 (blue) and TAZ2 (yellow) structures, represented as backbone ribbons. The proteins are aligned on the backbone heavy atoms of helices α1 and α2. The structures are rotated ≈180° relative to the view of TAZ1 in Fig. 2 to reveal the similar packing arrangements of helices α1, α2, and α3, and the similar locations of the Zn1 and Zn2 sites. The arrows indicate the location of the Zn3 site in the two structures. (C) Close-up stereoview of the Zn3 site, showing the different position of the α4-helix in the TAZ1 (blue) and TAZ2 (yellow) structures and the different locations of Zn3 (blue and yellow spheres) and the zinc-binding loops. The backbone of the αC-helix of Hif-1α in the complex with TAZ1 is shown as a pink tube to illustrate how binding to TAZ2 would be occluded by the different arrangement of Zn3 and α4.
The TAZ1 domain is roughly tetrahedral in overall shape, and its surface is highly irregular. A pronounced groove, in which the Hif-1α CAD binds, cuts through three of the surfaces. The complex is more spherical in shape. Consistent with the observed spectral changes, the Hif-1α CAD undergoes local folding transitions upon binding to the TAZ1 motif, forming three short helices designated αA, αB, and αC (Fig. 2B). The CAD remains relatively extended in the complex and has no intramolecular hydrophobic core; interactions with TAZ1 are required for stabilization of the helical secondary structure. Hif-1α wraps almost entirely around the globular TAZ1 domain, burying ≈2,400 Å (2), or approximately 40%, of the CAD surface.
Structure of the TAZ1 Domain.
The overall fold of the TAZ1 (CH1) motif is similar to that of the TAZ2 domain of CBP (16) (Fig. 3B). Helices α1, α2, and α3 are arranged similarly in the two structures, and the positions of the first two zinc atoms are nearly superimposable. However, significant local structural differences undoubtedly contribute to the differing binding specificities of the TAZ1 and TAZ2 domains of CBP/p300. The most notable differences are in the length of loop 1, between helices α1 and α2, and in the location of helix α4 and the third zinc-binding site. In TAZ1, loop 1 contains an additional 7 residues that are accommodated through an extension of helix α1 (Fig. 3B). Despite the different length and conformation of this loop, the relative orientation of the α1- and α2-helices is similar in TAZ1 and TAZ2, with helix crossing angles of 83° and 81°, respectively. In contrast, the angles between the axes of helices α2 and α3 (82° in TAZ1, 110° in TAZ2) and helices α3 and α4 (115° in TAZ1, 64° in TAZ2) are very different in the two domains. The structural differences between TAZ1 and TAZ2 are intrinsic and do not arise from conformational changes associated with complex formation; NMR spectra of TAZ1 show little change in chemical shift in the α4 and Zn3 region upon Hif-1α binding (data not shown). The interhelix crossing angle is directly related to the number of amino acid residues in the loop between the third and fourth cysteine ligands (16). Crossing angles near 80° are characteristic of zinc sites with CX4C coordination, whereas the tighter turn between the Cys ligands in the CX2C coordination motif is accommodated by moving the axis of the helix, giving rise to a more obtuse crossing angle (ca. 110°) (16). Thus, the observed differences in the α2/α3- and α3/α4-helix crossing angles in the TAZ1 and TAZ2 structures are directly attributable to differences in loop length. In addition, the C-terminal end of the α3-helix of TAZ2 contains an extra residue, inserted immediately before the histidine ligand (Fig. 3A). The net effect of these sequence differences is that the location of the Zn3 site is significantly changed, and the α4-helix is positioned on the opposite side of α3 in the TAZ1 and TAZ2 structures (Fig. 3C). As discussed later, these changes have a major influence on the binding specificity of the two TAZ motifs.
Interactions Between the Hif-1α CAD and the TAZ1 Domain.
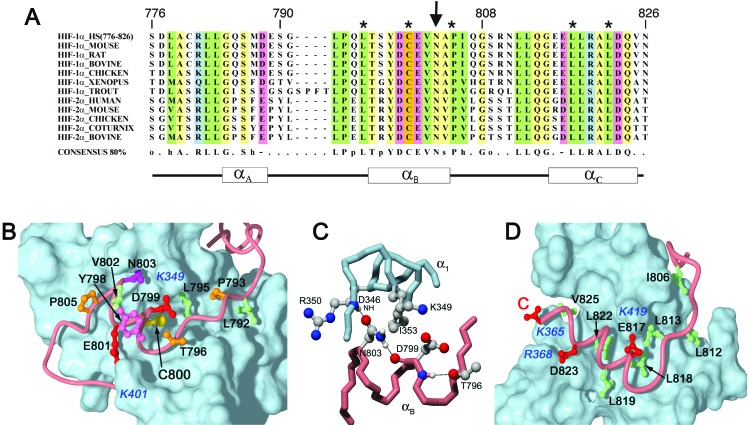
Residues 776–790 in the N-terminal region of the Hif-1α CAD are poorly defined in the structures. Nevertheless, intermolecular NOEs are observed which show that this region of the CAD does bind, albeit weakly, in a broad shallow groove formed by helices α1 and α4 of TAZ1. Residues 784–788 form a helix (αA) in most structures in the family. The intermolecular interactions in this region seem to be primarily hydrophobic, with Ala-779, Leu-783, and Met-787 of the Hif-1α CAD contacting Val-358 and Leu-359 on the TAZ1 α1-helix. Deletion of this region of the CAD impairs binding: ITC experiments show that a truncated form of the CAD (residues 790–826) binds about 5-fold more weakly than Hif-1α (776–826).
The structure of the bound Hif-1α CAD is better defined after Gly-791. The Gly-791 backbone carbonyl forms a hydrogen bond to the side-chain amide group of Gln-355 in helix α1. Residues 792–796 are highly conserved in Hif-1α and its homolog Hif-2α (Fig. 4A). These residues adopt an extended conformation and make numerous contacts with the surface of the TAZ1 domain (Fig. 4B). The side chain of Leu-795 projects into a deep hydrophobic cavity, lined by the side chains of Leu-352, Ile-353, Gln-356, and Ile-414 on helices α1 and α3 of TAZ1; substitution of Leu-795 by small or polar side chains impairs Hif-1α transcriptional activity (43). Additional hydrophobic contacts are made by Leu-792, which fits into a shallow depression at the α1/α2 interface, and by Pro-793, which fits neatly into a groove between Leu-352 and Gln-356 on helix α1 of TAZ1. Chemical shift mapping has established that the corresponding α1/α2/α3 surface of TAZ2 is involved in binding the p53 activation domain (16).
Figure 4.
(A) Alignment of Hif-1α and Hif-2α CAD sequences showing conserved residues. The asterisks indicate sites where mutations abrogate CBP/p300 binding. The arrow indicates Asn-803, which functions as a hypoxic switch. (B) Binding site for αB-helix and the preceding extended region of Hif-1α CAD (pink tube) in a groove on the surface of TAZ1 (blue). The side chain of Asn-803 is shown in magenta. The side chains of Cys-800 (yellow) and other critical residues that form the molecular interface are shown and labeled in black. The surfaces of interacting positively charged side chains from TAZ1 are labeled in blue. (C) Interactions involving Asn-803 of Hif-1α. Hydrogen bonds to the Hif-1α Asp-799 carbonyl and the TAZ1 Asp-346 backbone NH are drawn as black lines. The helix N-cap hydrogen bond formed by the Thr-796 Oγ is also shown. (D) Binding site for the amphipathic αC-helix and the extended region linking αC and αB. The hydrophobic side chains that form the molecular interface are shown in green. The position of the conserved Glu-817 and Asp-823 side chains and the carboxy terminus (labeled C) of Hif-1α are shown in red; the conserved basic residues on TAZ1 with which they interact are labeled in blue.
The extended region of the Hif-1α CAD leads directly into a short α-helix (residues 796–804), termed αB, which docks in a groove formed by the N-terminal end of α1, helix α3, and the Zn3 site of TAZ1 (Fig. 2B). Another helix, αC, is formed by residues 815–824. Helices αB and αC lie on opposite sides of helix α3 of TAZ1 (Fig. 2B); residues 804–813 form an irregular bridge over α3 and make numerous contacts with the surface of TAZ1. In particular, the conserved leucines at positions 812 and 813 (Fig. 4A) together with Ile-806 form a shallow hydrophobic groove that fits over two turns of helix α3 (Fig. 4D). The central residues (Gln-807–Asn-811) in this interhelix bridge are disordered in the present structures.
An unusual feature of the structure is that helix αB of the Hif-1α CAD is docked to TAZ1 through its more polar face, with Cys-800 and Asn-803 deeply buried in the molecular interface (Fig. 4B). This feature is consistent with mutagenesis data assigning Cys-800 and Asn-803 essential roles in CBP/p300 complex formation and hypoxic signal transduction (19, 43–45). The outer surface of the CAD helix is quite hydrophobic, and the conserved Tyr-798 and Val-802 side chains project into solvent and make little contact with TAZ1. The side-chain carboxyl groups of Asp-799 and Glu-801 are in close contact with K349 and K401, respectively. The helix is capped at the N terminus by the Oγ of Thr-796, which hydrogen bonds to the backbone NH of Asp-799. The invariant Pro-805 most likely functions as a helix stop signal.
Helix αC binds in a deep hydrophobic groove, with the charged carboxy terminus positioned between Lys-365 and Arg-368 on helix α1 of TAZ1 (Fig. 4D). The invariant Leu-818, Leu-819, and Leu-822 side chains form the hydrophobic face of αC and are docked deeply within the groove. Leu-818 and Leu-822 are critical for binding, and mutation of either of these residues impairs or abrogates transcriptional activation (43). Val-825 also makes hydrophobic contacts with TAZ1. A small side chain is required at residue 821 (invariant alanine) because of steric constraints imposed by the proximity of α3 of TAZ1, which forms the wall of the hydrophobic groove. Steric clash at the C-terminal end of α2, between the Cys and His ligands of Zn2, means that no side chain can be accommodated at residue 815; glycine is found at this position in all available sequences (Fig. 4A). The solvent-exposed surface of the CAD αC-helix is highly polar, and complementary electrostatic interactions occur with charged side chains on TAZ1 (Fig. 4D); the conserved Glu-817 and Asp-823 of Hif-1α are in close proximity to Lys-419 and Arg-368, respectively, in the CBP TAZ1 domain. Despite the critical contacts provided by helix αC, it binds TAZ1 only weakly (Kd > 500 μM) for Hif-1α (808–826) in the absence of stabilizing contacts involving other regions of the CAD.
The hydrophobic groove that binds the αC-helix of Hif-1α is absent from the TAZ2 structure (16), because of the shorter α1-helix and different packing arrangement of α4. The Zn3-binding loop in TAZ2 docks in the position occupied by the αC-helix of Hif-1α in the complex with TAZ1 (Fig. 3C). The location of the Zn3-binding loop and the α4-helix of TAZ2 would preclude binding of the αC-helix of the Hif-1α CAD. Thus, the pronounced differences in local structure in the Zn3 region are directly responsible for the changes in surface topology (i.e., the presence or absence of a hydrophobic groove) that impart different binding specificities to the TAZ1 and TAZ2 domains of CBP.
Structural Basis for the Function of Asn-803 as a Hypoxic Switch.
The conserved Asn-803 functions as a hypoxic switch; hydroxylation of Asn-803 under normoxic conditions results in abrogation of binding of the Hif-1α CAD to CBP (19). Asn-803 is located on the αB-helix and is deeply buried in the protein–protein interface, packed against Ile-353 and the hydrophobic part of the Lys-349 side chain (Fig. 4B). It also forms a network of side-chain hydrogen-bonding interactions that probably play an important role in stabilization of the αA-helix and of the complex (Fig. 4C). The Asn-803 side-chain carboxamide group is hydrogen-bonded to the Asp-799 backbone carbonyl through the Nδ2 atom and to the Asp-346 backbone NH through Oδ1. The site of Asn-803 hydroxylation under normoxic conditions is not yet known. Hydroxylation commonly occurs at the β-carbon, to yield the erythro isomer (46); although β-hydroxyasparagine can be accommodated in the molecular interface, introduction of a β-branched side chain would be expected to destabilize the αA-helix. In addition, the cost of desolvating the hydroxyl group could also disfavor complex formation. On the other hand, hydroxylation of the side-chain amide nitrogen could not be accommodated without disruption of both the side-chain hydrogen-bonding network and the protein–protein interface.
Conclusion
The structure of the complex of the Hif-1α CAD with the TAZ1 domain of CBP is of interest not only for the insights it provides into the molecular basis of the hypoxic response, but also as a potential target for design of antitumor agents (3, 4, 42). Hypoxia plays an important role in tumor progression and metastasis. Tumor growth leads to hypoxia, which in turn results in decreased therapeutic efficacy, stimulation of angiogenesis, and tumor progression (42); overexpression of Hif-1α has been observed in many common human cancers and is correlated with aberrant p53 accumulation and tumor progression (4). In mice, tumor growth is attenuated by disruption of the Hif-1α/p300 interaction (42). Our structure now reveals the details of the molecular interface between Hif-1α and the CBP/p300 TAZ1 domain and provides a basis for design of inhibitors. It is notable that the interface involves almost the entire Hif-1α CAD, and that high-affinity binding depends on synergistic interactions involving several Hif-1α structural elements. This involvement is probably a consequence of the coupled folding and binding transition undergone by the Hif-1α activation domain.
Acknowledgments
We thank Mindy Landes for yeast two-hybrid assays, Melissa Allen, Ted Foss, Linda Tennant, and Leonard Kaljevic for invaluable technical assistance, John Chung and Gerard Kroon for assistance with NMR data collection, David Case and Micah Gearhart for advice on the structure calculations, and Lucia Pappalardo for contributions in the early stages of this project. This work was supported by grants from the National Institutes of Health and by the Skaggs Institute for Chemical Biology. S.A.D. was supported by a postdoctoral fellowship from the Swiss National Science Foundation and the Novartis Foundation. R.N.D. is supported by a postdoctoral fellowship from the Cancer Research Institute. Plasmids containing the mouse CBP and human Hif-1α coding sequences were kind gifts of Marc Montminy and Gregg Semenza.
Abbreviations
- HIF-1
hypoxia-inducible factor 1
- CAD
carboxy-terminal activation domain of Hif-1α
- CBP
CREB (cyclic-AMP response element binding protein) binding protein
- TAZ
transcriptional adapter zinc-binding motif
- CH1
cysteine/histidine-rich domain-1 of CBP (equivalent to TAZ1 domain)
- CH3
cysteine/histidine-rich domain-3 of CBP (zz domain + TAZ2 domain)
- ITC
isothermal titration calorimetry
- NOE
nuclear Overhauser effect
Footnotes
Data deposition: Chemical shift assignments have been deposited in the BioMagResBank (accession no. 5327), and structural coordinates in the Protein Data Bank, www.rcsb.org (accession no. 1L8C).
References
- 1.Wang G L, Jiang B, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza G L. Genes Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- 3.Blagosklonny M V. Int J Oncol. 2001;19:257–262. doi: 10.3892/ijo.19.2.257. [DOI] [PubMed] [Google Scholar]
- 4.Zhong H, De Marzo A M, Laughner E, Lim M, Hilton D A, Zagzag D, Buechler P, Isaacs W B, Semenza G L, Simons J W. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 5.Iyer N V, Kotch L E, Agani F, Leung S W, Laughner E, Wenger R H, Gassmann M, Gearhart J D, Lawler A M, Yu A Y, Semenza G L. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza G L. Trends Mol Med. 2001;7:345–350. doi: 10.1016/s1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- 7.Semenza G L. Cell. 2001;107:1–3. doi: 10.1016/s0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 8.Jaakkola P, Mole D R, Tian Y M, Wilson M I, Gielbert J, Gaskell S J, Kriegsheim A, Hebestreit H F, Mukherji M, Schofield C J, et al. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 9.Ivan M, Kondo K, Yang H F, Kim W, Valiando J, Ohh M, Salic A, Asara J M, Lane W S, Kaelin W G., Jr Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 10.Yu F, White S B, Zhao Q, Lee F S. Proc Natl Acad Sci USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giordano A, Avantaggiati M L. J Cell Phys. 1999;181:218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 12.Blobel G A. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 13.Goodman R H, Smolik S. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 14.Ponting C P, Blake D J, Davies K E, Kendrick-Jones J, Winder S J. Trends Biochem Sci. 1996;21:11–13. [PubMed] [Google Scholar]
- 15.Shikama N, Lyon J, La Thangue N B. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01149-5. [DOI] [PubMed] [Google Scholar]
- 16.De Guzman R N, Liu H Y, Martinez-Yamout M, Dyson H J, Wright P E. J Mol Biol. 2000;303:243–253. doi: 10.1006/jmbi.2000.4141. [DOI] [PubMed] [Google Scholar]
- 17.Kallio P J, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, Poellinger L. EMBO J. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arany Z, Huang L E, Eckner R, Bhattacharya S, Jiang C, Goldberg M A, Bunn H F, Livingston D M. Proc Natl Acad Sci USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lando D, Peet D J, Whelan D A, Gorman J J, Whitelaw M L. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 20.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharya S, Michels C L, Leung M K, Arany Z P, Kung A L, Livingston D M. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida E, Nakajima T, Murakami K, Fukamizu A. Gene. 1998;208:307–314. doi: 10.1016/s0378-1119(98)00008-0. [DOI] [PubMed] [Google Scholar]
- 23.Demarest S J, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson H J, Evans R M, Wright P E. Nature (London) 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister G W, Guang Z, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Johnson B A, Blevins R A. J Chem Phys. 1994;29:1012–1014. [Google Scholar]
- 26.Wittekind M, Mueller L. J Magn Reson. 1993;101:201–205. [Google Scholar]
- 27.Grzesiek S, Bax A. J Magn Reson. 1992;96:432–440. [Google Scholar]
- 28.Grzesiek S, Bax A. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 29.Grzesiek S, Anglister J, Bax A. J Magn Reson Series B. 1993;101:114–119. [Google Scholar]
- 30.Bax A, Clore G M, Gronenborn A M. J Magn Reson. 1990;88:425–431. [Google Scholar]
- 31.Grzesiek S, Kuboniwa H, Hinck A P, Bax A. J Am Chem Soc. 1995;117:5312–5315. [Google Scholar]
- 32.Vuister G W, Bax A. J Biomol NMR. 1992;2:401–405. doi: 10.1007/BF01874818. [DOI] [PubMed] [Google Scholar]
- 33.Zwahlen C, Legault P, Vincent S J F, Greenblatt J, Konrat R, Kay L E. J Am Chem Soc. 1997;119:6711–6721. [Google Scholar]
- 34.Duggan B M, Legge G B, Dyson H J, Wright P E. J Biomol NMR. 2001;19:321–329. doi: 10.1023/a:1011227824104. [DOI] [PubMed] [Google Scholar]
- 35.Wishart D S, Sykes B D. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 36.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 37.Pearlman D A, Case D A, Caldwell J W, Ross W S, Cheatham T E, III, DeBolt S, Ferguson D, Seibel G, Kollman P. Comp Phys Commun. 1995;91:1–41. [Google Scholar]
- 38.Tsui V, Case D A. J Am Chem Soc. 2000;122:2489–2498. [Google Scholar]
- 39. Xia, B., Tsui, V., Case, D. A., Dyson, H. J. & Wright, P. E. (2002) J. Biomol. NMR, in press. [DOI] [PubMed]
- 40.Laskowski R A, Rullmann J A C, MacArthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 41.Pugh C W, O'Rourke J F, Nagao M, Gleadle J M, Ratcliffe P J. J Biol Chem. 1997;272:11205–11214. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- 42.Kung A L, Wang S, Klco J M, Kaelin W G, Livingston D M. Nat Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]
- 43.Gu J, Milligan J, Huang L E. J Biol Chem. 2001;276:3550–3554. doi: 10.1074/jbc.M009522200. [DOI] [PubMed] [Google Scholar]
- 44.O'Rourke J F, Tian Y M, Ratcliffe P J, Pugh C W. J Biol Chem. 1999;274:2060–2071. doi: 10.1074/jbc.274.4.2060. [DOI] [PubMed] [Google Scholar]
- 45.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenflo J, Lundwall A, Dahlback B. Proc Natl Acad Sci USA. 1987;84:368–372. doi: 10.1073/pnas.84.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]