Abstract
Type III polyketide synthases (PKS) generate an array of natural products by condensing multiple acetyl units derived from malonyl-CoA to thioester-linked starter molecules covalently bound in the PKS active site. One strategy adopted by Nature for increasing the functional diversity of these biosynthetic enzymes involves modifying polyketide assembly by altering the preference for starter molecules. Chalcone synthase (CHS) is a ubiquitous plant PKS and the first type III PKS described functionally and structurally. Guided by the three-dimensional structure of CHS, Phe-215 and Phe-265, which are situated at the active site entrance, were targeted for site-directed mutagenesis to diversify CHS activity. The resulting mutants were screened against a panel of aliphatic and aromatic CoA-linked starter molecules to evaluate the degree of starter molecule specificity in CHS. Although wild-type CHS accepts a number of natural CoA thioesters, it does not use N-methylanthraniloyl-CoA as a substrate. Substitution of Phe-215 by serine yields a CHS mutant that preferentially accepts this CoA-thioester substrate to generate a novel alkaloid, namely N-methylanthraniloyltriacetic acid lactone. These results demonstrate that a point mutation in CHS dramatically shifts the molecular selectivity of this enzyme. This structure-based approach to metabolic redesign represents an initial step toward tailoring the biosynthetic activity of plant type III PKS.
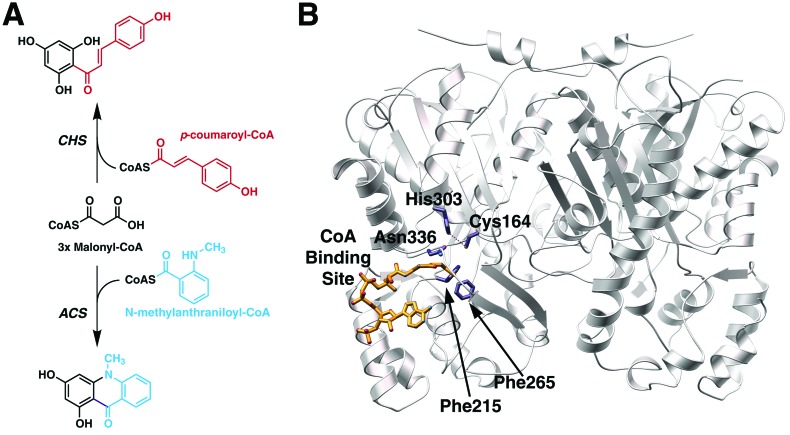
Plant anthocyanin floral pigments, anti-microbial phytoalexins, and regulators of Rhizobium nodulation genes derive from polyketides formed by the condensation of multiple acetyl units to a specific starter molecule (1). Chalcone synthase (CHS, EC 2.3.1.74) and other type III polyketide synthases (PKS), including acridone synthase (ACS), catalyze the formation of enzyme-bound polyketide intermediates that undergo regio-specific cyclization reactions yielding unique products (2). CHS uses p-coumaroyl-CoA as a starter molecule and three malonyl-CoAs to form a tetraketide intermediate that undergoes cyclization to 4,2′,4′,6′-tetrahydroxychalcone (chalcone), whereas, ACS uses N-methylanthraniloyl-CoA as a starter molecule together with three malonyl-CoAs to produce 1,3-dihydroxy-N-methylacridone (acridone; Fig. 1A). Unlike the larger 6-deoxyerythronolide and actinorhodin PKS of bacterial origin (3), the type III PKS are homodimeric enzymes that orchestrate a series of acyltransferase, decarboxylation, condensation, cyclization, and aromatization reactions at two functionally independent active sites (4). Crystallographic and protein engineering studies on Medicago sativa (alfalfa) CHS2 and Gerbera hybrida 2-pyrone synthase demonstrate that the structure of the type III PKS active site dictates starter molecule preference, polyketide chain-length, and regio-specificity of polyketide cyclization (5–7). This architecturally simple molecular organization forms an attractive three-dimensional scaffold both for understanding the structural basis for rapid diversification of biosynthetic activities in plants and for guiding structure-assisted engineering experiments aimed at altering the biosynthetic activities of these PKS (8, 9).
Figure 1.
Overview of plant type III polyketide synthases. (A) Reactions catalyzed by CHS and ACS. Only the starter molecules are shown. Both enzymes use three molecules of malonyl-CoA during the elongation of the starter molecule. Chalcone is on top and acridone is on the bottom. (B) Ribbon diagram of the CHS homodimer showing the catalytic residues (Cys-164, His-303, and Asn-336) and the two phenylalanines (Phe-215 and Phe-265) at the boundary between the CoA (gold) binding site and the active site of one monomer. Figure prepared with molscript (35) and pov-ray [POV-Team (1997) pov-ray, Persistence of Vision Ray-Tracer; http://www.povray.org].
The catalytic residues of CHS sit at the intersection of the CoA binding tunnel and a large internal cavity that accommodates the growing polyketide chain (ref. 5; Fig. 1B). Mechanistic and crystallographic studies confirm the importance of Cys-164 as the polyketide attachment site and His-303 and Asn-336 as key catalytic residues during decarboxylation of malonyl-CoA and polyketide chain extension (10–12). Conservation of these residues among all type III PKS implies that these enzymes use a core set of reactions for starter molecule loading, malonyl-CoA decarboxylation, and polyketide chain elongation (11, 13).
In vitro, CHS accepts disparate CoA starter molecules, including aromatic and aliphatic CoA-thioesters of different lengths (14–18). A tunnel extending from the exterior of CHS to an interior cavity provides access to the buried catalytic residues. Two phenylalanines (Phe-215 and Phe-265) sit at the juncture of the CoA binding tunnel and the entrance to the active site (Fig. 1B). Phe-215 is conserved in all type III PKS except for benzalacetone synthase, which has a leucine at this position and catalyzes the condensation of a single acetate unit to p-coumaroyl-CoA (19). Given the different starter molecule preferences of type III PKS, the identity of residue 215 in PKS sequences does not appear at first glance to influence starter molecule selection. On the other hand, residue 265 is a phenylalanine in CHS and almost all type III PKS, but is valine in both ACS isoforms from Ruta graveolens (20, 21). This difference in CHS and ACS suggests that Phe-265 may play a role in determining starter molecule specificity.
Here, we examine the role of these two phenylalanines on starter molecule selectivity. Contrary to our expectations, the F265V mutant does not alter selectivity for starter molecules whereas the F215S mutant of CHS preferentially uses N-methylanthraniloyl-CoA as a starter molecule to yield a tetraketide lactone. To date, this natural variant has not been reported. Structural analysis of the F215S mutant suggests that widening the active site entrance allows the productive binding of the shorter and bulkier N-methylanthraniloyl-CoA. The success of this structure-guided approach indicates that specific point mutations selected for proximity to the PKS catalytic machinery can expand the biosynthetic repertoire of type III PKS. This initial step toward tailoring the biosynthetic activity of CHS suggests that further modification of polyketide assembly may be achieved through either rational mutagenesis or directed randomization of residues on or near the catalytic cavity.
Materials and Methods
Mutant Generation, Protein Expression, Protein Purification, and CoA-Thioester Synthesis.
Site-directed mutants were generated by using the QuikChange (Stratagene) protocol. R. graveolens ACS2 cDNA was provided by R. Lukacin (Philipps-Universität, Marburg, Germany). ACS was PCR-amplified from the ACS2-pTZ19R vector (20) and subcloned into the pHIS8 expression vector (11). All recombinant proteins were expressed in Escherichia coli BL21 (DE3) and purified to homogeneity as previously described (11). Synthesis of p-coumaroyl-CoA, m-coumaroyl-CoA, o-coumaroyl-CoA, 3-phenylpropionyl-CoA, cinnamoyl-CoA, 4-methylcinnamoyl-CoA, feruloyl-CoA, and N-methylanthraniloyl-CoA used the method of Stoeckigt and Zenk (22). Electrospray mass spectrometry confirmed the identity of the reaction products.
Enzyme Assays.
Standard assay conditions for screening were 100 mM potassium phosphate (pH 7.0), 30 μM [2-14C]malonyl-CoA (50,000 cpm), and 15 μM starter CoA in a 100-μl reaction volume at 25°C. Reactions were quenched with 5% (vol/vol) acetic acid and extracted with ethyl acetate. Steady-state kinetic constants were determined from initial velocity measurements (11). Inhibition of wild-type CHS and the F265V mutant was investigated by using the standard assay system with increasing concentrations of N-methylanthraniloyl-CoA (0–162 μM).
Liquid-Chromatography Coupled Mass-Spectrometry (LC/MS/MS) Product Analysis.
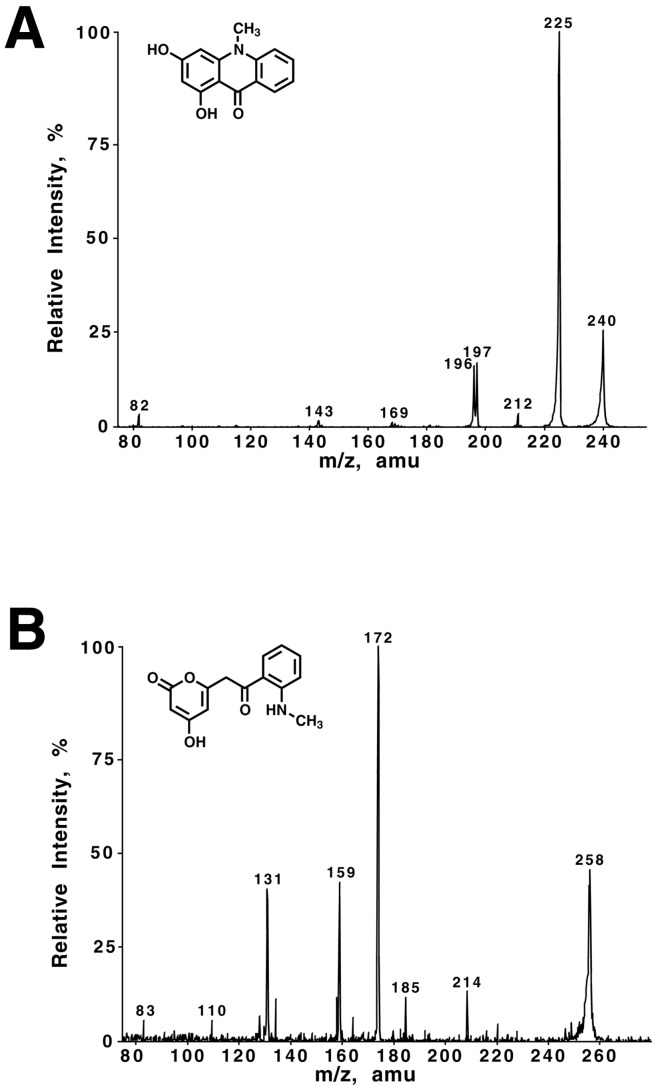
Assays (5-ml reaction volume; 100 μg protein) were performed under standard conditions with 50 μM starter-CoA and 120 μM malonyl-CoA. Reactions were quenched with 5% (vol/vol) acetic acid and extracted with ethyl acetate. Product analysis was performed in the mass spectrometry facility of the Scripps Research Institute. Extracts were analyzed on a Hewlett–Packard HP1100 MSD single quadrupole mass spectrometer coupled to a TSK-Gel ODS-80TS (TosoHaas, Montgomeryville, PA) column (5 μm, 4.6 mm × 150 mm). HPLC conditions were as follows: gradient from 30 to 70% (vol/vol) methanol both in 0.2% (vol/vol) acetic acid over 30 min; flow rate 0.8 ml⋅min−1. See Supporting Text (which is published as supporting information on the PNAS web site, www.pnas.org) for fragmentation data. ACS major product with N-methylanthraniloyl-CoA: acridone, [M-H]− 240(28); [M-H-CH3]− 225(100); 212(3); 197(14). F215S major product with N-methylanthraniloyl-CoA: tetraketide lactone, [M-H]− 258(47); [M-H-CO2]− 214(11); 185(10); [M-H-CO2-CH2-CO]− 172(100); 159(39); 131(37).
Crystallography.
Crystals of the F215S mutant (P3221; a = 97.81 Å; c = 65.73 Å) were obtained under the same conditions used for wild-type CHS (5). Diffraction data were collected at 105 K at beamline 7-1 (1.07 Å wavelength) of the Stanford Synchrotron Radiation Laboratory on a 34.5-cm MAR imaging plate. Data processing was conducted with denzo/scalepack (23). The structure of the F215S mutant was determined by the difference Fourier method with refmac (24). The starting model was wild-type CHS minus the side-chain of Phe-215. Inspection of the electron density maps and model building were performed with o (25). Water molecules were introduced with Automated Refinement Procedure (ARP; ref. 26). Refinement converged to the R factors shown in Table 1.
Table 1.
Data collection and refinement statistics for the CHS F215S mutant
| Resolution, Å | 26.0–1.63 |
| Total reflections/unique reflections | 226,775/43,804 |
| Completeness of data (highest shell), % | 95.9 (81.6) |
| I/σ (highest shell) | 27.5 (6.1) |
| Rsym* (highest shell), % | 3.5 (12.0) |
| Rcryst†/Rfree,‡ % | 19.0/22.5 |
| No. of protein atoms | 3,108 |
| No. of water molecules | 347 |
| Rmsd–ideal bond lengths, Å | 0.013 |
| Rmsd–ideal bond angles, deg | 2.1 |
| Avg. B-factor–protein, Å2 | 18.5 |
| Avg. B-factor–solvent, Å2 | 28.8 |
Rsym = ∑|Ih − 〈Ih〉|/∑Ih, where 〈Ih〉 is the average intensity over symmetry equivalent reflections.
Rcryst = ∑|Fobs − Fcalc|/∑Fobs, where summation is over the data used for refinement.
Rfree is the same definition as for Rcryst, but includes only 5% of data excluded from refinement.
Results
Functional Screen of CHS Starter Molecule Specificity.
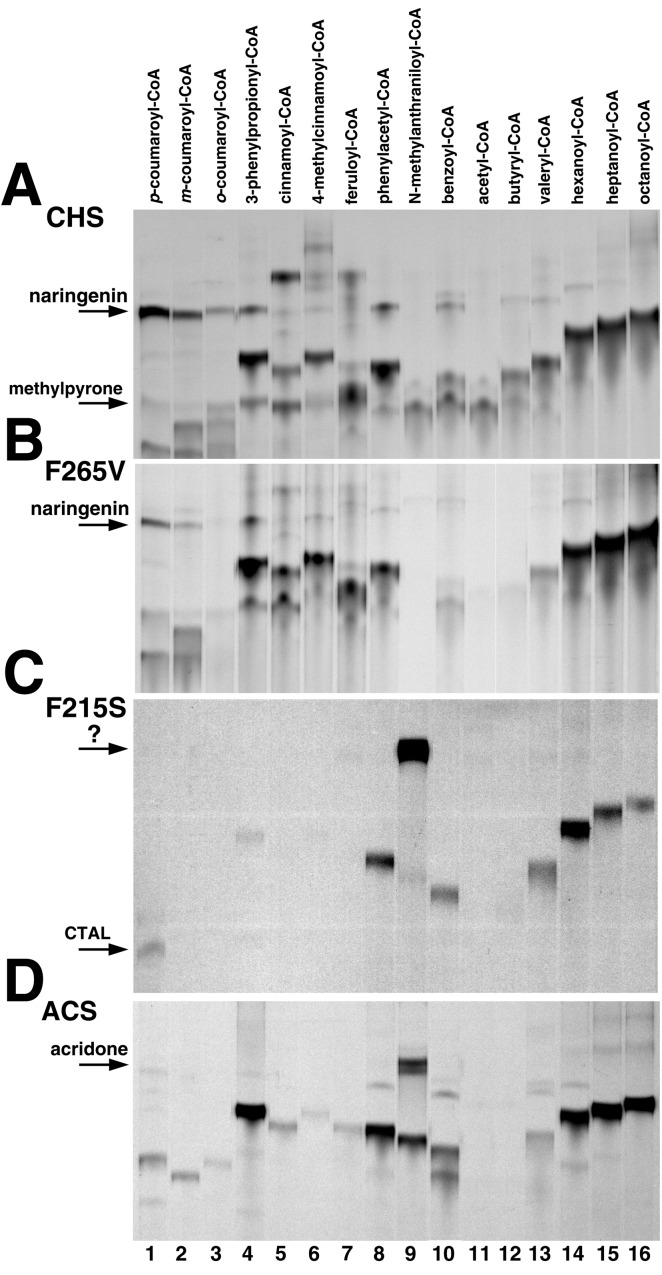
We first identified a CoA-thioester not used by the wild-type enzyme as a substrate. By screening a panel of sixteen aromatic and aliphatic CoA-thioesters, we qualitatively and quantitatively examined the substrate tolerance of wild-type CHS and showed that N-methylanthraniloyl-CoA was the only CoA-thioester not accepted as a starter (Fig. 2A). Analysis by LC/MS/MS verified that reactions with p-coumaroyl-CoA, m-coumaroyl-CoA, o-coumaroyl-CoA, and cinnamoyl-CoA yield the corresponding flavanones after spontaneous cyclization of the chalcones produced by CHS. The major product of the 3-phenylpropionyl-CoA reaction is a chalcone-like compound lacking the α,β-unsaturated double bond. Use of 4-methylcinnamoyl-CoA produces a triketide styrylpyrone. With feruloyl-CoA, benzoyl-CoA, phenylacetyl-CoA, and aliphatic CoA-thioesters, CHS generates tetraketide and triketide lactones (7, 17). The kcat/Km values for different starter molecules confirm that alfalfa CHS2 prefers cinnamoyl-derived CoAs and aliphatic CoAs of 6–8 carbons (Table 2). In the presence of N-methylanthraniloyl-CoA, CHS produces methylpyrone solely from malonyl-CoA (6), as confirmed by LC/MS/MS (see supporting information). UV/Vis spectroscopy did not indicate the presence of the N-methylanthraniloyl-group in the product (not shown). Overnight incubation of wild-type CHS (100 μg) also did not yield a product incorporating the N-methylanthraniloyl-CoA starter molecule. Given the detection limit of the assay (≈5 pmol radiolabeled product), any potential activity of CHS with this CoA-thioester would be 105-fold less than that with p-coumaroyl-CoA.
Figure 2.
Thin-layer chromatography screen for activity with different starters. By using standard assay conditions, 10 μg of either wild-type CHS (A), F265V mutant (B), F215S mutant (C), or ACS (D) was incubated for 30 min with various starters and [2-14C]malonyl-CoA. The starter molecule for each reaction is indicated above the lanes. The position of naringenin in lane 1 is indicated for CHS and the F265V mutant. The positions of p-coumaroyltriacetic acid lactone in lane 1 and the new product in lane 9 are indicated for the F215S mutant. The position of acridone in lane 9 is indicated for ACS.
Table 2.
Steady-state kinetic parameters
|
CHS
|
F265V
|
F215S
|
ACS
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat | Km | kcat/Km | kcat | Km | kcat/Km | kcat | Km | kcat/Km | kcat | Km | kcat/Km | |
| p-Coumaroyl-CoA | 5.14 | 6.1 | 14,043 | 2.01 | 5.0 | 6,700 | 0.02 | 4.0 | 83 | 0.02 | 16.3 | 20.4 |
| m-Coumaroyl-CoA | 3.50 | 4.5 | 12,963 | 1.67 | 5.5 | 5,060 | — | — | — | 0.08 | 14.7 | 89.6 |
| o-Coumaroyl-CoA | 2.32 | 3.8 | 10,175 | 0.47 | 4.0 | 1,958 | — | — | — | 0.02 | 20.5 | 19.5 |
| 3-Phenylpropionyl-CoA | 4.95 | 4.9 | 16,836 | 4.40 | 13.8 | 5,314 | — | — | — | 1.62 | 30.0 | 900 |
| Cinnamoyl-CoA | 4.76 | 5.0 | 15,866 | 1.67 | 17.1 | 1,628 | — | — | — | 0.07 | 10.2 | 106 |
| 4-Methylcinnamoyl-CoA | 1.57 | 3.4 | 7,696 | 0.82 | 7.4 | 1,846 | — | — | — | 0.01 | 9.5 | 24.6 |
| Feruloyl-CoA | 1.04 | 5.2 | 3,333 | 2.43 | 9.0 | 4,500 | — | — | — | 0.01 | 16.7 | 15.0 |
| Phenylacetyl-CoA | 2.17 | 5.1 | 7,092 | 4.37 | 5.7 | 12,778 | 0.04 | 4.5 | 145 | 0.59 | 4.9 | 2,010 |
| N-methylanthraniloyl-CoA | — | — | — | — | — | — | 0.09 | 3.0 | 500 | 0.68 | 29.4 | 386 |
| Benzoyl-CoA | 1.73 | 2.2 | 13,106 | 1.28 | 4.7 | 7,907 | 0.04 | 3.8 | 175 | 0.55 | 15.6 | 590 |
| Acetyl-CoA | 0.19 | 8.4 | 376 | 0.39 | 8.6 | 755 | — | — | — | — | — | — |
| Butyryl-CoA | 0.91 | 4.2 | 3,611 | 0.56 | 5.2 | 1,794 | — | — | — | — | — | — |
| Valeryl-CoA | 1.86 | 4.5 | 6,889 | 1.33 | 6.1 | 3,634 | 0.04 | 4.5 | 151 | 0.11 | 9.9 | 180 |
| Hexanoyl-CoA | 2.52 | 4.1 | 10,243 | 3.56 | 5.0 | 11,866 | 0.07 | 4.5 | 272 | 1.59 | 3.8 | 6,970 |
| Heptanoyl-CoA | 3.03 | 4.0 | 12,625 | 6.58 | 7.8 | 14,060 | 0.05 | 5.4 | 154 | 1.06 | 4.4 | 4,020 |
| Octanoyl-CoA | 2.74 | 3.8 | 12,017 | 9.82 | 5.6 | 29,226 | 0.04 | 4.8 | 150 | 0.78 | 3.9 | 3,340 |
Values for kcat, Km, and kcat/Km are min−1, μM, M−1⋅s−1, respectively. Values shown are means (n = 3) with standard errors less than 15%.
Functional Screen for Starter Molecule Specificity of the F265V and F215S Mutants.
Based on sequence comparison with ACS, we predicted that the F265V mutant would accept N-methylanthraniloyl-CoA as a starter molecule. Conversely, because Phe-215 is conserved in nearly all type III PKS, even those with starter molecule specificity that differs from CHS, we predicted that mutation of this residue would not alter starter choice. Therefore, we chose a previously generated mutation of Phe-215 (F215S; ref. 11) as a starting point for comparative examination of substrate selectivity.
Unexpectedly, the F265V mutant accepted each CoA-thioester tested except N-methylanthraniloyl-CoA and yielded a product profile similar to wild-type CHS (Fig. 2B). Substitution of a valine for Phe-265 causes 2- to 3-fold changes in kcat/Km for most of the CoAs tested, although the kcat/Km values for o-coumaroyl-CoA and cinnamoyl-CoA were 5- to 10-fold lower, respectively (Table 2). As with wild-type CHS, overnight incubations with the F265V mutant did not yield any N-methylanthraniloyl-CoA-derived products.
Surprisingly, replacement of Phe-215 with a serine, which lowers the malonyl-CoA decarboxylation activity of CHS (11), does alter starter molecule specificity (Fig. 2C and Table 2). With p-coumaroyl-CoA, the F215S mutant exhibits a 170-fold reduction in kcat/Km compared with wild-type CHS and forms p-coumaroyltriacetic acid lactone as the major tetraketide product instead of chalcone. Other cinnamoyl-derived CoA-thioesters are poor starter molecules for the F215S mutant. This mutant also accepts benzoyl-CoA, phenylacetyl-CoA, and 5–8 carbon length aliphatic CoA-thioesters with 1–3% the catalytic efficiency of wild-type CHS. Most notably, the F215S mutant employs N-methylanthraniloyl-CoA as a starter molecule to generate a product not observed with either wild-type CHS, the F265V mutant, or wild-type ACS. Omission of N-methylanthraniloyl-CoA from the assay mixture with CHS F215S yielded no detectable reaction products.
Activity Comparison of ACS and the CHS F215S Mutant.
For comparison to the CHS F215S mutant, R. graveolens ACS2 was screened against the sixteen CoAs (Fig. 2D). ACS produces acridone from N-methylanthraniloyl-CoA and three malonyl-CoAs via a Claisen-condensation with kcat and Km values (Table 2) similar to those reported elsewhere (20, 21, 27). LC/MS/MS analysis (Fig. 3A) confirms that acridone (λmax = 394 nm) is the major reaction product of ACS with the observed fragmentation pattern matching literature values (28). 3-Phenylpropionyl-CoA, phenylacetyl-CoA, benzoyl-CoA, hexanoyl-CoA, heptanoyl-CoA, and octanoyl-CoA are efficient starter molecules for ACS, with kcat/Km values better than that of the physiological substrate, but short aliphatic CoA-thioesters and the cinnamoyl-derived aromatic CoA-thioesters are poor starter molecules (Table 2). For example, the catalytic efficiency of ACS with p-coumaroyl-CoA is 700-fold lower than that of CHS with the same substrate. The 20-fold kcat/Km preference of ACS for N-methylanthraniloyl-CoA over p-coumaroyl-CoA mirrors the reported specific activities of the enzyme (20, 21).
Figure 3.
LC/MS/MS analysis of the major reaction products of ACS and CHS F215S. (A) Fragmentation pattern and chemical structure of the major product acridone obtained by using N-methylanthraniloyl-CoA and malonyl-CoA as substrates in a large scale reaction with ACS. (B). The analogous LC/MS/MS analysis as carried out in A but now using the CHS F215S mutant.
Kinetic characterization of the CHS F215S mutant demonstrates that it accepts N-methylanthraniloyl-CoA with a kcat/Km comparable to that of ACS (Table 2). Comparison of the catalytic efficiencies of the F215S mutant for N-methylanthraniloyl-CoA and p-coumaroyl-CoA shows a 6-fold preference for the ACS starter molecule. Benzoyl-CoA, phenylacetyl-CoA, and aliphatic CoAs longer than five carbons are slightly less efficient starter molecules than N-methylanthraniloyl-CoA. LC/MS/MS analysis of the N-methylanthraniloyl-CoA reaction with the F215S mutant indicates that the major product (λmax = 392 nm) corresponds to N-methylanthraniloyltriacetic acid lactone (Fig. 3B). Thus, although the F215S mutant accepts a new starter molecule of natural origin, the regio-specific Claisen cyclization reaction that ultimately leads to acridone formation does not occur in this mutant.
To evaluate the possibility that wild-type CHS and the F265V mutant bind N-methylanthraniloyl-CoA in a catalytically unproductive manner, the enzymatic activity of CHS and the F265V mutant were determined in the presence of increasing concentrations of N-methylanthraniloyl-CoA. N-methylanthraniloyl-CoA inhibited chalcone formation of wild-type CHS and the F265V mutant with IC50 values of 22.5 ± 0.3 μM and 3.8 ± 0.5 μM, respectively, demonstrating that this CoA-thioester binds unproductively to both wild-type CHS and the F265V mutant.
Structural Analysis of the F215S Mutant.
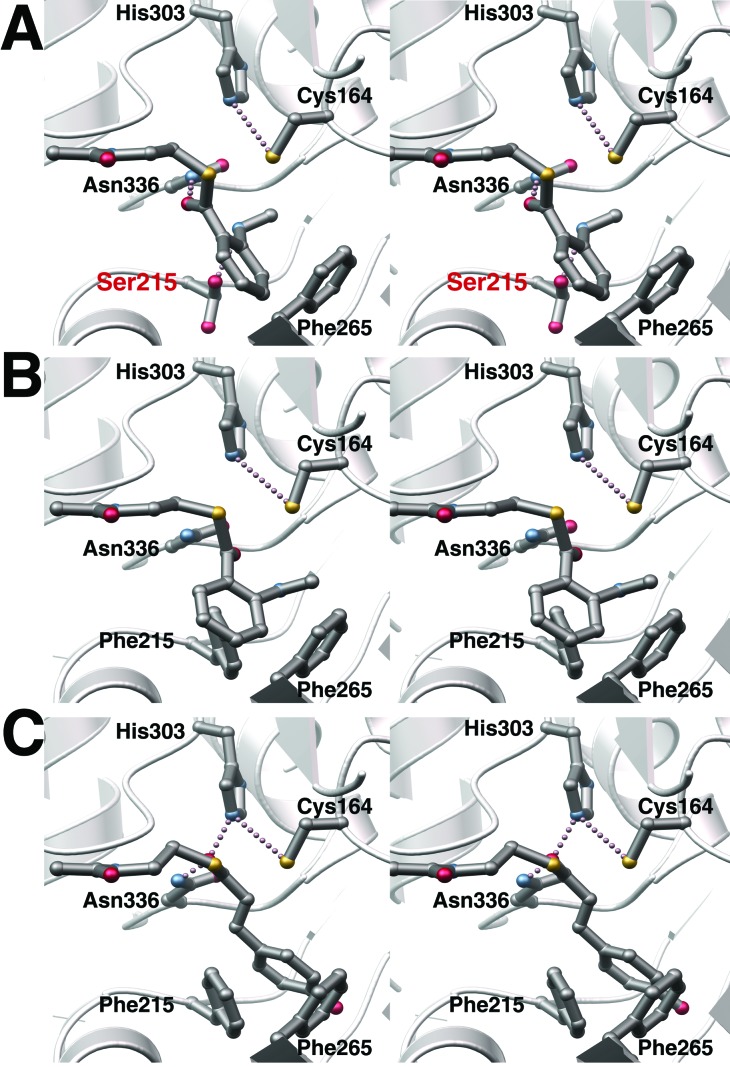
The ability of the F215S mutant, but not wild-type CHS, to use N-methylanthraniloyl-CoA as a starter molecule suggested that the three-dimensional structure of the F215S mutant might provide a structural explanation for the observed difference in substrate selectivity. The x-ray crystal structure of the mutant confirms that the overall three-dimensional structure resembles wild-type CHS (0.17 Å rms deviation of the Cα-atoms). Within the active site cavity, the catalytic residues (Cys-164, His-303, and Asn-336) maintain nearly identical positions as in the wild-type structure (Fig. 4A). The F215S mutant structure confirms a serine substitution at position 215 and shows the serine side-chain in two distinct conformations. In one rotamer, the Cβ carbon and the terminal hydroxyl group of Ser-215 are isosteric with the side-chain Cβ and Cγ carbons of Phe-215 in the wild-type CHS structure. In the second rotamer, the serine hydroxyl group rotates nearly 180o away and now forms a hydrogen bond (2.7 Å) with the backbone carbonyl oxygen of Gly-211. In this later conformation, the hydrogen bond linking the coumaroyl-derived portion of chalcone with CHS would be interrupted (5). Sterically, in both conformations, introduction of a serine at position 215 widens the entrance to the active site by 4–5 Å and positions a new hydrogen bond donor-acceptor moiety near the catalytic machinery (Fig. 4A). Attempts to determine the structure of either wild-type or mutant CHS with a starter molecule bound have thus far proved unsuccessful. Therefore, we undertook a modeling approach to explain the functional results with the F215S mutant.
Figure 4.
Structure of the F215S mutant active site and model of starter molecule binding. (A) Stereo-view illustrates the active site of the F215S mutant, including the catalytic residues (Cys-164, His-303, and Asn-336) and Phe-265. Both conformers of Ser-215 are shown. The CoA-thioester extends from the left side of the view. N-methylanthraniloyl-CoA has been modeled by applying the structural restraints discussed in the text. (B) Stereo-view of the wild-type CHS active site with N-methylanthraniloyl-CoA modeled at the entrance. Steric clashes with Phe-215 prevent the CoA-thioester from adopting the conformation depicted in A. (C) Stereo-view of the wild-type CHS active site with p-coumaroyl-CoA modeled at the entrance highlighting the ability of the propanoid moiety to extend the phenolic ring deep into the active site.
The structures of CHS complexed with CoA, acetyl-CoA, and hexanoyl-CoA define the overall placement of CoA in the enzyme (5, 11). Because Cys-164 is the catalytic nucleophile required for tethering of the starter group and the growing polyketide chain, the CoA-thioester carbonyl carbon must be accessible to the cysteine Sγ. Interaction between the thioester carbonyl oxygen and the side-chain of Asn-336 and the side-chain of His-303 facilitates substrate binding and stabilizes the transition state during nucleophilic attack of Cys-164 on the thioester carbonyl (5, 11, 12). With these constraints, we modeled the binding of N-methylanthraniloyl-CoA to both the F215S mutant and wild-type CHS and the binding of p-coumaroyl-CoA to wild-type CHS (Fig. 4).
Comparison of the three-dimensional structures of both CHS and the F215S mutant with N-methylanthraniloyl-CoA modeled at the active site entrance suggest a structural basis for altered starter molecule specificity. Substitution of a serine for Phe-215 opens a space at the cavity entrance to accommodate the methylamine moiety of N-methylanthraniloyl-CoA and allows for positioning of the thioester carbonyl moiety next to Cys-164, His-303, and Asn-336 (Fig. 4A). Widening of the active site entrance in the F215S mutant also provides space for narrower CoA-thioesters to sample alternate orientations at the catalytic site that impair loading of these starter molecules, thus negatively impacting substrate selectivity.
The serine hydroxyl group at position 215 may also provide a hydrogen bond to the methylamine moiety of the starter molecule to ensure proper positioning of the starter molecule. To test this possibility, we generated the CHS F215A mutant. The product profile of this mutant with p-coumaroyl-CoA and N-methylanthraniloyl-CoA is identical to that of the F215S mutant (not shown). The F215A mutant also exhibits a 7-fold increase in Km for N-methylanthraniloyl-CoA (kcat = 0.10 min−1, Km = 20.7 μM, kcat/Km = 80 M−1⋅s−1), supporting the possibility of an enzyme-substrate hydrogen bond and resultant catalytic improvement in the F215S mutant.
In wild-type CHS, Phe-215 prevents the N-methylanthraniloyl moiety from adopting the same conformation (Fig. 4B). Steric interference between the N-methylanthraniloyl group and Phe-215 results from both the expanded width and the shorter length of the N-methylanthraniloyl moiety that places the starter molecule directly between Phe-215 and Phe-265. Considering that CHS accepts benzoyl-CoA as a substrate, the methylamine group of N-methylanthraniloyl-CoA appears crucial in preventing binding in a catalytically productive manner. With p-coumaroyl-CoA (Fig. 4C), the longer propanoid linker positions the coumaroyl ring deeper in the active site cavity behind the two phenylalanines. Phe-215 thus steers the rotation of the thioester carbonyl into an optimal position for nucleophilic attack by Cys-164.
Discussion
The three-dimensional structure of alfalfa CHS2 and sequences of diverse plant type III PKS suggested that the identity of residues at the juncture of the active site and the CoA binding tunnel influence substrate and product specificity. As gatekeepers to the active site of CHS, Phe-215 and Phe-265 offered attractive targets for structure-guided enzyme engineering studies. The results reported here show that mutation of Phe-265 to the valine found in ACS does not alter the starter molecule preference of alfalfa CHS2. Unexpectedly, the F215S CHS mutant, which is not a naturally occurring type III PKS variation, accepts N-methylanthraniloyl-CoA as a substrate but yields a novel alkaloid, namely N-methylanthraniloyltriacetic acid lactone, instead of the naturally occurring acridone. Comparison of the three-dimensional structures of wild-type CHS and the F215S mutant suggests that substitution of a serine for Phe-215 allows for productive binding of N-methylanthraniloyl-CoA by allowing the bulkier starter molecule to reside in the wider active site entrance.
Aromatic CoA-thioesters smaller than p-coumaroyl-CoA, such as benzoyl-CoA and phenylacetyl-CoA, can efficiently access the CHS active site. In contrast, N-methylanthraniloyl-CoA projects a methylamine substituent at the ortho ring position causing steric hindrance with Phe-215 or Phe-265. Thus, the active site architecture of CHS filters out certain CoA-thioesters as potential starter molecules. A similar use of size-exclusion for selection of starter molecules occurs in 2-pyrone synthase, where three amino acid differences from CHS constrict the active site entrance and reduce the volume of the internal cavity (6). In 2-pyrone synthase, p-coumaroyl-CoA is excluded whereas acetyl-CoA or benzoyl-CoA initiate polyketide chain extension, which is limited to two instead of three acetyl additions (29).
Although this model accounts for how N-methylanthraniloyl-CoA binds at the F215S active site, it does not explain why this CHS mutant does not form acridone or how ACS effectively catalyzes acridone formation. Characterization of the reaction product generated by the F215S mutant with N-methylanthraniloyl-CoA and malonyl-CoA demonstrates that this mutant forms a tetraketide reaction intermediate identical to that required for acridone production by ACS. However, the regio-specific Claisen condensation reaction that occurs during acridone formation does not proceed in the F215S mutant. This observation indicates that the conformation of the tetraketide intermediate differs from that found in either the CHS or ACS active site. Recent experiments demonstrate that mutations of residues lining the active site cavity or use of unnatural starter molecules result in production of polyketide lactones by type III PKS (7, 16–18, 30). Because the overall three-dimensional folds of CHS and the F215S mutant are identical, the structural differences between the N-methylanthraniloyl- and the p-coumaroyl-moieties of the tetraketide likely alter how the starter molecule fits within the active site. In turn, this difference in starter molecule binding changes the orientation of the extended polyketide and prevents adoption of the conformation leading to the regio-specific Claisen condensation reaction. Our results indicate that further mutations in the CHS F215S active site cavity are required to successfully manipulate the cyclization reaction.
The failure of the F265V mutant to modify the starter molecule specificity of alfalfa CHS2 is striking in light of recently published work describing the transformation of R. graveolens ACS2 into a CHS (31). By using ACS, which displays 10% CHS activity, as the starting enzyme, replacement of Val-265 with a phenylalanine yielded a mutant enzyme exhibiting equal ACS and CHS activities with a 4-fold reduction in overall catalytic activity. The V265F mutation and two additional substitutions of residues lining the active site cavity convert ACS to a functional CHS with 10% ACS activity remaining. Lukacin et al. (31) note “conversion of ACS to CHS appears easier than the opposite, because CHSs including the one from Ruta, do not show any ACS side activity.” Our results reinforce this statement. In addition, examination of a CHS F215S/F265V mutant failed to result in acridone formation, although this mutant produces the same product as the F215S mutant. Even though N-methylanthraniloyl-CoA binds unproductively to the F265V mutant, substitution of a valine for Phe-265 does not introduce any detectable ACS activity in CHS. Valine retains sufficient steric bulk to allow other starter molecules access to the CHS active site without altering substrate specificity. Without structural knowledge of ACS, the molecular determinants governing the observed differences between CHS and ACS remain unclear. Additional steric differences in the ACS cavity, possibly mediated by second tier interactions, may modulate starter molecule binding and polyketide folding, and warrant structural examination.
Earlier studies on stilbene synthase showed that naturally occurring mutations subtly alter substrate specificity (p-coumaroyl-CoA vs. cinnamoyl-CoA) while preserving the regiochemistry of the cyclization reaction (32, 33). Clearly, introduction of mutations not found in nature can cause unexpected changes in the activity of these PKS. As demonstrated, the CHS F215S mutant represents an alternative solution to that of ACS for accepting N-methylanthraniloyl-CoA as a starter molecule, but differs in the cyclization of the tetraketide intermediate. This outcome fits the model for evolution of CHS-like enzymes in which the enzymatic activities of the plant-specific type III PKS evolved independently from CHS at different times in various organisms, with each enzyme exhibiting a unique activity (34).
With further experimentation, redesign of the type III PKS scaffold could yield new products or generate compounds usable as substrates for other enzymes involved in secondary metabolism in vivo. Ultimately, efforts to produce type III PKS with novel properties, for example, in the biosynthesis of phytoalexins with new pathogen-defense activity or the production of anthocyanin pigments of different shades coupled with metabolic engineering, may supply novel metabolites for new plant natural products.
Supplementary Material
Acknowledgments
We gratefully acknowledge the gift of the ACS gene from the laboratory of Professor Ulrich Matern. This work was supported by a grant from the National Science Foundation (MCB9982586) to J.P.N. J.M.J. was a National Institutes of Health Postdoctoral Research Fellow (CA80396).
Abbreviations
- ACS
acridone synthase
- CHS
chalcone synthase
- PKS
polyketide synthase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1JWX).
References
- 1.Bohm B A. Introduction to Flavonoids. Singapore: Harcourt; 1998. [Google Scholar]
- 2.Schröder J. Trends Plant Sci. 1997;2:373–378. [Google Scholar]
- 3.Staunton J, Weissman K J. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 4.Tropf S, Kärcher B, Schröder G, Schröder J. J Biol Chem. 1995;270:7922–7928. doi: 10.1074/jbc.270.14.7922. [DOI] [PubMed] [Google Scholar]
- 5.Ferrer J-L, Jez J M, Bowman M E, Dixon R A, Noel J P. Nat Struct Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 6.Jez J M, Austin M B, Ferrer J-L, Bowman M E, Schröder J, Noel J P. Chem Biol. 2000;7:919–930. doi: 10.1016/s1074-5521(00)00041-7. [DOI] [PubMed] [Google Scholar]
- 7.Jez J M, Bowman M E, Noel J P. Biochemistry. 2001;49:14829–14838. doi: 10.1021/bi015621z. [DOI] [PubMed] [Google Scholar]
- 8.Dixon R A, Steele C L. Trends Plant Sci. 1999;4:394–400. doi: 10.1016/s1360-1385(99)01471-5. [DOI] [PubMed] [Google Scholar]
- 9.Dixon R A. Nature (London) 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- 10.Lanz T, Tropf S, Marner F-J, Schröder J, Schröder G. J Biol Chem. 1991;266:9971–9976. [PubMed] [Google Scholar]
- 11.Jez J M, Ferrer J-L, Bowman M E, Dixon R A, Noel J P. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 12.Jez J M, Noel J P. J Biol Chem. 2000;275:39640–39646. doi: 10.1074/jbc.M008569200. [DOI] [PubMed] [Google Scholar]
- 13.Funa N, Ohnishi Y, Fujii I, Shibuya M, Ebizuka Y, Horinouchi S. Nature (London) 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- 14.Hrazdina G, Kreuzaler F, Hahlbrock K, Grisebach H. Arch Biochem Biophys. 1976;175:392–399. doi: 10.1016/0003-9861(76)90526-9. [DOI] [PubMed] [Google Scholar]
- 15.Schüz R, Heller W, Hahlbrock K. J Biol Chem. 1983;258:6730–6734. [PubMed] [Google Scholar]
- 16.Zuurbier K W M, Leser J, Berger T, Hofte A J, Schröder G, Verpoorte R, Schröder J. Phytochemistry. 1998;49:1945–1951. doi: 10.1016/s0031-9422(98)00346-x. [DOI] [PubMed] [Google Scholar]
- 17.Morita H, Takahashi Y, Noguchi H, Abe I. Biochem Biophys Res Commun. 2000;279:190–195. doi: 10.1006/bbrc.2000.3920. [DOI] [PubMed] [Google Scholar]
- 18.Abe I, Morita H, Nomura A, Noguchi H. J Am Chem Soc. 2000;122:11242–11243. [Google Scholar]
- 19.Abe I, Takahashi Y, Morita H, Noguchi H. Eur J Biochem. 2001;268:3354–3359. doi: 10.1046/j.1432-1327.2001.02255.x. [DOI] [PubMed] [Google Scholar]
- 20.Lukacin R, Springob K, Urbanke C, Ernwein C, Schröder G, Schröder J, Matern U. FEBS Lett. 1999;448:135–140. doi: 10.1016/s0014-5793(99)00355-5. [DOI] [PubMed] [Google Scholar]
- 21.Springob K, Lukacin R, Ernwein C, Gröning I, Matern U. Eur J Biochem. 2000;267:6552–6559. doi: 10.1046/j.1432-1327.2000.01746.x. [DOI] [PubMed] [Google Scholar]
- 22.Stoeckigt J, Zenk M H. Z Naturforsch, C. 1975;30:352–358. doi: 10.1515/znc-1975-5-609. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G N, Vagin A A, Dodson E J. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr D. 1993;49:148–157. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Lamzin V S, Wilson K S. Acta Crystallogr D. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 27.Baumert A, Maier W, Groeger D, Deutzmann R. Z Naturforsch, C. 1994;49:26–32. doi: 10.1515/znc-1994-1-205. [DOI] [PubMed] [Google Scholar]
- 28.Baumert A, Schneider G, Groeger D. Z Naturforsch, C. 1986;41:187–192. [Google Scholar]
- 29.Eckermann S, Schröder G, Schmidt J, Strack D, Edrada R A, Helariutta Y, Elomaa P, Kotilainen M, Kilpeläinen I, Proksch P, Teeri T H, Schröder J. Nature (London) 1998;396:387–390. [Google Scholar]
- 30.Suh D Y, Fukuma K, Kagami J, Yamazaki Y, Shibuya M, Ebizuka Y, Sankawa U. Biochem J. 2000;350:229–235. [PMC free article] [PubMed] [Google Scholar]
- 31.Lukacin R, Schreiner S, Matern U. FEBS Lett. 2001;508:413–417. doi: 10.1016/s0014-5793(01)03061-7. [DOI] [PubMed] [Google Scholar]
- 32.Schröder G, Schröder J. J Biol Chem. 1992;267:20558–20560. [PubMed] [Google Scholar]
- 33.Raiber S, Schröder G, Schröder J. FEBS Lett. 1995;361:299–302. doi: 10.1016/0014-5793(95)00199-j. [DOI] [PubMed] [Google Scholar]
- 34.Tropf S, Lanz T, Rensing S A, Schröder J, Schröder G. J Mol Evol. 1994;38:610–618. doi: 10.1007/BF00175881. [DOI] [PubMed] [Google Scholar]
- 35.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.