Abstract
The cis-acting mRNA elements that promote programmed −1 ribosomal frameshifting present a natural target for the rational design of antiretroviral chemotherapies. It has been commonly accepted that the HIV-1 frameshifting signal is special, because its downstream enhancer element consists of a simple mRNA stem loop rather than a more complex secondary structure such as a pseudoknot. Here we present three lines of evidence, bioinformatic, structural, and genetic, showing that the biologically relevant HIV-1 frameshift signal contains a complex RNA structure that likely includes an extended RNA triple-helix region. We suggest that the potential intramolecular triplex structure is essential for viral propagation and viability, and that small molecules targeted to this RNA structure may possess antiretroviral activities.
The HIV type 1 (HIV-1) is the causative agent of AIDS (1–3), and understanding the HIV-1 life cycle at the molecular level has played an important role in the development of antiviral agents with therapeutic properties. A successful outgrowth of such studies has been the identification of compounds that inhibit the activity of viral proteins, leading to dramatic reductions in the HIV loads in patients (reviewed in refs. 1 and 4). Although these drugs have proven successful, drug-resistant strains are emerging rapidly, and they do not eliminate HIV completely in infected patients (1, 5–8). Thus, there is an urgent need to develop new drug targets that ultimately will increase the repertoire of antiviral agents that can be used to eliminate or reduce HIV loads in patients. The HIV particle consists of a lipoprotein envelope surrounding a core composed of a protein capsid shell, within which are reverse transcriptase, integrase, protease, and two copies of the RNA genome (2). The structural and enzymatic components of the viral core are synthesized as polyproteins with a common N terminus (reviewed in ref. 1). The ORF encoding the major viral structural 55-kDa Gag protein is located at the 5′ end of the mRNA. The pol ORF, which encodes the viral protease, reverse transcriptase, and integrase, is 3′ of and out-of-frame with respect to the gag ORF (3), and these HIV-1 enzymatic proteins are translated only as a result of a programmed −1 ribosomal frameshifting (PRF) event (9–11). In viruses that use PRF, the majority of translational events result in the production of the Gag protein, and a minority yield viral enzymatic proteins. Consequently, the frameshift efficiency determines the ratio of structural to enzymatic proteins available for viral particle assembly. The ratio of Gag to Gag-pol synthesized in retroviruses as a consequence of programmed frameshifting varies between a narrow window of 20:1 to 60:1 (reviewed in ref. 12). The importance of maintaining this precise ratio on viral propagation has been demonstrated for many different viruses from the simple L-A totivirus of yeast to HIV-1 (reviewed in ref. 13). In all such viruses examined to date, even small alterations in frameshifting efficiencies inhibit virus propagation. Thus, PRF presents a potential target for antiviral therapeutics.
The bipartite −1 PRF signal consists of a heptameric “slippery site” followed by a downstream secondary RNA structure. The slippery site (X XXY YYZ, where the gag reading frame is indicated by spaces) does not have a precise sequence, but exhaustive analyses have determined that X can be any three identical nucleotides, Y can be three A or U residues, and Z is A, U, or C (14–17). The downstream RNA secondary structure is typically an RNA pseudoknot located ≈6 nucleotides 3′ of the slippery site. This structure is thought to position elongating ribosomes to pause with their A- and P-site tRNAs over the slippery site (15, 18–22), where they can simultaneously slip in the 5′ direction by one base, repairing their nonwobble bases to the respective −1 frame codons after which translation resumes in the new reading frame (14). These rules governing −1 PRF are universal in eukaryotes, because PRF signals from mammalian viruses function in yeast cells (9, 10, 23). A number of parameters can influence frameshifting efficiencies including the sequence of the slippery site and its distance from the RNA secondary structure, the thermodynamic stability of the RNA secondary structure, the interactions between ribosomes and tRNAs and those between the ribosome-associated tRNAs and the mRNA template, and the interactions between the translational apparatus and numerous host-encoded trans-acting factors (reviewed in refs. 12, 24, and 25).
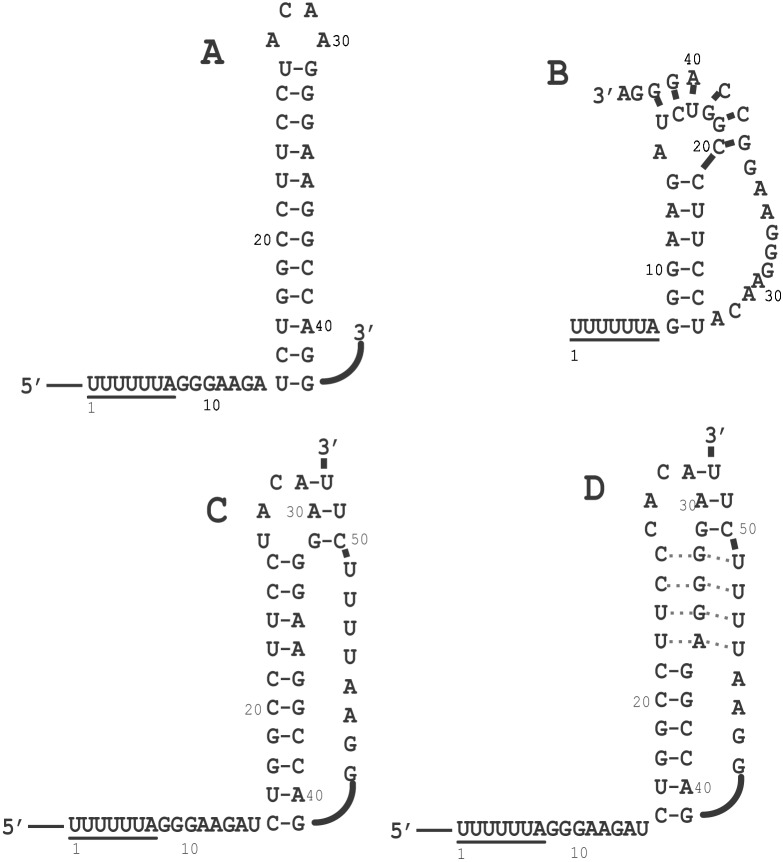
Although no data exist proving the universal requirement for an RNA pseudoknot, frameshift-stimulating pseudoknots cannot be replaced by simple stem-loop structures of equal or greater thermodynamic stability (26). In the −1 PRF literature, however, the HIV-1 frameshift signal is noted as a prominent exception. The original reports characterizing the HIV-1 frameshift signal suggested that only the UUUUUUA slippery site heptamer was required to promote −1 PRF (10, 11). Subsequent studies demonstrated that a simple RNA stem-loop structure 3′ of the slippery site was required to promote “efficient” frameshifting (see Fig. 1A and refs. 27 and 28). However, there has never been a correlation between “efficient” frameshifting and the actual efficiency required by HIV-1 for optimal viral particle assembly. Two possible RNA pseudoknot structures 3′ of the HIV-1 slippery site have been suggested. One of these structures (Fig. 1B; ref. 29) eliminates the “spacer region” between the slippery site and the RNA pseudoknot, presenting the close juxtaposition between these two components of the frameshift signal that is known to interfere with efficient −1 PRF (16, 18, 19, 24, 28, 30, 31). A subsequent analysis demonstrated that this structure is not real (32). The short A-U-rich stem 2 of the other suggested that RNA pseudoknot conformer probably is too unstable to be biologically relevant (Fig. 1C; ref. 33). However, closer examination of this structure reveals that a range of potential RNA pseudoknot conformers may be capable of forming either by stem 2 becoming larger at the expense of stem1 or even one containing a potential triple-helix structure (Fig. 1D). No experimental analyses have been conducted that would either prove or disprove such RNA structural conformers. For example, the synthetic RNA that was used in the physical and biochemical studies disproving the pseudoknot shown in Fig. 1B stopped at nucleotide 44 of the HIV-1 frameshift signal and thus did not contain all of the sequence required for formation of the structures shown in Fig. 1 C and D (32). Similarly, the constructs used by two other groups (23, 34) stop at nucleotide 41. Interestingly, the potential to form the entire range of alternative RNA pseudoknot or RNA triplex conformers is conserved in a protease-resistant HIV-1 variant (35). Thus, as a potential target for antiviral therapies it is critical to define the biologically relevant mRNA secondary structure that enhances −1 PRF in HIV-1. This report addresses this problem, presenting three lines of evidence supporting the involvement of an intramolecular triplex RNA structure in the HIV-1 −1 PRF signal.
Figure 1.
Different representations of the HIV-1 frameshift signal. (A) Simple stem loop proposed by Jacks et al. (11) and investigated by Wilson et al. (10) and Parkin et al. (27). (B) RNA Pseudoknot proposed by Du et al. (29). (C) RNA pseudoknot proposed by Taylor et al. (33). (D) Intramolecular triplex structure proposed in this study.
Materials and Methods
Database Analysis.
A database containing all of the 20,946 reported HIV-1 sequences was compiled using a search of the GenBank nucleotide sequence database at the National Center for Biotechnology Information (NCBI). Said search was done on the web site of the NCBI-developed Entrez retrieval system, selecting for HIV-1 matches in the “organism” field (http://www.ncbi.nlm.nih.gov/Entrez/). This set of sequences was analyzed by using a PERL script program that selected for the HIV-1 frameshift signal. This set was extracted by searching for TTTTTTA (the slippery site), followed by 10 bases with no specific sequence requirement (the spacer between the slippery site and the RNA secondary structure), followed by the highly conserved sequence GG (at the beginning of stem 1). From this data set, the program extracted the relevant sequence from each reported sequence beginning at the HIV-1 slippery site TTTTTTA and extending an additional 45 nucleotides. The output file (Fasta format) containing 715 sequences was analyzed by the CLUSTAL W 1.7 program (36) to produce a global alignment of all the sequences. This program analyzes identities between these sequences and weights each variation at each of the 52 nucleotides in the target sequence, allowing us to define the allowed variations in the HIV-1 frameshift signal. Subsequently, each unique sequence was analyzed manually for its ability to form the structure depicted in Fig. 1D.
Nuclease Mapping.
All RNAs were purchased from Dharmacon Research (Lafayette, CO). RNAs were 5′ end-labeled with 0.5 μM [γ-32P]ATP (6,000 Ci/mmol; 1 Ci = 37 GBq) (ICN) per 100 pmol of nucleic acid by incubation with 16 units of T4 polynucleotide kinase (NEB, Beverly, MA) in the provided buffer. The end-labeled RNAs were purified through 20% denaturing gels, visualized by autoradiography, eluted out of the acrylamide, and desalted on a reverse-phase cartridge. The sequences of the RNAs were verified by alkaline hydrolysis and nuclease digestion. For the RNase protection assay, 2 pmol of labeled RNA in 10 μl of final reaction volume were incubated with: 3–7 units of S1 nuclease (Amersham Pharmacia) in S1 nuclease cleavage buffer (30 mM sodium acetate, pH 4.6/100 mM NaCl/1 mM ZnSO4) for 4 min at 4°C or 2 min at room temperature; 1 milliunit of RNase T1 (United States Biochemical) in 100 mM Tris⋅HCl, pH 8.0, for 4 min at 4°C or 2 min at room temperature; or 3 microunits of RNase A (United States Biochemical) in 50 mM sodium acetate, pH 5.2, for 4 min at 4°C, or 2 min at room temperature. RNA ladders were obtained by incubation of 2 pmol of RNA in hydrolysis buffer (50 mM Na2CO3/NaHCO3, pH 9.2) at 85°C for 10 and 12 min. All reactions were stopped by addition with 5 μl of sample loading buffer (95% formamide/0.1% bromophenol blue/20 mM EDTA) and immediate freezing on dry ice. Samples then were separated through 20% denaturing gels at 90 W (33 mA) for 1.5 and 3 h and visualized by phosphorimagery.
Plasmids and Frameshift Assays.
For yeast-based assays, the plasmid pJD160.0 was used as the 0-frame control (37). The synthetic oligonucleotides (Integrated DNA Technologies, Coralville, IA) 5′-CCCCGGATCCATTTTTTAGGGAAGATCTGGCCTTCCCACAAGGGGAGGCCAGGGAATTTTCTTCAGGTACCCCCC-3′ (forward) and 5′-GGGGGGTACCT-GAAGAAAATTCCCTGGCCTCCCCTTGTGGGAAG-GCCAGATCTTCCCTAAAAAATGGATCCGGGG-3′ (reverse) were used to construct the −1 frameshift reporter pJD160.HIV-wild type (wt). The complementary oligonucleotides were annealed with one another, digested with BamHI and KpnI, and cloned into similarly digested pJD160.0 such that the lacZ reporter gene was in the −1 reading frame with respect to the translational start site, and thus could be translated only as a result of a −1 PRF. The reporter plasmids pJD160.HIV-c1, pJD160.HIV-c3, and pJD160.HIV-c2, harboring mutant RNA pseudoknot, were constructed similarly. To measure −1 PRF efficiencies, these plasmids were introduced into yeast cells [JD111: MATα ura3-52 lys2-801 trp1Δ leu2=his3= (L-AHNB M1)] by lithium-acetate transformation, transformants were selected on H-trp medium, and β-galactosidase activities and PRF efficiencies were determined as described (15). All assays were performed in triplicate, and each assay was repeated at least three times.
Frameshifting in HeLa cells was monitored by using a dual luciferase reporter plasmid system (34). Synthetic reporter mRNAs were transiently transfected into HeLa cells, cells were cultured for 24 h and lysed, and the activities of Renilla (first ORF, Luc-R) versus firefly luciferase (second ORF, Luc-F) were determined by using a dual luciferase kit (Promega) and a Turner (Palo Alto, CA) 20/20 luminometer. The construct in which Luc-F is in the same frame as Luc-R, p2luci (34), serves to generate a 0-frame baseline of translation of the second ORF. In the pHIVluc-wt and pHIVluc-m2 constructs, the two different HIV-1-derived frameshift signals were positioned between Luc-R and Luc-F, and Luc-F is in the −1 frame with regard to Luc-R. Thus, the firefly luciferase can be produced only as a consequence of a −1 PRF event. Synthetic mRNAs generated by p2luc (Luc-F in the −1 frame with respect to Luc-R with no intervening sequence; ref. 34) serve to control for undirected frameshift events. With these reporter mRNAs, the ratio of Luc-F to Luc-R activities provides a normalized measurement of translation of the second ORF. Further, comparison of this ratio as generated from the test mRNAs to that of the 0-frame control (p2luci) yields a measure of directed (by pHIVluc-wt or pHIVluc-m2) or nondirected (by p2luci) −1 ribosomal frameshift efficiencies.
Results
The Potential RNA Triple-Helix Structure Is Highly Conserved.
If HIV-1 frameshifting requires the RNA triplex, then this structure should be highly conserved among all isolates of HIV-1. Examination of all of the 715 frameshift signal-containing HIV-1 sequences in our database shows that the overwhelming majority of the sequences were able to form an RNA triplex structure (Table 1). Eighty six percent (618 sequences total) conserved the ability to form the triplex at all seven positions. A further 5% of the sequences contained single changes that would affect base pairing at either positions 46 or 52, i.e., at the ends of strand 3 of the triplex. Thus, in 91% of the sequences in the database, a contiguous stretch of six of the seven possible base triples are conserved. Indeed, less than 8% of the sequences contained one or more internal mismatches between strands 2 and 3, with the remainder perturbing the strand 1/strand 2 base pairing. Of particular relevance, the identity of the nucleotide at position 34 was distributed almost evenly between G and A. Although the base at position 47 was U in over 90% of the sequences, in all but one of the instances where nucleotide 47 was a C the nucleotide at position 34 was a G. This analysis supports the notion that the ability to form a stable intramolecular RNA triple-helix structure has been evolutionarily conserved in the HIV-1 frameshift signal.
Table 1.
Evolutionary conservation of the intramolecular triple helix
| No affect on triple helix formation (618 of 715 = 86.4%) |
| Consensus sequence, 106 |
| Changes do not affect the ability to form potential triplex, 512 |
| Single changes to the ends of strands that minimally impact on triplex formation (36 of 715 = 5%) |
| Bottom of strand 1/strand 2 base pairing region, 6 |
| Bottom or top of strand 2/strand 3 base pairing region, 30 |
| Two simultaneous changes only at nucleotides 46 and 52, 5 |
| Internal changes that disrupt strand 1/strand 2 base pairing region, 1 |
| One or more internal mismatches in strand 2/strand 3 base pairing region, 55 (7.7%) |
Of 20,946 reported HIV-1 sequences in the NCBI sequence database, 715 contained the sequence TTTTTTA plus at least 45 nucleotides. A global alignment of these sequences was produced using clustal w 1.7 (36), and the resulting output file was analyzed for the impact of each variant on intramolecular triplex structure formation.
Biochemical Analyses Indicate the Presence of an RNA Triplex Structure.
Formation of triple helices requires the third strand to hydrogen-bond to purine bases present in the underlying Watson–Crick duplex. In the wt HIV-1 frameshift signal sequence, there is a stable stem-loop structure that is followed by a pyrimidine-rich sequence. We reasoned that a pyrimidine⋅purine⋅pyrimidine triplex structure can exist in this RNA sequence, and this structure could have functional significance in frameshifting. To test this hypothesis, we synthesized two RNA sequences, HIV-wt and HIV-m2 (Fig. 2A). To map single- and double-stranded regions within the sequence, secondary structure-specific RNA cutting enzymes were used; nuclease S1 nonspecifically cuts all single-stranded nucleotides, RNase T1 cuts at single-stranded G residues, and RNase A cleaves preferentially at single-stranded pyrimidine bases. The RNA cleavage pattern of the wt sequence was compared with mutant sequences.
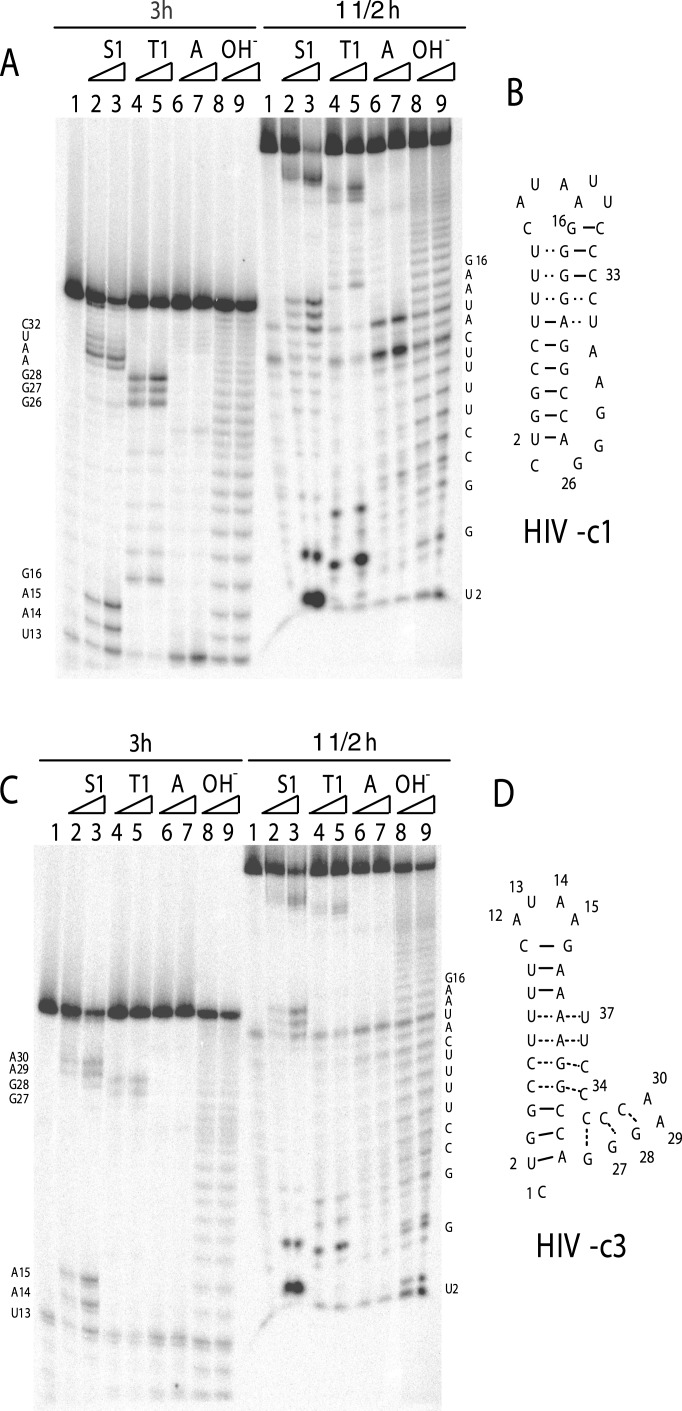
Figure 2.
Secondary structure mapping of RNA. (A) Sequences and proposed secondary structures for the wt and mutant RNA sequences. The numbering of nucleotides in the RNA corresponds to their positions in the wt HIV-1 frameshift signal. (B) The results of nuclease cleavage of wt and mutant RNA. RNase S1, RNase T1, and RNase A were used in these experiments. RNA were 5′ end-labeled with 32P and subjected to enzymatic digestion as described in Materials and Methods. Lanes 1 and 6, control lanes containing RNA without the addition of ribonucleases; lanes 2 and 7, RNase S1 digestion; lanes 3 and 8, RNase T1 digestion; lanes 4 and 9, RNase A digestion; lanes 5 and 10, RNA hydrolysis ladder.
Three different ribonucleases were used, and in each case the HIV-wt and HIV-m2 RNAs displayed different patterns of hydrolysis (Fig. 2B). We performed ribonuclease reactions at various pH values (6.8–8.0), temperatures (4–37°C), and salt conditions (50 mM NaCl with and without 5 mM MgCl2). Digestion with RNase S1 enzyme highlights the absence of hydrogen bonding of strand 3 with the duplex region of the stem-loop RNA structure (m2), because all nucleotides at the 3′ end, starting from position 42, are cleaved efficiently (Fig. 2, lane 8). In contrast, a minor cleavage of the third strand in HIV-wt RNA indicates a conformational equilibrium existing between the triplex and nontriplex form (Fig. 2, lane 2). As expected, both RNAs are cleaved equally by RNase S1 nuclease in the loop 1 region. Digestion of HIV-m2 by nuclease T1 again shows the presence of the single-stranded region in strand 3 beyond nucleotide 42 (lane 9). Cleavage of HIV-wt RNA by nuclease T1 shows the presence of a duplex RNA and loop 2; however, because there are no Gs at the 3′ end of HIV-wt, no evidence of stem 2 formation can be inferred.
RNase A digestion further confirms the single-stranded structure of HIV-m2 RNA after nucleotide 42. RNase A cleavage of HIV-wt, the putative strand 3 of which in triplex structure is very pyrimidine-rich, is very interesting. As shown in Fig. 2B, there is a single weak cleavage site at position 49 in the strand 3 region. Mutant RNA was cleaved efficiently at U47 and U49 by RNase A (lane 10). A second cleavage occurs at U28 in loop 1 as well as in loop 1 of HIV-m2 RNA. These comparative RNase cleavage results demonstrate the presence of a triplex RNA structure in HIV-wt, whereas only the stem-loop duplex RNA is present in HIV-m2. It is also notable that the bases in loop 2 (G42–A45) are relatively well protected from nucleolytic attack, suggesting that they actively participate in a complex RNA tertiary structure that promises to be interesting and unique. Further, we did not observe any ribonuclease cleavage at the C22–U25 region, supporting the idea that this strand was part of triplex-like structure and was not single-stranded region.
To examine the formation of a triplex RNA further, we designed two more constructs, c1 and c3, in which C-G-U base triples were either substituted with U-G-C (construct c1, Fig. 3 A and B) to preserve the intramolecular triplex RNA structure or disrupt it (construct c3, Fig. 3 C and D). To resolve two regions of the RNA structure at high resolution, we ran gels for various periods of time. As shown in Fig. 3, the 5′-end regions of the RNAs were better resolved when gels were run for 1.5 h. The 3′-end regions were visualized by running gels for 3 h. For the c1 construct (Fig. 3A), two loop regions (G26–A30 and U13–A15) were hydrolyzed by ribonucleases (3 h gel, lanes 2–8), indicating that the overall triplex-like structure was similar to the wt sequence. However, it is interesting to note that although the major cleavage by S1 was observed at U14–A15 (lanes 2 and 3, 1½ h gel), there was some minor cleavage at three U residues below the loop. These results suggest that strands 1 and 3 are in a dynamic equilibrium with the strand 2 sequence. No such dynamic situation was observed in the wt sequence, possibly because of the U-rich third strand instead of U-rich strand 1. In contrast, structural mapping of construct c3 revealed that this RNA sequence adopted a stable secondary structure that did not resemble wt or m2 mutant RNA (Fig. 4 C and D). As discussed below, these structural dynamics also could explain the higher frameshift efficiencies for construct c1. Taken together, these results strongly support the notion that an intramolecular triplex RNA structure is formed in the HIV-wt frameshift signal sequence.
Figure 3.
Secondary structure mapping of constructs c1 and c3 RNAs. Ribonuclease mapping of c1 (A) and c3 (C) and the proposed secondary structures for c1 (B) and c3 (D) are shown. RNase S1, RNase T1, and RNase A were used in these experiments. RNA were 5′ end-labeled with 32P and subjected to enzymatic digestion as described in Materials and Methods. To resolve 5′ and 3′ ends unambiguously, gels were run for 1.5 and 3 h. Two temperature conditions (4 min at 4°C or 2 min at room temperature) for each enzyme reaction were used.
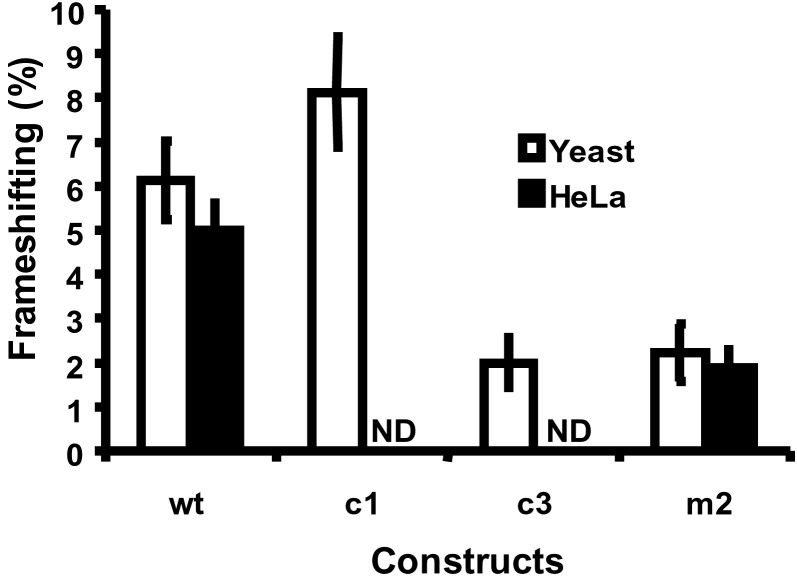
Figure 4.
Enhancement of −1 frameshifting by the intramolecular triplex. −1 PRF efficiencies were determined in yeast and HeLa cell systems as described in Materials and Methods. ND, not done. Error bars denote the percent error.
The Intramolecular Triplex Is Required for Maximal Enhancement of −1 PRF in Intact Yeast and HeLa Cells.
If the HIV-1 RNA triplex structure is biologically significant, then disrupting it should impact negatively on frameshift efficiencies. To test this hypothesis, wt and mutant frameshift signals differing only in their abilities to form the intramolecular triplex were cloned into both yeast- and HeLa cell-based frameshift reporters, and−1 PRF efficiencies were determined. The results are shown in Fig. 4. In yeast cells, the wt sequence promoted frameshifting with an efficiency of 6.13 ± 0.89%. In contrast, disruption of the triplex resulted in an approximately two-thirds decrease in frameshifting efficiencies (m2 = 2.23 ± 0.65%, and c3 = 2.00 ± 0.56%). In HeLa cells, the frameshift efficiency generated by pHIVluc-wt (5.0 ± 0.6%) was consistent with previously measured frameshift efficiencies and the 20:1 ratio of Gag to integrase in mature viral particles and, similar to the yeast system disruption of stem 2 (pHIVluc-m2), reduced frameshifting to less than 40% of wt levels (1.9 ± 0.4%). Undirected −1 frameshifting (0.06%) was nearly 2 orders of magnitude less than wt levels in both yeast and HeLa systems (data not shown). Further investigations using the yeast system revealed that substitution of C-G-U base triples with U-G-C sequence in construct c1 enhanced frameshifting (8.12 ± 1.25%). Although actual frameshift efficiencies varied between different experiments, in all seven repetitions the c1 construct always promoted −1 PRF to a significantly greater extent than the wt sequence in individual experimental repetitions. The significance of this finding is discussed below. These results clearly show that high efficiency frameshifting by HIV-1 depends on the presence of a unique and stable RNA structure that potentially folds into an intramolecular triplex.
Discussion
Rational drug design requires a full understanding of the target molecule, and in the case of the HIV-1 frameshift signal, the existing literature falls short of this requirement. Specifically, it has been accepted as common knowledge that HIV-1 had dispensed with the requirement for an RNA pseudoknot as the downstream enhancer element, because its stem-loop structure was very thermodynamically stable. This belief has led many other researchers in the field to delete sequences downstream of the presumed stem loop from reporter constructs, which in at least one published case probably invalidated the results of a high-throughput drug screen (38). The findings presented here seek to address this issue, demonstrating that the downstream enhancer element in the HIV-1 frameshift signal is a complex RNA structure that likely contains an extended RNA triple-helix region. Our findings that simple stem loops can only promote approximately one-third of wt frameshifting efficiencies are significant with regard to antiviral therapeutic approaches. Previous studies examining the effects of changes in −1 PRF in other viral systems show that the viral propagation is significantly more sensitive to small decreases in −1 PRF efficiencies than they are to increases of similar magnitude. For example, similar decreases in −1 PRF efficiencies caused by the presence of anisomycin (39) or to mutations in 5S rRNA (40) were sufficient to cure yeast cells of the L-A virus completely. Similarly, a 50% decrease in +1 frameshifting efficiency inhibited Ty1 retrotransposition in yeast by 98% (41). Thus, our findings suggest that the potential intramolecular triplex structure is essential for viral propagation and viability, and small molecules targeted to this RNA structure may possess antiretroviral activities.
Structure/Function: Why Not a Stem Loop?
Although a stem loop does promote levels of frameshifting that are significantly greater than normal, they still are not sufficient to meet the demands of the virus. A more complex secondary structure, typically an RNA pseudoknot but in this case the intramolecular triplex, is required as a specific enhancer of −1 PRF. We have proposed previously a “torsional resistance” model to explain the requirement for such structures (25). By this model, the specific location on the mRNA at which a ribosome stalls is determined by distance and resistance: how far the ribosome can elongate into the secondary structure before it is stopped in the slippery site proximal region of the structure (i.e., strand 1/strand 2) by the negative supercoiling forces imposed as a consequence of the distal region's (i.e., the strand 2/strand 3 interactions) limiting the rotational freedom of loop 1. By enforcing a specific equilibrium point between forward and reverse forces, these structures specifically direct ribosomes to pause with their A- and P-site tRNAs positioned directly over the slippery site, thus increasing the proportion of ribosomes that can slip. The results presented here support this view. In cases where loop 1 was torsionally restrained (wt and c1 constructs), −1 PRF efficiencies were stimulated relative to those where it had greater rotational freedom (m2 and c3). In particular, although the RNA structure of the c3 construct is quite complex, loop 1 is not torsionally constrained; as predicted by the model, the increased rotational freedom of loop 1 resulted in decreased −1 PRF efficiencies. A second point of interest with regard to these data addresses the notion that, whereas the slippery site proximal region is unwound by the ribosome in cis, unwinding of the distal region of the structure must be in trans (either passively by simple thermodynamic “breathing” or actively by a distributive RNA helicase, e.g., Upf1p; see refs. 42 and 43). In either case, stabilization of the distal region of the structure would increase the amount of time that elongating ribosomes would be paused at the frameshift signal, increasing the chance of slippage. Thus, although the distal region of the structure should not affect where the ribosome is directed to stall, its stability should influence the amount of time that it pauses. Our observation that increasing the thermodynamic stability of the strand 2/strand 3 interaction serves to stimulate programmed frameshifting further is consistent with this notion (compare the HIV-c1 to the HIV-wt constructs in Fig. 4).
In addition to forming well documented double-helix structures, nucleic acids have been found also to form three- and four-stranded complexes under some circumstances (44–50). Formation of triple helices requires the third strand to hydrogen-bond to purine bases present in the underlying Watson–Crick duplex. Triplexes can be subdivided into intramolecular versus intermolecular complexes. Intramolecular triplexes occur in H⋅DNA structure (51–53). Our results strongly suggest the formation of an intrastrand RNA triplex structure in the HIV-1 frameshift signal where a C-G repeat in duplex RNA provides a site for hydrogen bonding with a U-rich third strand. It is interesting to note that these are not typical C+-G-C or T-A-T triplet bases known to form stable triplex structures. Given the flexibility of RNA to form unusual structures and well known G-U interactions in RNA, we propose that stable C-G-U triplets are present under physiological conditions in HIV-1 frameshift signal. It is intriguing that although isolated base triplets occur in tRNA, ribozyme, and transactivation response element RNA structures (54–57), no intrastrand triple helices with consecutive stacked triplets have yet been observed in the folded structures of natural RNAs. Our results presenting the notion of short intramolecular triplex RNA involvement in HIV-1 frameshift signal suggest that such structures could play important roles in other biological functions.
Acknowledgments
This work was supported by National Institutes of Health Grants GM 58859 (to J.D.D.) and AI 45466 and AI 43198 (to T.M.R.).
Abbreviations
- PRF
programmed ribosomal frameshifting
- wt
wild type
References
- 1.Coffin J M. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 2.Levy J A. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaishnav Y N, Wong-Staal F. Annu Rev Biochem. 1991;60:577–630. doi: 10.1146/annurev.bi.60.070191.003045. [DOI] [PubMed] [Google Scholar]
- 4.Balzarini J, Pelemans H, Karlsson A, De ClercQ E, Kleim J P. Proc Natl Acad Sci USA. 1996;93:13152–13157. doi: 10.1073/pnas.93.23.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Q, Yang G, Goff S P, Prasad V R. J Virol. 1992;66:7568–7571. doi: 10.1128/jvi.66.12.7568-7571.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kew Y, Qingbin S, Prasad V R. J Biol Chem. 1994;269:15331–15336. [PubMed] [Google Scholar]
- 7.Moutouh L, Corbeil J, Richman D D. Proc Natl Acad Sci USA. 1996;93:6106–6111. doi: 10.1073/pnas.93.12.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, et al. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 9.Bidou L, Stahl G, Grima B, Liu H, Cassan M, Rousset J-P. RNA. 1997;3:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson W, Braddock M, Adams S E, Rathjen P D, Kingsman S M, Kingsman A J. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 11.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Nature (London) 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 12.Farabaugh P J. Programmed Alternative Reading of the Genetic Code. Austin, TX: Landes; 1997. [Google Scholar]
- 13.Dinman J D, Ruiz-Echevarria M J, Peltz S W. Trends Biotechnol. 1998;16:190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacks T, Madhani H D, Masiraz F R, Varmus H E. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinman J D, Icho T, Wickner R B. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinman J D, Wickner R B. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brierley I A, Jenner A J, Inglis S C. J Mol Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brierley I A, Dingard P, Inglis S C. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morikawa S, Bishop D H L. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somogyi P, Jenner A J, Brierley I A, Inglis S C. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu C, Tzeng T-H, Bruenn J A. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopinski J D, Dinman J D, Bruenn J A. Mol Cell Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahl G, Bidou L, Rousset J-P, Cassan M. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brierley I. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 25.Dinman J D. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brierley I A, Rolley N J, Jenner A J, Inglis S C. J Mol Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkin N T, Chamorro M, Varmus H E. J Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kollmus H, Hentze M W, Hauser H. RNA. 1996;2:316–323. [PMC free article] [PubMed] [Google Scholar]
- 29.Du Z, Giedroc D P, Hoffman D W. Biochemistry. 1996;35:4187–4198. doi: 10.1021/bi9527350. [DOI] [PubMed] [Google Scholar]
- 30.Kollmus H, Honigman A, Panet A, Hauser H. J Virol. 1994;68:6087–6091. doi: 10.1128/jvi.68.9.6087-6091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honda A, Nakamura T, Nishimura S. Biochem Biophys Res Commun. 1995;213:575–582. doi: 10.1006/bbrc.1995.2170. [DOI] [PubMed] [Google Scholar]
- 32.Kang H. Biochim Biophys Acta. 1998;1397:73–78. doi: 10.1016/S0167-4781(98)00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor E W, Ramanathan C S, Jalluri R K, Nadimpalli R G. J Med Chem. 1994;37:2637–2654. doi: 10.1021/jm00043a004. [DOI] [PubMed] [Google Scholar]
- 34.Grentzmann G, Ingram J A, Kelly P J, Gesteland R F, Atkins J F. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 35.Doyon L, Payant C, Brakier-Gingras L, Lamarre D. J Virol. 1998;72:6146–6150. doi: 10.1128/jvi.72.7.6146-6150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammell A B, Taylor R L, Peltz S W, Dinman J D. Genome Res. 1999;9:417–427. [PMC free article] [PubMed] [Google Scholar]
- 38.Hung M, Patel P, Davis S, Green S R. J Virol. 1998;72:4819–4824. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinman J D, Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Proc Natl Acad Sci USA. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith M W, Meskauskas A, Wang P, Sergiev P V, Dinman J D. Mol Cell Biol. 2001;21:8264–8275. doi: 10.1128/MCB.21.24.8264-8275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harger J W, Meskauskas A, Nielsen N, Justice M C, Dinman J D. Virology. 2001;286:216–224. doi: 10.1006/viro.2001.0997. [DOI] [PubMed] [Google Scholar]
- 42.Czaplinski K, Weng Y, Hagan K, Peltz S W. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 43.Cui Y, Dinman J D, Peltz S W. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang C, Zhang X, Ratliff R, Moyzis R, Rich A. Nature (London) 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- 45.Chastain M, Tinoco I., Jr Nucleic Acids Res. 1992;20:315–318. doi: 10.1093/nar/20.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sen D, Gilbert W. Nature (London) 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 47.Maher L J., III BioEssays. 1992;14:807–815. doi: 10.1002/bies.950141204. [DOI] [PubMed] [Google Scholar]
- 48.Guschlbauer W, Chantot J F, Thiele D. J Biomol Struct Dyn. 1990;8:491–511. doi: 10.1080/07391102.1990.10507825. [DOI] [PubMed] [Google Scholar]
- 49.Moser H E, Dervan P B. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 50.Rettberg C C, Prere M F, Gesteland R F, Atkins J F, Fayet O. J Mol Biol. 1999;286:1365–1378. doi: 10.1006/jmbi.1999.2546. [DOI] [PubMed] [Google Scholar]
- 51.Lyamichev V I, Mirkin S M, Danilevskaya O N, Voloshin O N, Balatskaya S V, Dobrynin V N, Filippov S A, Frank-Kamenetskii M D. Nature (London) 1989;339:634–637. doi: 10.1038/339634a0. [DOI] [PubMed] [Google Scholar]
- 52.Htun H, Dahlberg J E. Science. 1989;243:1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- 53.Htun H, Dahlberg J E. Science. 1988;241:1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- 54.Goddard J P. Prog Biophys Mol Biol. 1977;32:233–308. [PubMed] [Google Scholar]
- 55.Westhof E, Romby P, Romaniuk P J, Ebel J P, Ehresmann C, Ehresmann B. J Mol Biol. 1989;207:417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- 56.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 57.Puglisi J D, Tan R, Calnan B J, Frankel A D, Williamson J R. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]