Abstract
Los1p, the Saccharomyces cerevisiae exportin-t homologue, binds tRNA and functions in pre-tRNA splicing and export of mature tRNA from the nucleus to the cytosol. Because LOS1 is unessential in yeast, other pathways for tRNA nuclear export must exist. We report that Cca1p, which adds nucleotides C, C, and A to the 3′ end of tRNAs, is a multicopy suppressor of the defect in tRNA nuclear export caused by los1 null mutations. Mes1p, methionyl-tRNA synthetase, also suppresses the defect in nuclear export of tRNAMet in los1 cells. Thus, Cca1p and Mes1p seem to function in a Los1p-independent tRNA nuclear export pathway. Heterokaryon analysis indicates that Cca1p is a nucleus/cytosol-shuttling protein, providing the potential for Cca1p to function as an exporter or an adapter in this tRNA nuclear export pathway. In yeast, most mutations that affect tRNA nuclear export also cause defects in pre-tRNA splicing leading to tight coupling of the splicing and export processes. In contrast, we show that overexpressed Cca1p corrects the nuclear export, but not the pre-tRNA-splicing defects of los1∷Kanr cells, thereby uncoupling pre-tRNA splicing and tRNA nuclear export.
In eukaryotic cells most RNAs are transcribed in the nucleus, but function in the cytosol, whereas karyophilic proteins are translated in the cytosol, but function in the nucleus. Movement of macromolecules between the nucleus and the cytoplasm is a signal-mediated process with localization signals within cargoes. Transport of many macromolecules across the nuclear pore complex (NPC) requires Ran, a small GTPase, and its regulators, members of the RanGTPase-binding importin-β family, as well as functional NPCs (for review, see ref. 1). Importin-β family members, also known as importins, exportins, or karyopherins, in addition to binding to Ran, interact with NPC components and cargo and shuttle between the nucleus and cytosol. The Saccharomyces cerevisiae importin-β family has 14 members and the human genome has more than 20 (1). According to a simple model, a given importin-β family member would provide unidirectional nucleus/cytosol movement for a specific subset of cargoes (1). However, departures from this simple model occur. For example, Msn5p/Kap142p mediates nuclear import of some proteins and export of others (2); nuclear import of histones H2A, H2B, and some ribosomal proteins are also mediated by multiple importin-β family members (3, 4). Moreover, macromolecules, such as mRNAs, can be exchanged across the nuclear membrane by both Ran-independent (for review, see ref. 5) and Ran-dependent mechanisms (6, 7).
Exportin-t, a member of the importin-β family (8), binds tRNAs directly by a Ran-GTP-dependent mechanism and it functions to export tRNAs from the nucleus to the cytosol (9, 10). Los1p is the yeast exportin-t homologue (8, 11–13). Because LOS1 is unessential (14), alternative pathway(s) for tRNA nuclear export must exist, at least in budding yeast. Aminoacylation of tRNAs in nuclei facilitates tRNA nuclear export in Xenopus oocytes (15) and S. cerevisiae (16, 17), and a priori nuclear tRNA aminoacylation could provide a yeast Los1p-independent tRNA nuclear export pathway. However, our previous studies failed to resolve whether nuclear tRNA aminoacylation functions in series or in parallel with Los1p (18).
To address this problem we sought to identify gene products that participate in tRNA nuclear export. We used fluorescence in situ hybridization (FISH) and screened a collection of yeast thermosensitive (ts) mutants (19) for candidates with specific defects in tRNA nuclear export at the nonpermissive temperature. This screen uncovered ts352, which possesses a mutation of CCA1 (20). CCA1 is an essential gene encoding ATP(CTP):tRNA nucleotidyltransferase (Cca1p) that catalyzes addition of C, C, and A nucleotides to the tRNA 3′ termini in nuclei, cytosol, and mitochondria (21). Cca1p was previously implicated in tRNA nuclear export (15–17, 22, 23). Here, we report that Cca1p can function in a Los1p-independent tRNA nuclear export pathway. We also show that Cca1p shuttles between the nucleus and the cytosol, providing the potential for it to function as an exporter or an adapter for tRNA nuclear export.
Recently it was shown that mRNA nuclear export and mRNA splicing are coupled (24, 25). A similar situation exists in yeast for tRNA splicing and tRNA nuclear export. Most mutations that should affect only the export process, in fact, also affect pre-tRNA intron removal. Examples include mutations of LOS1, RNA1 encoding RanGAP, PRP20 encoding RanGEF, and several genes encoding components of the NPC (26–29). Here we show that unlike the Los1p-dependent pathway, the Los1p-independent nuclear export pathway is uncoupled from pre-tRNA processing.
Materials and Methods
Strains and Media.
The yeast strains used were: SS328 [MATα ade2–101 his3Δ200 lys2–801 ura3–52; 19); SS330 (MATa ade2–101 his3Δ200 tyr1 ura3–52 (19)]; ts352 [MATa cca1–1 ade2–101 his3Δ200 tyr1 ura3–52 (20)]; X2316–3C [MATα LOS1 SUP4 ade2–1 can1–100 lys1–1 his5–2 trp5–48 ura3–1 (26)]; 201–1-5 [MATα los1–1 SUP4 his5–2 lys1–1 can1–100 trp5–48 ade2–1 ura3–1 (26)]; SS700 [MATα los1∷Kanr SUP4 ade2–1 can1–100 lys1–1 his5–2 trp5–48 ura3–1; constructed by a Kanr disruption of LOS1 in strain X2316–3C by the methods described (18)]; SWY27 (MATα ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 nup116Δ∷HIS3) from S. Wente, Washington University School of Medicine, St. Louis (30); YM6154 (MATa MSN5 ura3–52 ADE2 his3–200 lys2–801 met− gal80− GAL1-LacZ∷LEU2) and YM6141 (MATa msn5∷HIS3 ura3–52 ADE2 his3–200 lys2–801 met− gal80− GAL1-LacZ∷LEU2), from M. Johnston, Washington University School of Medicine; Y1706 (MATα ura3–52 ade2–101 his3–11 trp1-Δ901 CSE1) and Y1709 (MATa ura3–52 ade2–101 his3–11 trp1-Δ901 cse1–1), both from M. Fitzgerald-Hayes, University of Massachusetts Amherst (31); PSY1200 (MATa sxm1∷HIS3 leu2Δ1 his3Δ200 trp1Δ63 ura3–52), PSY1201 (MATa pse1–1 ura3–52 trp1Δ63 leu2Δ1), and Kap123Δ (MATa kap123∷HIS3 lys2–801 leu2–3, 2–112 his3Δ200 trp1–1), all from P. Silver, Dana Farber Cancer Institute, Boston (32); DF5α (MATα MTR10 lys2–801 leu2–3, 2–112 ura3–52 his3Δ200 trp1–1), mtr10Δ (MATα mtr10∷HIS3 lys2–801 leu2–3, 2–112 ura3–52 his3Δ200 trp1–1), kap123 (MATα kap123∷URA3 lys2–801 leu2–3, 2–112 ura3–52 his3Δ200 trp1–1), and kap104–16 [MATα kap104∷ura3∷HIS3 lys2–801 leu2–3, 2–112 ura3–52 his3Δ200 trp1–1 and containing pRS314kap104–16 (ts allele)], all from L. Pemberton, University of Virginia Medical School, Charlottesville (33, 34); W303 XPO1 (MATa xpo1∷LEU2 ade2–1 ura3–1 his3–1,115 trp1–1 leu2–3,2–112 can1–100 ura3–1 and containing plasmid pKW440 encoding XPO1) and W303 xpo1–1 (MATa xpo1∷LEU2 ade2–1 ura3–1 his3–1,115 trp1–1 leu2–3, 2–112 can1–100 ura3–1 ura3–1 and containing plasmid pKW457 encoding xpo1–1) from C. Guthrie, University of California Medical School, San Francisco (35); MS739 [MATα ura3–52 leu2–3, 112 ade2–101 kar1–1 (36)] from M. Rose, Princeton University, Princeton, NJ.
Yeast extract/peptone glucose or synthetic defined media lacking appropriate nutritional ingredients were used to grow yeast. For growth assays, cells were grown to saturation in liquid media, and then 10 μl of serial dilutions of cells was applied to solid media and incubated for 3–4 days at the indicated temperatures.
CCA1 Constructs.
Escherichia coli DH5α was used to propagate recombinant DNA. To generate nuclear localization sequence (NLS)-CCA1-III-GFP-pRS416GAL1/10F, a 0.7-kb DNA encoding the green fluorescent protein (GFP) ORF was excised from pSEY181GAL1–10/GFP (provided by R. Y. Tsien, University of California, San Diego) by BamHI digestion and fused into pRS426NLS-CCA1 (21) at the BclI site located at the 3′ end of CCA1. The 2.6-kb fragment of CCA1-GFP was excised from pRS426NLS-CCA1-GFP by EcoRV digestion and ligated into pRS416GAL1/10F (from G. Peng, Penn State University, Hershey) at the SmaI site. NLS-Cca1p-III-GFP in this plasmid is controlled by the GAL1 promoter.
FISH.
FISH was performed according to published procedures (12) with the following modifications: prehybridized and hybridized temperatures were 39°C and washing with 2× SSC was performed at 50°C for tRNA probes.
Heterokaryon Analysis.
The heterokaryon assay was a modification of published procedures (37, 38). SS330 cells transformed with SV40NLS-CCA1-III-GFP-pRS416GAL1/10F or pGAL-H2B-GFP (37, 38) were grown to midlog phase in media containing 2% glucose as the carbon source. The cultures were diluted and grown overnight to ≈5 × 106 cells per ml in media containing 2% raffinose as the carbon source. NLS-Cca1p-GFP synthesis was induced by addition of 2% galactose for 2 h. Switching the carbon source from galactose to glucose terminated further NLS-Cca1p-GFP synthesis. Matings were initiated by mixing 3 × 106 cells with an equal number of MS739 cells (relevant genotype: kar1–1), concentrating the cells on a 25-mm-diameter, 0.45-μm-pore nitrocellulose filter, and placing the filter on a yeast extract/peptone glucose plate at 23°C. After ≈1.5 h about 30% of the cells were zygotes. At this time the subcellular location of GFP was determined in live cells by microscopy. For indirect immunofluorescence, ≈107 cells were mixed with an equal number of MS739 cells and concentrated on an 82-mm-diameter filter. After ≈1.5 h, the cells were washed off the filter and immunofluorescence was performed (39).
Microscopy.
Fluorescence images were observed by using a Nikon Microphot-FX microscope and were captured with a SenSys charge-coupled device camera (Photometrics, Tucson, AZ) with QED software (QED Imaging, Pittsburgh, PA). Images were assembled with Adobe PHOTOSHOP 5.0.
Preparation of RNA and Northern Blot Analysis.
RNA was isolated from log-phase yeast cells by phenol extraction as described (26). Northern blot analysis was performed according to Sarkar and Hopper (12). Blots were probed with tRNAIleUAU and each band was quantitated by phosphoimaging. The percentages of end-matured intron-containing pre-tRNA relative to the total of precursors and mature tRNAs were calculated.
Results
CCA1 Is a Multicopy Suppressor of the los1∷Kanr Defect of tRNA Nuclear Export.
We screened ≈200 ts mutants (19) by FISH with oligonucleotides specific to the intronless tRNAs, tRNAMet, and tRNAIleAAU, to identify variants that accumulate tRNA, but not mRNA, in the nucleus at the nonpermissive temperature of 37°C. Three mutants were identified and mutant ts352 was analyzed in detail. SS328 cells, one of two parents of this ts collection, do not accumulate either mRNA or tRNA (Fig. 1A) in the nucleus. Cells with a deletion of the nucleoporin, Nup116p, accumulate both tRNA and mRNA in the nucleus at 37°C (refs. 12 and 30; Fig. 1A). In contrast, ts352, accumulates only tRNA in the nucleus at 37°C (Fig. 1A). As determined by tetrad analyses ts352 possesses a single ts mutation that cosegregates with the defect in tRNA nuclear export (data not shown). Multicopy and single-copy yeast genomic libraries (40, 41) were transformed into ts352 to clone the wild-type counterpart of the mutated gene by complementation of the ts growth defect. Plasmid DNA was isolated from transformants showing concomitant loss of plasmid and temperature resistance and restoration of tRNA nuclear export. Sequence alignment showed that the identified plasmids had only one gene in common, CCA1. Thus, as reported, the gene mutated in ts352 is CCA1 (20).
Figure 1.
Subcellular distribution of tRNAMet determined by FISH. (A) tRNAMet accumulates in the nuclei of cca1–1 cells. Wild-type (SS328; 1, 1′), nup116–5∷HIS3 (SWY27; 2, 2′) or cca1–1 (ts352; 3, 3′), were prepared for FISH and the RNAs were hybridized with tagged oligonucleotides specific to tRNAMet or poly(A) to locate tRNA (1′, 2′, 3′) or mRNAs (1, 2, 3). (B) Overexpressed Cca1p suppresses the tRNA nuclear export defect caused by the los1∷Kanr mutation. los1∷Kanr cells containing YEp24 (1, 1′), YEpCCA1 (2, 2′), YEpLOS1-ET (3, 3′). Cells in 1, 2, and 3 were hybridized with an oligonucleotide specific to tRNAMet; 1′, 2′, and 3′, the same cells stained with 4′,6-diamidino-2-phenylindole.
CCA1 was previously implicated in tRNA nuclear export because Cca1p catalyzes addition of C, C, and A nucleotides to the tRNA 3′ end and exportin-t preferentially binds tRNAs with mature CCA 3′ termini (22, 23). Also, the 3′ CCA nucleotides are required for tRNA aminoacylation and tRNA nuclear aminoacylation facilitates tRNA nuclear export (15–17). Moreover, in Xenopus oocytes tRNAs lacking 3′ CCA nucleotides are not exported to the cytosol (15, 22). Finally, the cca1–1 mutant has been reported to accumulate tRNA in nuclei (16, 17). However, it was unknown whether CCA addition and/or tRNA aminoacylation functions in the yeast Los1p-dependent tRNA nuclear export pathway and/or in Los1-independent pathways (18). To learn whether CCA addition and/or tRNA aminoacylation function in series or in parallel with Los1p we tested whether overexpressed Cca1p would suppress los1 mutations. Overexpression of Cca1p should not suppress a los1 null mutation if they function in an obligatory series, but it could if they function in parallel.
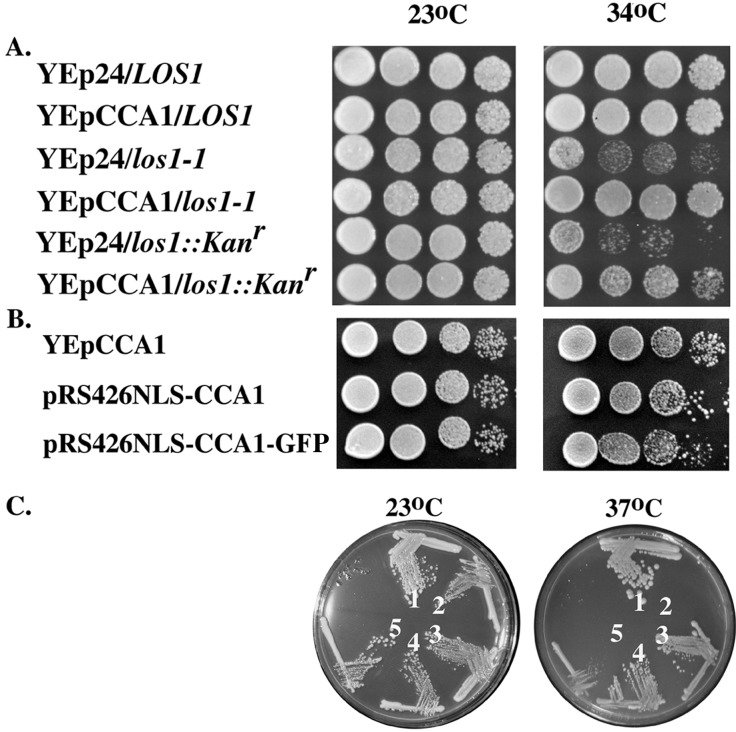
We first tested whether multicopy plasmids encoding CCA1 affect growth of cells with LOS1 mutations. Although LOS1 is unessential, mutations of it cause detectable phenotypes (14, 26). At elevated temperatures (34–38°C), los1 cells grow slower than the parental LOS1 strain, X2316–3C. The growth characteristics of LOS1, los1–1, and los1∷Kanr with vector alone or with the multicopy vector carrying CCA1 (YEpCCA1) were determined. Los1–1 and los1∷Kanr cells with YEpCCA1 grew better than the same strains with vector alone (Fig. 2A). Therefore, Cca1p functions as a multicopy suppressor of the growth defect caused by los1 mutations.
Figure 2.
(A) Overexpressed Cca1p suppresses the los1 growth defect. LOS1, los1–1, and los1∷Kanr cells containing YEp24 or YEpCCA1 were grown to saturation and serially diluted; aliquots were spotted onto solid rich media and incubated at 23°C or 34°C for 3 days. (B) Overexpressed nucleus-located Cca1p suppresses los1. Los1∷Kanr cells containing YEpCCA1, pRS426NLS-CCA1, or pRS426NLS-CCA1-GFP we tested for los1 suppression as in A. (C) pRS426NLS-CCA1 and pRS426NLS-CCA1-GFP encode active Cca1 proteins. LOS1 (designated 1) or cca1–1 (designated 2–5) cells containing pRS426 (designated 1, 2), YEpCCA1 (designated 3), pRS426NLS-CCA1 (designated 4), or pRS426NLS-CCA1-GFP (designated 5) were streaked for single colonies and were grown on rich media at 23°C and 37°C.
To determine whether suppression of the los1 growth defect relates to tRNA nuclear export, we determined the consequences of Cca1p overexpression upon tRNA nuclear export by using FISH. Los1ΔV (12) and los1∷Kanr (Fig. 1B, 1 and 1′) cells have defects in export of the intronless tRNAMet from the nucleus to the cytosol. The defect is suppressed by Cca1p overexpression provided by YEpCCA1 (Fig. 1B, 2 and 2′). The defect is also corrected by a multiple-copy vector containing LOS1 [YEpLOS1-ET (13); Fig. 1B, 3 and 3′]. Thus, Cca1p is a multicopy suppressor of the defect in tRNA nuclear export caused by a los1 null mutation, consistent with its suppression of the growth defect described above. Together the data support the model that Cca1p functions in a Los1p-independent pathway for tRNA nuclear export.
To address the mechanism by which this Los1p-independent tRNA nuclear export pathway operates, we tested whether it uses a member of the importin-β family. We screened yeast with mutations of genes encoding eight members including all known exportins and those importins implicated in RNA metabolism (Transportin/Kap104p, Yrb4p/Kap123p, Pse1p/Kap121p, Sxm1p/Kap108p, Mtr10p, Crm1p/Xpo1p, Cse1p and Msn5p; see Materials and Methods for strains) for defects in tRNA nuclear export. Only the xpo1–1/crm1–1 mutant showed tRNA nuclear accumulation, and it was weak and detectable only after 2 h at the nonpermissive temperature (data not shown). Because under these conditions Rna1p accumulates in yeast nuclei, the effect of xpo1–1/crm1–1 on tRNA nuclear export may be a secondary consequence of the predicted Ran-GTP nucleus/cytosol gradient collapse (42).
MES1 Is a Multicopy Suppressor of the los1∷Kanr Defect of tRNA Nuclear Export.
Aminoacyl tRNA synthetases (aaRS) catalyze addition of amino acids to the 3′ CCA termini of tRNAs. Because Cca1p seems to function in a Los1p-independent pathway for tRNA nuclear export, it is possible that tRNA nuclear aminoacylation also functions in a Los1p-independent pathway. As cells have 20 different aaRS, it is not expected that overexpression of a single one would suppress the growth defect of a los1 null mutation. However, overexpression of a single aaRS might suppress a nuclear export defect of its cognate tRNA.
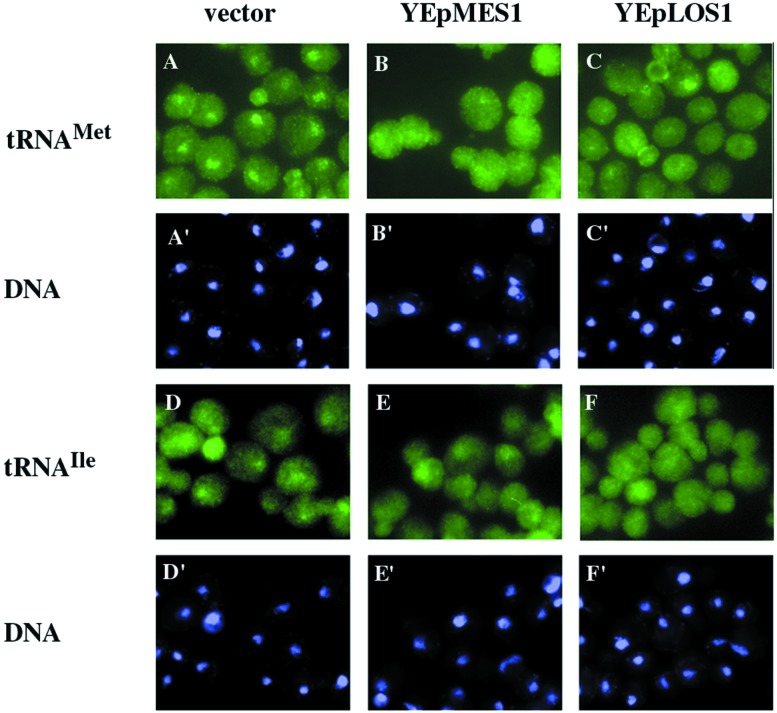
We tested whether overexpressed MES1, encoding methionyl-tRNA synthetase (43), suppresses the defect of intronless tRNAMet nuclear export in los1∷Kanr cells. As expected, los1∷Kanr cells with vector alone accumulate tRNAMet in the nucleus (Fig. 3 A and A′). In contrast, los1∷Kanr cells with YEpMES1 (43) show little tRNAMet nuclear accumulation (Fig. 3 B and B′) and seem similar to los1∷Kanr cells with YEpLOS1-ET (Fig. 3 C and C′). Overexpression of Mes1p suppresses the defect in tRNAMet nuclear export of a los1 null mutation. If suppression is caused by methionine addition to tRNA rather than to a nonspecific affect on cellular metabolism, then suppression may be specific to the cognate tRNAMet. To test this hypothesis, we assessed the location of another intronless tRNA, tRNAIleAAU. tRNAIleAAU accumulates in the nucleus of los1∷Kanr cells with YEp24 (Fig. 3 D and D′) and in nuclei of los1∷Kanr with YEpMES1 (Fig. 3 E and E′). However, little nuclear accumulation of tRNAIleAAU occurs in los1∷Kanr cells with YEpLOS1-ET (Fig. 3 F and F′). Therefore, MES1 seems to function as a cognate-specific multicopy suppressor of the los1∷Kanr defect of tRNAMet nuclear export. The data indicate that Mes1p may operate in a Los1p-independent pathway for tRNA nuclear export. Cca1p and Mes1p could either function in series with each other in a single Los1p-independent pathway for tRNA nuclear export, or they could function in parallel in separate Los1p-independent pathways.
Figure 3.
Mes1p is a cognate tRNA-specific multicopy suppressor of the los1∷Kanr tRNAMet nuclear export defect. FISH for los1∷Kanr cells containing YEp24 (A-A′ and D-D′), YEpMES1 (B-B′ and E-E′), or YEpLOS1-ET (C-C′ and F-F′). Cells in A–C were hybridized with an oligonucleotide specific to tRNAMet; D–F were hybridized with an oligonucleotide specific for tRNAIleAAU. A′–F′, the same cells stained with 4′,6-diamidino-2-phenylindole.
CCA1 Is Not a Multicopy Suppressor of Defective pre-tRNA Splicing Caused by los1 Mutations.
LOS1 was initially identified as a gene involved in pre-tRNA splicing (26). It is not clear why a tRNA exportin should cause defects in pre-tRNA splicing that precedes nuclear export. Nevertheless, the data are consistent with other reports showing that mutations of the RanGTPase pathway [rna1–1 (27) and prp20 (28)] and alterations of genes encoding NPC components (29) affect pre-tRNA splicing as well as tRNA nuclear export. Because Cca1p seems to function in a Los1p-independent pathway for tRNA nuclear export, we were interested in determining whether pre-tRNA splicing and tRNA nuclear export are coupled in this pathway as they are in the Los1p-dependent pathway. If so, multicopy Cca1p should correct the splicing defect of los1∷Kanr cells; if pre-tRNA splicing and tRNA nuclear export are not coupled, multicopy Cca1p should not correct the splicing defect of los1∷Kanr cells.
We performed Northern blot analyses to determine the ratios of end-matured, intervening sequence (intron) (IVS)-containing pre-tRNA (IVS-tRNAs) to total tRNAs by using probes specific to the intron-containing tRNAIleUAU family (12). LOS1 and los1∷Kanr cells bearing YEp24, YEpCCA1, or YEpLOS1-ET were shifted to elevated temperatures (34°C, 37°C, and 38°C) for 1–2 h before RNA isolation. LOS1 cells with YEp24, YEpCCA1, or YEpLOS1, or los1∷Kanr cells with YEpLOS1 have low levels of IVS-tRNAs (≈10%; Fig. 4, data shown only for 37°C). As predicted, los1∷Kanr cells with YEp24 accumulate greater amounts of IVS-tRNAs (30%). No significant difference existed in the relative levels of IVS-tRNAs between los1∷Kanr cells with YEp24 and los1∷Kanr cells with YEpCCA1 (34%; Fig. 4); so, Cca1p overexpression does not correct the los1∷Kanr pre-tRNA-splicing defect. The data indicate that pre-tRNA splicing and tRNA nuclear export are uncoupled in the Los1p-independent pathway.
Figure 4.
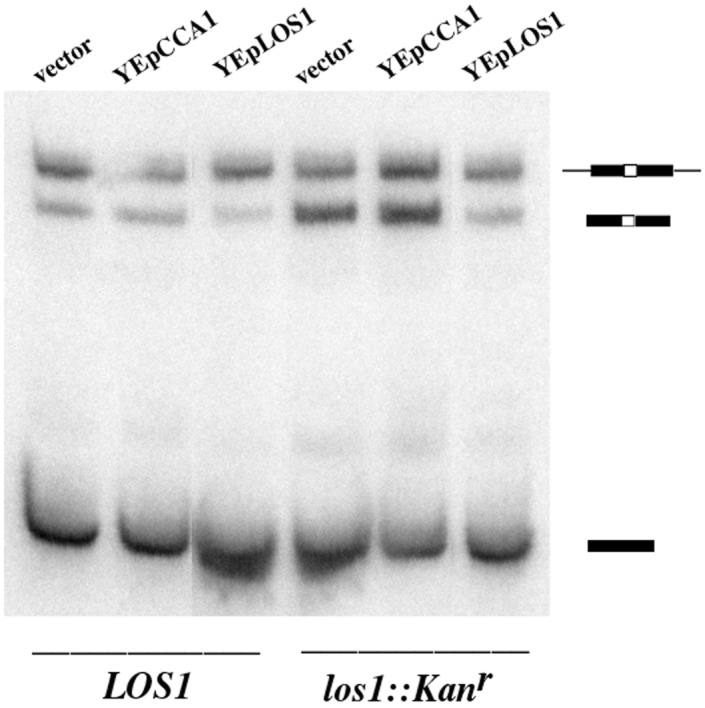
Multicopy Cca1p does not suppress the los1 pre-tRNA-splicing defect. RNAs were prepared from LOS1 (X2316–3C) and los1∷kanr (SS700) cells containing YEp24, YEpCCA1, or YEpLOS1-ET. The RNAs were transferred to membranes and hybridized with an oligonucleotide specific for tRNAIleUAU. The identity of the initial transcripts, IVS-tRNAs and mature tRNAs are indicated in cartoon form.
Cca1p Seems to Be a Nucleus/Cytosol-Shuttling Protein.
We considered possible mechanisms by which Cca1p could participate in a Los1p-independent tRNA nuclear export pathway. It could act catalytically, preparing tRNA for interaction with an unknown exportin. Alternatively, Cca1p could function as an “exportin” to move tRNA to the cytosol or as an adapter for an unidentified “exportin.” The latter possibilities predict that Cca1p would shuttle between the nucleus and the cytosol.
To test whether Cca1p is a nucleus/cytosol-shuttling protein we used a heterokaryon assay (37) by using the kar1–1 mutation that prevents nuclear fusion in zygotes. Movement of a reporter protein from a KAR1 nucleus to the kar1 nucleus, in the absence of new synthesis, is indicative of nucleus/cytosol shuttling. This assay requires that the reporter protein be primarily nucleoplasmic so that movement of reporter cytosolic pools from the joint cytoplasm to the second nucleus cannot occur. Assessment of Cca1p nucleus/cytosol shuttling by this assay is complicated by the fact that Cca1p is a sorting isozyme residing in the nucleus, cytoplasm, and mitochondria (21). CCA1 has three in-frame ATGs; Cca1p-I, translated from AUG1, is primarily located in mitochondria, whereas Cca1p-II and III, translated from AUG2 and AUG3, respectively, are primarily located in the cytosol with small functional nuclear pools (21). To perform the heterokaryon analysis for Cca1p, we first created a CCA1 allele encoding a modified Cca1p-III residing primarily in the nucleus. Addition of the simian virus 40 (SV40) large T antigen NLS to Cca1p-III (NLS-Cca1p) generated a nucleus located catalytically active Cca1p able to complement cca1–1 and suppress los1 when expressed from pRS426 (ref. 21; Fig. 2 B and C; data not shown). Addition of GFP at the NLS-Cca1p carboxyl terminus generated a tagged version of Cca1p efficiently located to nuclei with slightly reduced ability to complement cca1–1 (Fig. 2 B and C). GFP tagged NLS-Cca1p was inserted into pRS416GAL1/10F (James E. Hopper, personal communication) generating a protein tightly controlled by the GAL1 promoter (data not shown) allowing manipulation of conditions to prevent synthesis of Cca1p during mating.
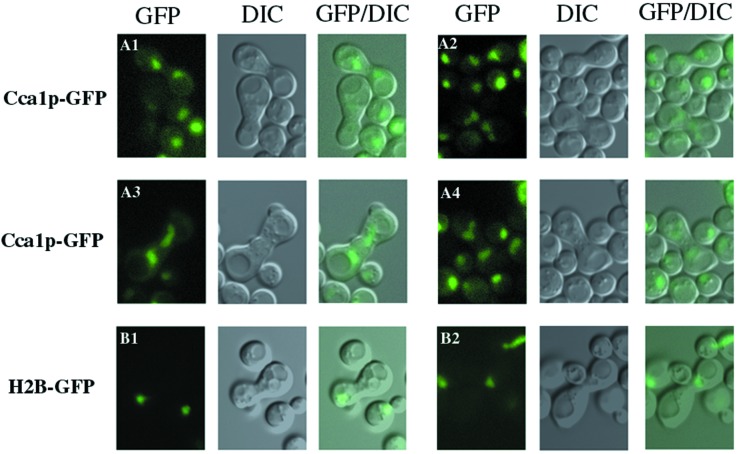
NLS-Cca1p-GFP synthesis was induced for 2 h in SS330 cells; synthesis was terminated by change in the carbon source and then the cells were mated to the kar1–1 cells. As a control, the location of histone 2B (H2B)-GFP (37) was monitored. As H2B-GFP is not a shuttling protein, it was expected to remain in the KAR1 nucleus. For heterokaryons generated by mating SS330 cells containing H2B-GFP to kar1–1 cells, the GFP signal was in only one nucleus in all observed zygotes as viewed in live cells (Fig. 5 B1 and B2) or by immunofluorescence (data not shown). In contrast, for heterokaryons generated by mating SS330 cells containing NLS-Cca1p-GFP with kar1–1 cells, the GFP signal distributed equally between both nuclei (Fig. 5 A1–A4; data not shown). Thus, NLS-Cca1p can move between the KAR1 and the newly introduced kar1–1 nucleus, supporting the notion that Cca1p may be a nucleus/cytosol-shuttling protein, thereby meeting a requirement for a direct role in tRNA nuclear export.
Figure 5.
Cca1p can shuttle to an introduced nucleus in heterokaryons. Wild-type (SS330) cells containing SV40NLS-CCA1-III-GFP-pRS416GAL1/10F or pGAL-H2B-GFP pregrown in media containing galactose were transferred to media containing glucose and were mated to kar1–1 cells (MS739). After zygotes were formed (≈1.5 h), the locations of the GFP-tagged proteins in live cells were visualized. (Left) A1–A4 and B1–B2, images of NLS-Cca1p-III-GFP and H2B-GFP, respectively, viewed in the GFP channel; (Center) differential interference contrast microscopy of the same cells; (Right) converged GFP/differential interference contrast microscopy images.
Discussion
Redundant Pathways for tRNA Nuclear Export.
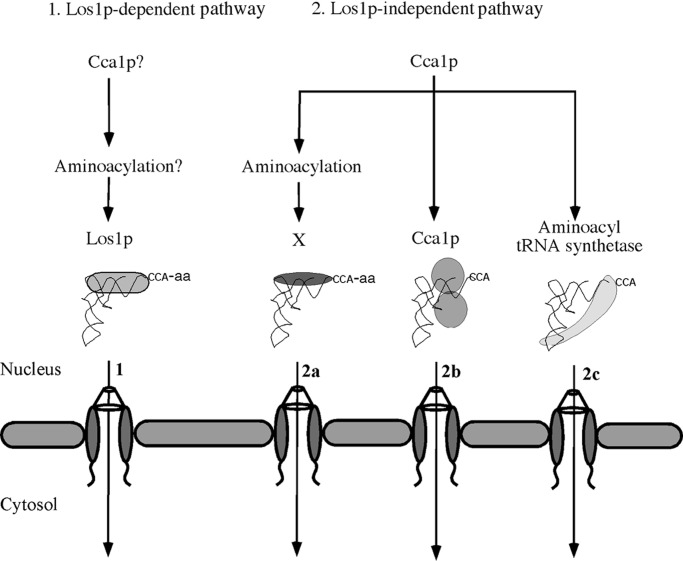
We report that the cca1–1 mutation causes defects in tRNA nuclear export and that CCA1 and MES1 (in a cognate tRNA-specific fashion) function as multicopy suppressors of the los1∷Kanr defect of tRNA nuclear export. The data support the notion that Cca1p and tRNA nuclear aminoacylation can function in a Los1p-independent tRNA nuclear export pathway, leading to the model that yeast cells possess at least two tRNA nuclear export pathways for intronless tRNAs (Fig. 6). Do vertebrate cells also possess exportin-t-independent tRNA nuclear export pathways? It is unknown whether exportin-t is unessential for life in vertebrates, like it is for yeast. However, the existing data indicate that exportin-t provides the major export pathway for tRNAs in Xenopus oocytes because anti-exportin-t injection into nuclei reduced tRNA export by >80% (22, 23). Nevertheless, residual tRNA export, regardless of the amount of introduced antibody, could indicate that Xenopus, like yeast, possesses an alternative tRNA nuclear export pathway(s).
Figure 6.
Models for tRNA nuclear export (1). Los1p-dependent pathway. It is unknown whether Cca1p and nuclear tRNA aminoacylation function in this pathway in yeast (2). Three possible mechanisms for the Los1p-independent pathway. (2a) Cca1p and nuclear aminoacylation function in series to prepare tRNAs for interaction with an unknown “exportin”; (2b) Cca1p interacts with tRNA, remains bound after catalysis, and ferries tRNA to the cytosol; (2c) a mechanism similar to 2b in which putative shuttling of aaRS serves to export tRNA via stable aaRS/tRNA complexes.
Cca1p and aaRS may function in both the Los1p-dependent and the Los1p-independent pathways. It is unknown whether Los1p, like exportin-t, preferentially binds to tRNAs with mature CCA 3′ termini (22, 23). Also, although exportin-t binds uncharged tRNAs in vitro (22, 23), no data address the relative affinities of exportin-t or Los1p to aminoacylated versus nonaminoacylated tRNAs. Our genetic studies provide no evidence for synthetic interactions between los1 null and cca1–1 ts mutations (data not shown). Lack of synthetic interactions is compatible with either the model that Cca1p functions in both Los1p-dependent and Los1p-independent pathways or with the model that there are more than two export pathways. Nevertheless, we showed that most yeast nuclear tRNAs seem to be aminoacylated when nuclear export is inhibited (17), indicating that under these conditions there are sufficient nuclear aaRS pools to charge most nuclear tRNAs. Therefore, most yeast tRNAs encountered by all export pathways likely possess 3′ CCA nucleotides and are aminoacylated.
The most straightforward mechanism for the Los1p-independent pathway would be for Cca1p and aaRS to act catalytically, modifying tRNA transcripts for efficient binding to an unknown importin-β family member as it does for exportin-t in vertebrate cells (Fig. 6 2a). However, we were unable to identify a member of this family other than Los1p that affects tRNA nuclear export. Vertebrate cells may possess a minor tRNA nuclear export pathway that is Ran-independent, because tRNA nuclear export is less sensitive to perturbations of Ran cycle than are other RNAs, such as snRNAs in Xenopus oocytes (44, 45). If the yeast Los1p-independent pathway is Ran-independent, CCA addition to nuclear tRNAs may prepare them for interactions with a RNA-binding protein that mediates tRNA export. In this case, this tRNA export pathway would resemble the currently favored model for Ran-independent mRNA nuclear export (5).
On the other hand, we show that Cca1p is capable of nucleus/cytosol shuttling. If Cca1p remained bound to tRNAs long enough, it might export tRNA in tow upon exiting the nucleus (Fig. 6 2b). However, because genome wide two-hybrid analyses provide no evidence for Cca1p interactions with nucleoporins (46), Cca1p could function as an adapter for an unknown protein that interacts with the NPC. These possibilities require the unusual behavior for an enzyme (Cca1p) to remain associated with its product after catalysis. The Sulfolobus shibatae CCA-adding enzyme forms stable complexes with mature tRNA-CCA (47) providing precedence for this behavior. Alternatively, Cca1p shuttling may function solely for its appropriate nuclear and cytosolic subcellular distribution. These alternatives could be distinguished by using Cca1p mutants that fail to shuttle. Unfortunately, such mutants are not yet available because the Cca1p NLS seems not to be one of the characterized motifs (21) and the nuclear export sequence is not sensitive to Crm1p (data not shown).
We showed that small nuclear pools of tyrosyl-tRNA synthetase, Tys1p, exist. The Tys1p NLS is sufficient to target a large passenger protein into nuclei even though most endogenous Tys1p is cytosolic (18). Thus, either Tys1p is tethered in the cytosol as has been recently reported for methionyl- and glutamyl-tRNA synthetases (48) or it can shuttle between the nucleus and the cytosol. In the latter case, Tys1p and possibly other aaRS theoretically could bind and export tRNA in tow, thereby generating numerous parallel export pathways (Fig. 6 2c).
Coupling of tRNA Splicing and tRNA Nuclear Export.
tRNA nuclear export in yeast is thought to be tightly coupled to pre-tRNA splicing because mutations affecting Los1p-dependent tRNA nuclear export cause IVS-tRNA accumulation. Nevertheless, this coupling cannot be absolute because spliced cytosolic pools of functional tRNA are essential and Los1p is unessential. The situation for yeast is in contrast to Xenopus oocytes for which depletion of exportin-t or the Ran-GTP nucleus/cytosol gradient has no affect upon pre-tRNA splicing (15, 22). The differences in processing pathways between these systems might account for the differences in the presence/absence of the coupling of splicing and nuclear export. In Xenopus, tRNA splicing usually precedes end-processing (15), whereas in yeast end-processing usually precedes splicing, although spliced tRNAs with unprocessed termini can be detected (26, 27, 49). Lund and Dahlberg (15) proposed that combinations of the tRNA kinetic processing pathway and proofreading of tRNA end-processing by aaRS assures that most tRNAs exported to the cytosol are correctly matured. We report that although overexpressed Cca1p does not suppress the pre-tRNA splicing defect of los1∷Kanr cells, it mediates export of intronless tRNAs to the cytosol. Moreover, we reported that cca1–1 mutants do not accumulate unspliced pre-tRNAs (17). Thus, the Los1p-independent pathway resembles the Xenopus exportin-t pathway in that it appears to be uncoupled from pre-tRNA splicing. Perhaps, tRNA aminoacylation serves a proofreading role for this yeast pathway as it does for Xenopus oocytes.
Curiously, Cca1p can catalyze CCA 3′ addition to IVS-tRNAs (21), even though overexpressed Cca1p fails to suppress the los1 pre-tRNA-splicing defect. A possible mechanism for this would be for overexpressed Cca1p to cause nuclear export of IVS-tRNAs, separating them from the splicing endonuclease located in the inner nuclear membrane (50), similar to the consequences of overproduction of exportin-t or tRNAs in vertebrate cells (15, 22). However, because we were unable to detect decreased nuclear pools or increased cytosolic pools of IVS-tRNAs in los1∷Kanr cells containing YEpCCA1 (data not shown), we think this possibility is unlikely. Another possibility is that the Los1p-independent pathway is dedicated solely to export of tRNAs encoded by genes lacking IVSs, which, however, is inconsistent with the ability of overexpressed Cca1p to suppress the los1 null growth defect. Perhaps, Cca1p also functions to export tRNAs encoded by IVS-containing genes that are processed by the splicing-first pathway.
How are pre-tRNA splicing and tRNA nuclear export coupled in the Los1p-dependent pathway? Considering the recently proposed model for coupling pre-mRNA splicing and nonsense mediated decay (ref. 51 and references therein), we propose a similar model for the spliced last kinetic pathway whereby tRNA introns are marked by a tRNA nuclear retention protein(s) that is removed by components of the Los1p-dependent tRNA pathway when IVS-tRNAs reach the inner nuclear membrane. Accordingly, the Los1p-dependent tRNA nuclear export pathway would also provide a proofreading role for tRNAs whose termini are processed before intron removal, assuring that only mature tRNAs are exported to the cytosol.
Acknowledgments
We thank J. Abelson for the ts yeast collection, N. C. Martin for SV40NLS-Cca1p-IIIpRS426, G. Peng for pRS416Gal1/10F, R. Y. Tsien for pSEY181GAL1-10/GFP, and G. R. Fink for YEpMES1. We acknowledge M. Fitzgerald-Hayes, C. Guthrie, M. Johnston, L. Pemberton, M. Rose, S. Sarkar, P. Silver, and S. Wente for providing yeast strains. We thank A. Weiner for insightful discussions, A. Azad and S. Sarkar for comments on the manuscript, and D. Stanford and other members of the A.K.H. laboratory for numerous scientific interactions and help in manuscript preparation. This work was supported by Public Health Service Grant GM27930 from the National Institutes of Health to A.K.H.
Abbreviations
- NPC
nuclear pore complex
- FISH
fluorescence in situ hybridization
- ts
thermosensitive
- NLS
nuclear localization sequence
- GFP
green fluorescent protein
- aaRS
aminoacyl tRNA synthetase
- IVS
intervening sequence or intron
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Görlich D, Kutay U. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida K, Blobel G. J Cell Biol. 2001;152:729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rout M P, Blobel G, Aitchison J D. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 4.Mosammaparast N, Jackson K R, Guo Y, Brame C J, Shabanowitz J, Hunt D F, Pemberton L F. J Cell Biol. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole C N. Nat Cell Biol. 2000;2:E55–E58. doi: 10.1038/35008681. [DOI] [PubMed] [Google Scholar]
- 6.Gallouzi I E, Steitz J A. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Bogerd H P, Wang P J, Page D C, Cullen B R. Mol Cell. 2001;8:397–406. doi: 10.1016/s1097-2765(01)00303-3. [DOI] [PubMed] [Google Scholar]
- 8.Görlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arts G J, Fornerod M, Mattaj I W. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 10.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartmann E, Görlich D. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 11.Hellmuth K, Lau D M, Bischoff F R, Kunzler M, Hurt E, Simos G. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar S, Hopper A K. Mol Biol Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen W C, Selvakumar D, Stanford D R, Hopper A K. J Biol Chem. 1993;268:19436–19444. [PubMed] [Google Scholar]
- 14.Hurt D J, Wang S S, Lin Y H, Hopper A K. Mol Cell Biol. 1987;7:1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund E, Dahlberg J E. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 16.Grosshans H, Hurt E, Simos G. Genes Dev. 2000;14:830–840. [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar S, Azad A K, Hopper A K. Proc Natl Acad Sci USA. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azad A K, Stanford D R, Sarkar S, Hopper A K. Mol Biol Cell. 2001;12:1381–1392. doi: 10.1091/mbc.12.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vijayraghavan U, Company M, Abelson J. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 20.Aebi M, Kirchner G, Chen J Y, Vijayraghavan U, Jacobson A, Martin N C, Abelson J. J Biol Chem. 1990;265:16216–16220. [PubMed] [Google Scholar]
- 21.Wolfe C L, Hopper A K, Martin N C. J Biol Chem. 1996;271:4679–4686. doi: 10.1074/jbc.271.9.4679. [DOI] [PubMed] [Google Scholar]
- 22.Arts G J, Kuersten S, Romby P, Ehresmann B, Mattaj I W. EMBO J. 1998;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipowsky G, Bischoff F R, Izaurralde E, Kutay U, Schäfer S, Gross H J, Beier H, Görlich D. RNA. 1999;5:539–549. doi: 10.1017/s1355838299982134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo M L, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Nature (London) 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 25.Sträβer K, Hurt E. Nature (London) 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 26.Hopper A K, Schultz L D, Shapiro R A. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- 27.Hopper A K, Banks F, Evangelides V. Cell. 1978;14:211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 28.Kadowaki T, Goldfarb D, Spitz L M, Tartakoff A M, Ohno M. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma K, Fabre E, Tekotte H, Hurt E C, Tollervey D. Mol Cell Biol. 1996;16:294–301. doi: 10.1128/mcb.16.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wente S R, Blobel G. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Z, McGrew J T, Schroeder A J, Fitzgerald-Hayes M. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seedorf M, Silver P A. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aitchison J D, Blobel G, Rout M P. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 34.Pemberton L F, Rosenblum J S, Blobel G. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 36.Vallen E A, Hiller M A, Scherson T Y, Rose M D. J Cell Biol. 1992;117:1277–1287. doi: 10.1083/jcb.117.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins D A, Silver P A. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng G, Hopper J E. Mol Cell Biol. 2000;20:5140–5148. doi: 10.1128/mcb.20.14.5140-5148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hopper A K, Traglia H M, Dunst R W. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlson M, Botstein D. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 41.Kuo C-L, Campbell J L. Mol Cell Biol. 1983;3:1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng W, Benko A L, Lee J-H, Stanford D R, Hopper A K. J Cell Sci. 1999;112:339–347. doi: 10.1242/jcs.112.3.339. [DOI] [PubMed] [Google Scholar]
- 43.Meussdoerffer F, Fink G R. J Biol Chem. 1983;258:6293–6299. [PubMed] [Google Scholar]
- 44.Cheng Y, Dahlberg J E, Lund E. Science. 1995;267:1807–1810. doi: 10.1126/science.7534442. [DOI] [PubMed] [Google Scholar]
- 45.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Görlich D. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature (London) 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 47.Shi P-Y, Maizels N, Weiner A M. EMBO J. 1998;17:3197–3206. doi: 10.1093/emboj/17.11.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galani K, Groβhans H, Deinert K, Hurt E C, Simos G. EMBO J. 2001;20:6889–6898. doi: 10.1093/emboj/20.23.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Connor J P, Peebles C L. Mol Cell Biol. 1991;11:425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peebles C L, Gegenheimer P, Abelson J. Cell. 1983;32:525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- 51.Ishigaki Y, Li X, Serin G, Maquat L E. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]