Abstract
The Suppressor of Fused [Su(fu)] protein plays a conserved role in the regulation of Gli transcription factors of the hedgehog (Hh) signaling pathway that controls cell fate and tissue patterning during development. In both Drosophila and mammals, Su(fu) represses Gli-mediated transcription, but the mode of its action is not completely understood. Recent evidence suggests that Su(fu) physically interacts with the Gli proteins and, when overexpressed, sequesters Gli in the cytoplasm. However, Su(fu) also traverses into the nucleus under the influence of a serine-threonine kinase, Fused (Fu), and has the ability to form a DNA-binding complex with Gli, suggesting that it has a nuclear function. Here we report that the mouse homolog of Su(fu) [mSu(fu)] specifically interacts with SAP18, a component of the mSin3 and histone deacetylase complex. In addition, we demonstrate that mSu(fu) functionally cooperates with SAP18 to repress transcription by recruiting the SAP18-mSin3 complex to promoters containing the Gli-binding element. These results provide biochemical evidence that Su(fu) directly participates in modulating the transcriptional activity of Gli.
The Gli transcription factors are key intracellular mediators of the secreted hedgehog (Hh) protein that controls cell growth and tissue patterning during development of both vertebrates and invertebrates (1–3). All members of the Gli family proteins contain a highly conserved zinc-finger DNA-binding domain that recognizes a common DNA element (4). However, Gli proteins can either activate or repress transcription of Hh target genes, depending on whether they are stabilized as full-length transcription activators in the presence of Hh or proteolytically cleaved into truncated transcription repressors in the absence of Hh (5–7). Balancing the activation and repressive activity of Gli both spatially and temporally is an essential function of Hh signaling in shaping tissue pattern formation and cell fate induction during development.
In the receiving cells, Hh signaling is mediated by two transmembrane receptors, Patched (Ptc) and Smoothened (Smo). Some evidence suggests that Hh is capable of direct binding to Ptc (8, 9), which is believed to be in a complex with Smo and thereby inhibits the function of Smo (3). Binding of Hh to Ptc releases the otherwise active Smo from inhibition by Ptc. Many proteins, including Costal2 (Cos2), Fused (Fu), protein kinase A, and Slimb, participate in the signaling events downstream of Smo and control the function of Gli (10–13). Genetic and biochemical evidence from Drosophila has indicated that in the absence of an Hh signal, the Drosophila Gli, Cubitus interruptus (Ci), forms a large microtubule-binding complex with the kinesin-like protein, Cos2, and the serine-threonine kinase, Fu (10, 11). The precise function of this complex is not clear but it may be involved in proteolytic cleavage of Ci (6). The cleavage of Ci is also dependent on phosphorylation by protein kinase A and requires the action of a ubiquitin ligase, Slimb (13). In the presence of an Hh signal, the cleavage of Ci is blocked and the stabilized full-length Ci transverses into the nucleus to activate gene transcription (6). Although some counterparts of this complex have been found in mammals (14), the vertebrate Hh pathway is complicated by the presence of at least three Gli genes. Like Drosophila Ci, Gli3 undergoes a similar proteolytic cleavage that is subject to regulation by Hh; in contrast, Gli1 is not cleaved and plays primarily the role of a transcriptional activator (7). It remains to be determined whether Gli2 is subject to proteolytic cleavage in vertebrate cells, although Gli2 shares both overlapping and nonredundant functions with Gli3 (7, 15).
The Suppressor of Fused [Su(fu)] gene was originally identified in Drosophila based on its ability to rescue the mutant phenotypes of the fu alleles that cause undergrowth of wing tissue (16). This gene encodes another protein that binds Gli/Ci, but its mode of function remains enigmatic (17, 18). During Drosophila wing development, preventing the proteolytic cleavage is essential for activating Ci in response to Hh signaling but not sufficient, because the full-length Ci protein is still kept in a stable, inactive form by the action of Su(fu) (19). Fu is required to release the inhibition of Su(fu) and restore the full transcriptional activity of Ci (14, 20, 21). Several recent studies with Drosophila as well as cultured mammalian cells indicated that, when overexpressed, Su(fu) causes cytoplasmic sequestration of Gli/Ci (14, 20–22). However, the fact that Su(fu) physically interacts with the N-terminal repressor domain of Gli and forms a DNA-binding complex by means of this interaction (17, 18) raises the possibility that Su(fu) directly participates in modulating the transcriptional activity of Gli.
Here we report that SAP18, a member of the mSin3-histone deacetylase (HDAC) corepressor complex (23), is an interacting partner of mouse Suppressor of Fused [mSu(fu)]. In addition, we demonstrate that Gli, mSu(fu), SAP18, and mSin3 are capable of forming protein complex on a DNA oligo containing the Gli-binding element. Our data showed that mSu(fu) represses Gli-mediated transcription by recruiting the mSin3-HDAC corepressor complex to promoters containing the Gli-binding elements. Thus, our results provide biochemical evidence for a nuclear role of Su(fu) in repressing the transcriptional activity of Gli.
Materials and Methods
Construction of Mammalian Expression Plasmids.
BLAST searches were performed with the Drosophila Su(fu) protein sequence against a database of expressed sequence tags to obtain partial cDNA sequences for mouse and human Su(fu). I.M.A.G.E. Consortium cDNA clones (513730, 650817) were obtained from Research Genetics (Huntsville, AL) and sequenced. Full-length cDNAs for mouse Su(fu) were obtained by screening a mouse embryo (E11) cDNA library. Myc-tagged mSu(fu) was generated by PCR and subcloned into the cytomegalovirus promoter-driven mammalian expression vector.
Hemagglutinin (HA)-tagged Gli1 and Gli3 plasmids were constructed by inserting the full-length human Gli1 and Gli3 cDNA into pRKHA, a pRK5 derivative with the HA epitope tag (amino acids CYPYDVPDYASL) under a cytomegalovirus promoter. FLAG-SAP18 (23) was a generous gift from D. Reinberg (University of Medicine and Dentistry of New Jersey). A mammalian expression vector for sonic hedgehog was obtained from H. Roelink (University of Washington, Seattle); a constitutively active protein kinase A construct was from Stratagene.
Yeast Two-Hybrid Library Screen and Interaction Assay.
The Lex-A-based yeast two-hybrid system was used (24). Bait plasmid LexA-mSu(fu), which encodes the LexA DNA-binding domain fused to full-length mouse mSu(fu), was constructed by subcloning into pEG202. A HeLa cell cDNA library fused to B42 acidic activation domain on pJG4–5 was a gift from R. Brent (Molecular Science Institute, Berkeley, CA). A library screen was performed with yeast strain EGY48/pSH18–4. EGY48/pSH18–4 cells were cotransformed with LexA-mSu(fu) and 100 μg of library plasmids, and plated to Ura−His−Trp− glucose plates. Colonies were replated to Ura−His−Trp−Leu− galactose plates, and positives were picked after 2–4 days. DNA from clones that were positive for β-galactosidase on X-Gal plates in the presence of galactose were isolated from yeast and transformed into Escherichia coli KC8 cells for recovering the plasmid.
For the protein interaction assay, yeast strain EGY48/pSH18–4 was transformed with combinations of a bait plasmid [mSu(fu), or transforming growth factor-β receptor in pEG202], and a prey (SAP18 or FKBP12 in pJG4–5). Yeast transformants were selected on Ura−His−Trp− plates, and protein–protein interactions were determined by scoring for β-galactosidase activity.
Glutathione S-Transferase (GST)-Fusion Proteins and in Vitro Protein-Binding Assays.
The GST fusion proteins were expressed in E. coli and purified according to the protocol of Amersham Pharmacia. The Su(fu), mSin3A, or luciferase proteins were in a coupled in vitro transcription and translation reaction with the rabbit reticulocyte lysate (Promega) and labeled with [35S]methionine. The recombinant GST-fusion proteins that were preabsorbed to glutathione-Sepharose beads were then mixed with the in vitro translated and 35S-labeled protein. The binding reaction was performed for 1 h at 4°C in 20 mM Hepes, pH 7.95/100 mM NaCl/0.1 mM EDTA/2.5 mM MgCl2/1 mM DTT/0.05% Nonidet P-40/1% skimmed milk and protease inhibitors. Specific binding of proteins was then detected by SDS/PAGE and autoradiography.
Transfection, Immunoprecipitation, and Western Blotting.
HEK293T cells were maintained in DMEM containing 10% FBS and transfected by using Lipofectamine (Life Technologies, Grand Island, NY) according to the manufacturer's recommendation. Forty-eight hours after transfection, cells were lysed in 25 mM Tris⋅HCl, pH 8.0/300 mM NaCl/1% Triton X-100, and the lysates were then subjected to immunoprecipitation with anti-FLAG M2 antibody (Sigma), followed by adsorption to protein G-Sepharose (Amersham Pharmacia). Immunoprecipitated complexes were separated by SDS/PAGE and transferred to poly(vinylidene difluoride) membranes. Tagged proteins were detected by immunoblotting with either anti-FLAG M2 antibody or anti-Myc 9E10 antibody (Santa Cruz Biotechnology) and chemiluminescence (Amersham Pharmacia).
Transcriptional Response Assay.
To measure Gli-mediated transcriptional activation, the luciferase reporter construct, 8xGli-BS-Luc (25), that contains eight copies of Gli-binding elements was transfected into HEK293 cells in conjunction of Gli or various other expression plasmids. As an internal control for transfection efficiency and basal transcriptional response, a Renilla luciferase reporter, pTK-RL (Promega), whose expression is driven by the housekeeping thymidine kinase gene promoter was included in all transfection samples. For each transfection experiment, 0.5 μg of 8xGli-BS-Luc reporter, 0.01 μg of pTK-RL, and 0.1 μg of HA-Gli1 or HA-Gli3 plasmid or other relevant plasmids were used. Empty-vector DNA was added to keep the total amount of DNA in each sample constant. Forty-eight hours after transfection, the cells were lysed and luciferase and Renilla luciferase activities were measured by using the Promega dual luciferase assay kit. Whenever required, trichostatin A dissolved in 100% ethanol was added to the culture media 24 h before measuring the luciferase activity.
DNA Affinity Purification of Associated Proteins.
The nuclear extracts of the transfected HEK293 cells were prepared by harvesting cells in a hypotonic lysis buffer (20 mM Hepes, pH 7.9/0.1 mM EDTA/50 mM KCl/10% glucerol/2 mM DTT/0.15 mM spermine/0.5 mM spermidine and protease and phosphatase inhibitors). After Dounce homogenization, cell nuclei were collected by low-speed centrifugation. The nuclear proteins were extracted from the resuspended cell pellets on ice in a hypertonic buffer (20 mM Hepes, pH 7.9/0.1 mM EDTA/600 mM KCl/20% glycerol/2 mM DTT with protease and phosphatase inhibitors), followed by three cycles of freezing in liquid nitrogen and thawing on ice. After clearing off cell debris by high-speed centrifugation, the supernatant was taken as nuclear extract. To purify proteins that specifically bind the Gli-binding sequence, nuclear extracts were incubated with 200 ng of biotinylated wild-type (AGG CTA ACA AGC AGG GAC CAC CCA AGT AGA AGC TGG) or mutant (AGG CTA ACA AGC AGG GAC gtg ggA AGT AGA AGC TGG) DNA oligonucleotides. To block the nonspecific binding, 2 μg/ml poly(dI-dC) were added in the binding mixture. After incubation at 4°C for 1 h, the oligonucleotides were absorbed on streptavidin coated magnetic beads and washed extensively. The binding proteins retained on the beads were recovered by resuspending in SDS sample buffer and analyzed by protein blot. HA-tagged Gli1, myc-tagged mSu(fu), and Flag-tagged SAP18 were detected by immunoblotting, by using HA11 (Covance), myc 9E10 (Santa Cruz Biotechnology), or FLAG M2 antibodies (Sigma), respectively. The endogenous mSin3 protein was detected by using the mSin3A K20 antibodies (Santa Cruz Biotechnology).
Results
Physical Interaction Between Su(fu) and SAP18.
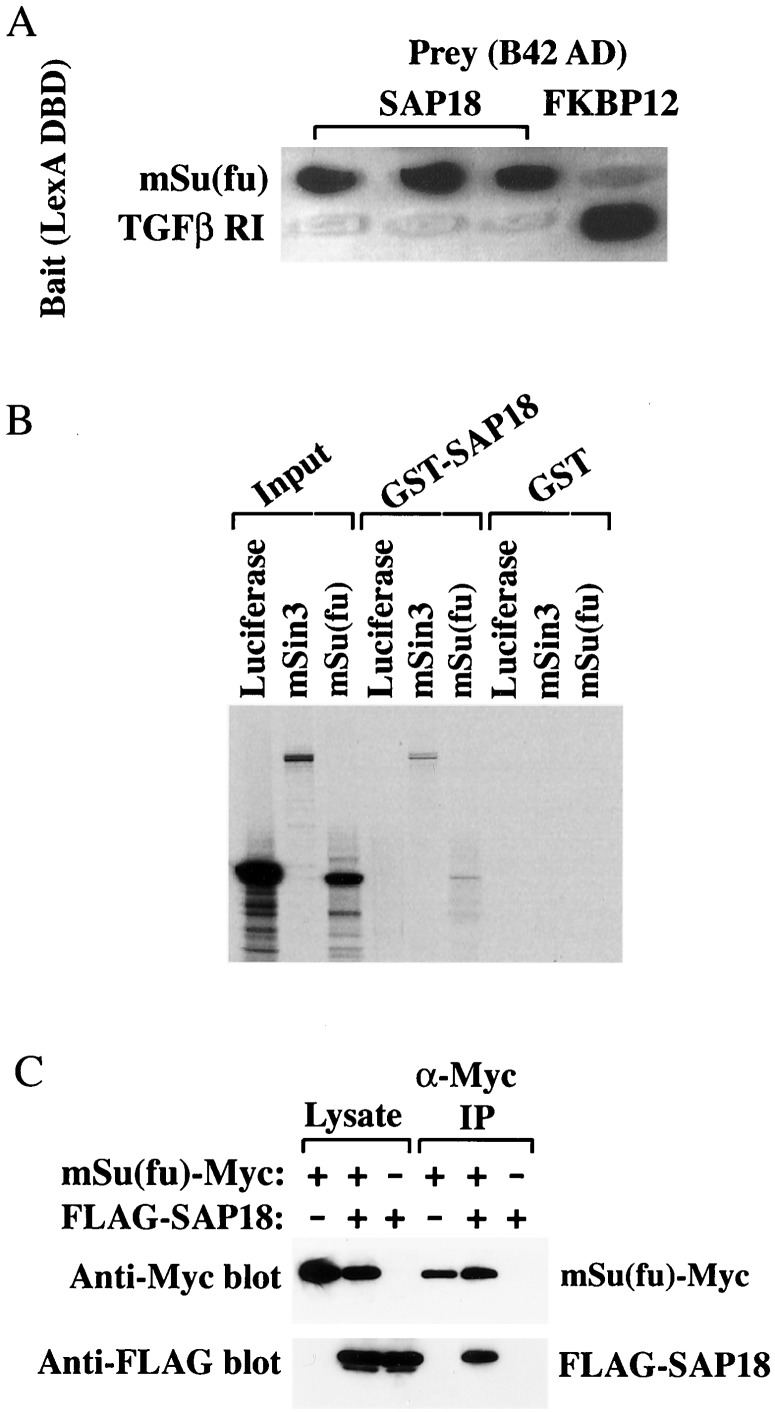
To gain insight into the function of Su(fu), we sought Su(fu)-interacting proteins by using a yeast two-hybrid screen. We isolated a full-length mSu(fu) cDNA from a mouse embryonic cDNA library based on the sequence of an expressed sequence tags entry in GenBank and fused this cDNA to the coding sequence of the DNA-binding domain of LexA to generate the “bait” construct. This fusion showed weak interaction with the Drosophila Fu and had a relatively low background activity in a yeast two-hybrid assay (data not shown). A library screen was performed by using a HeLa cell cDNA library fused to the B42 acidic activation domain on pJG4–5 in yeast strain EGY48/pSH18–4. Of 42 positive clones isolated, six contained sequences matched to that of SAP18, a component of the mammalian Sin3 (mSin3) transcription corepressor complex (23). mSu(fu) bound strongly to SAP18 (Fig. 1A), with an affinity similar to that of the interaction between transforming growth factor-β type I receptor and FK506-binding protein, FKBP12, a high-affinity interaction that served as a positive control (26) (Fig. 1A).
Figure 1.
Physical interaction between mSu(fu) and SAP18. (A) Association of mSu(fu) with SAP18 in yeast two-hybrid assays. The cDNAs of full-length mSu(fu) and the cytoplasmic domain of transforming growth factor-β (TGFβ) type I receptor were inserted downstream of the coding sequences for the LexA DNA-binding domain to generate the “bait” constructs. These plasmids were transformed into yeast strain EGY48/pSH18–4 along with SAP18, or FKBP12 cDNA that were fused in frame to the coding sequences of the activation domain B42. Yeast transformants were tested on galactose-containing X-Gal plates, and protein–protein interactions were determined by scoring for β-galactosidase activity. (B) Association of mSu(fu) with SAP18 in vitro. The translated, [35S]methionine-labeled mSin3A, mSu(fu), or luciferase was incubated with partially purified recombinant GST-SAP18 or GST alone immobilized on glutathione-Sepharose beads. The radioactively labeled proteins bound on beads were separated by SDS/PAGE and visualized by autoradiography. (C) Association of mSu(fu) with SAP18 in mammalian cells. HEK293T cells were transfected with Myc-tagged mSu(fu), FLAG-tagged SAP18 expression constructs individually or together. Total cell lysates and pellets from anti-Myc immunoprecipitation were separated by SDS/PAGE and followed by protein blot analysis by using anti-Myc 9E10 or anti-FLAG M2 antibodies.
To evaluate whether the interaction of mSu(fu) with SAP18 was direct or indirect, we incubated in vitro translated 35S-labeled mSu(fu) with purified GST or GST-SAP18 fusion immobilized on glutathione-agarose beads. In this affinity “pull-down” assay, mSu(fu) and the previously reported SAP18-binding protein mSin3A were retained on the GST-SAP18 beads but not the GST beads (Fig. 1B). In contrast, luciferase did not interact with either GST-SAP18 or GST alone. These results reinforce the conclusion that mSu(fu) and SAP18 interact directly.
To demonstrate that mSu(fu) and SAP18 can interact in mammalian cells, we transfected a myc-tagged mSu(fu) and a FLAG-tagged SAP18 into human embryonic kidney 293T cells. With immunoprecipitation analysis, we showed that the FLAG-tagged SAP18 could be efficiently coimmunoprecipitated with Myc-tagged mSu(fu) (Fig. 1C), thus demonstrating an mSu(fu)-SAP18 interaction in vivo.
Histone Deacetylase Activity Is Required for Su(Fu)-Mediated Transcription Repression.
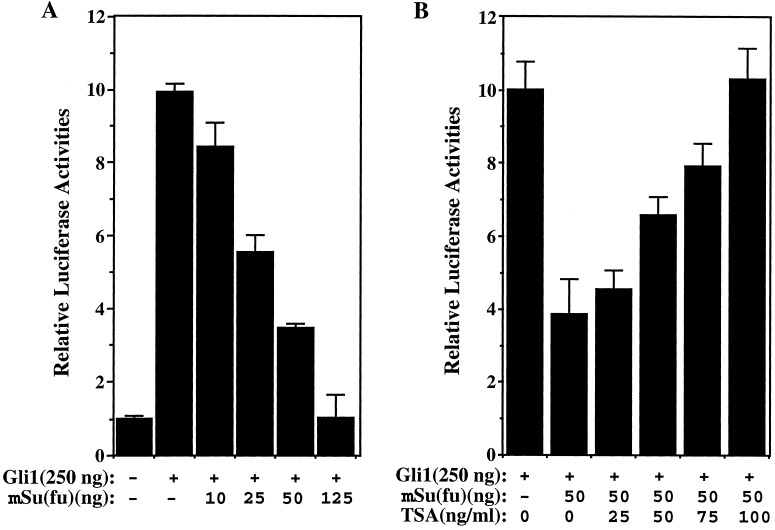
Although not well studied, the role of SAP18 in the mSin3-HDAC corepressor complex may be much the same as that of another mSin3-associated protein, SAP30 (23). A variety of evidence has demonstrated that SAP30 serves as an adapter protein that bridges mSin3-HDAC complex to the sequence specific, nuclear hormone corepressor N-CoR (27). In light of its ability to form a DNA-binding complex with the Gli proteins, mSu(fu) may recruit the mSin3-HDAC complex through interaction with SAP18 to repress the Gli-mediated transcription. To address this possibility, we first tested if histone deacetylase is involved in the mSu(fu)-mediated repression of transcription. We transfected HEK293T cells with Gli1, mSu(fu), and a luciferase reporter construct, 8xGliBS, that contains eight copies of Gli-binding sites derived from the HNF3β enhancer region (25). In agreement with the published results, transcription from this luciferase reporter was activated by Gli1 (Fig. 2A). The activation could be repressed by mSu(fu) in a dose-dependent manner (Fig. 2A). However, the mSu(fu)-mediated repression was reversed after addition of a known inhibitor of histone deacetylase, trichostatin A (TSA) (27) (Fig. 2B). All data reported here were normalized against the Renilla luciferase activity expressed from an internal control plasmid, pTK-RL. Neither mSu(fu) nor TSA had significant effect on the Renilla luciferase activity which was driven by the promoter of thymidine kinase gene. This result indicates that the histone deacetylase does play a role in transcription repression mediated by mSu(fu).
Figure 2.
Requirement for histone deacetylase activity in the repression of Gli1-mediated transcription by mSu(fu). To measure transcriptional activation from the Gli-dependent promoter, 8xGli-BS-Luc was used as the reporter. Plasmid pTK-RL (Promega), which expresses Renilla luciferase under the control of TK promoter, was included in all samples to normalize transfection efficiency. The total plasmid concentration was kept constant, and whenever needed, vector DNA was added. (A) Repression of Gli1-mediated transcription by mSu(fu). HEK293T cells were transfected with 8xGli-BS Luc reporter together with the indicated amount of plasmids encoding Gli1 or mSu(fu). (B) TSA, an inhibitor of histone deacetylase reversed repression by mSu(fu) on Gli1-mediated transcription. HEK293T cells were transfected with 8x Gli-BS Luc and Gli1 together with either a control-vector plasmid or a mSu(fu)-expression vector. Twenty-four hours before analysis, the indicated concentrations of TSA were added. The luciferase activity in the presence of cotransfected mSu(fu) relative to that obtained with the control plasmid was calculated for each concentration of TSA.
Functional Cooperation Between mSu(fu), SAP18, and mSin3.
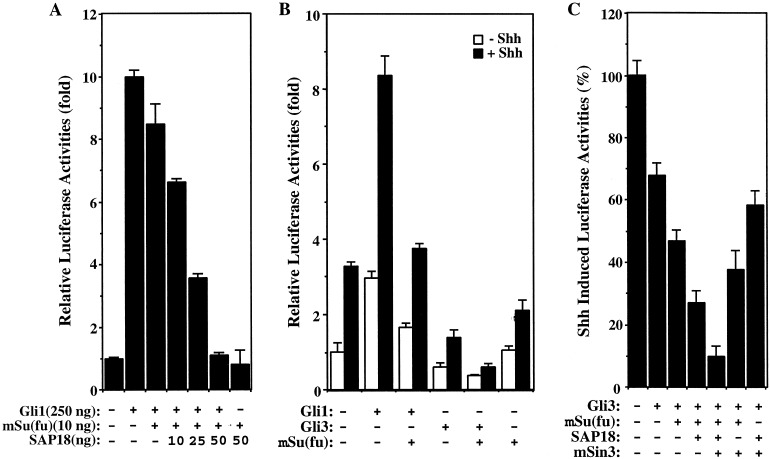
We then asked if mSu(fu), SAP18, and mSin3 collaborate in repressing Gli-mediated transcription in transfected HEK293T cells. At low levels, mSu(fu) caused a slight repression of Gli1-mediated transcription (Figs. 2A and 3A), which was augmented by addition of SAP18 in a dose-dependent manner (Fig. 3A). In the absence of Gli1, mSu(fu) and SAP18 had little effect on the basal transcription activity (Fig. 3A). The functional interaction between mSu(fu) and SAP18 was further tested on the transcriptional response of Gli3, which also has the ability to bind mSu(fu) through the conserved N-terminal domain (18, 28). In HEK293T cells, treatment of the sonic hedgehog (Shh) led to the induction of transcription from the 8xGliBS reporter (Fig. 3B). Although Gli1 augmented this Shh-induced transcription, Gli3 repressed it (Fig. 3B), which is consistent with Gli3 undergoing proteolytic cleavage and acting as a transcription repressor (7, 15). Expression of mSu(fu) in these cells inhibited the activator activity of Gli1 but enhanced the repressor activity of Gli3 (Fig. 3B). Coexpression of mSu(fu) together with SAP18 and mSin3 had an additive effect on the Gli3-mediated transcriptional repression, with maximum repression achieved when all four proteins are present (Fig. 3C). Taken together, these results indicate that mSu(fu), SAP18, and mSin3 functionally interact to repress Gli-mediated transcription.
Figure 3.
Functional cooperation of mSu(fu) and SAP18 in the repression of Gli-mediated transcription. Transcriptional assays were performed as in Fig. 2. (A) SAP18 cooperates with mSu(fu) to repress Gli1-mediated transcription. HEK293T cells were transfected with 8xGli-BS Luc reporter together with the indicated plasmids encoding Gli1, mSu(fu), or SAP18. (B) mSu(fu) potentiates repression of Shh-induced transcription by Gli3. In HEK293T cells, the transcription from Gli-dependent promoter, 8xGli-BS Luc, was induced by expression of Shh. Gli1 enhanced Shh-induced transcription, whereas Gli3 inhibited Shh-induced transcription. Expression of mSu(fu) inhibited transcription activated by Shh and Gli1, and potentiated repression of Shh-induced transcription by Gli3. (C) Coexpression of mSu(fu) with SAP18 and/or mSin3 enhanced Gli3-mediated repression of Shh-induced transcription.
Gli1, mSu(fu), SAP18, and mSin3 Form a DNA–Protein Ternary Complex.
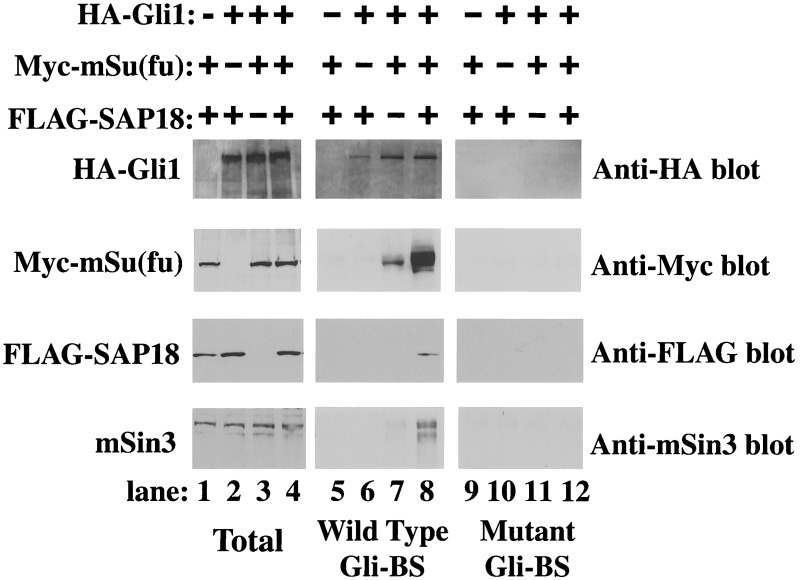
The functional interaction among mSu(fu), SAP18, and mSin3 raises the possibility that these three proteins may physically coexist in the same complex on promoters containing Gli-binding elements. To test this, we asked if mSin3, SAP18, and mSu(fu) can be copurified along with Gli1 from nuclear extracts by a biotinylated oligonucleotide containing a copy of the Gli-binding site (Gli-BS). Because Su(fu), SAP18, and mSin3 are not themselves DNA-binding proteins, in the absence of Gli1, none of these proteins bound to the DNA oligo-containing Gli-binding element (Fig. 4, lane 5). However, we were able to retain mSu(fu) along with Gli1 on the wild-type DNA oligo, but not on a mutant oligo-containing scrambled Gli-binding sequence (Fig. 4, compare lanes 7 and 11). We were also able to detect SAP18 and mSin3 in the oligonucleotide-bound protein complex when SAP18, mSu(fu), and Gli1 were all transfected into HEK293T cells (Fig. 4, lane 8), indicating the formation of a ternary complex on the Gli-binding oligo. The mSin3 detected in this experiment was of endogenous origin, which implies high affinity of Gli1-mSu(fu) and SAP18 complex toward the mSin3 corepressor. Our results also indicate that formation of this large DNA-bound ternary complex depends on a chain of interactions connecting mSin3, SAP18, mSu(fu), and Gli1. Disruption of any intermediate link prevents the formation of this complex (Fig. 4, lanes 5–7). Also, formation of this ternary complex may have stabilized DNA-bound Gli1-mSu(fu), as indicated by the elevated level of mSu(fu) detected in the complex (Fig. 4, lane 8).
Figure 4.
Recruitment of mSin3 to the consensus Gli DNA-binding sequence through interaction of mSu(fu) and SAP18. Nuclear extracts from HEK293T cells transfected with indicated plasmids were incubated with biotinylated DNA oligonucleotides containing either wild-type (lanes 5–8) or mutant (lanes 9–12) Gli-binding site (Gli-BS). DNA-bound proteins were selected by using streptavidin-coated magnetic beads (Promega) in the presence of 2 μg/ml poly(dI-dC) and subjected to Western blotting with corresponding antibodies. The expression levels of the transfected proteins or endogenous mSin3 were assessed by direct Western blot analysis of total nuclear extracts (lanes 1–4).
Discussion
We have identified SAP18, the 18-kDa mSin3-associated protein, as a Su(fu)-associated protein. We demonstrated that Su(fu) functionally cooperates with SAP18 to repress Gli-mediated transcription. Furthermore, we have shown that Gli1, mSu(fu), SAP18, and mSin3 can form a ternary DNA-binding complex on a Gli DNA-binding site. Our results provide a biochemical evidence for a direct nuclear function of Su(fu), and indicate that Su(fu) is able to repress Gli-mediated transcription by recruiting transcription corepressor mSin3-HDAC.
Several studies with Drosophila and cultured mammalian cells showed that overexpression of Su(fu) causes cytoplasmic sequestration of Gli and attributed the transcriptional repression by Su(fu) to this effect (14, 20–22). These findings do not preclude a nuclear function of Su(fu). In fact, several lines of evidence support a nuclear function. First, both Drosophila and vertebrate Su(fu) proteins are shown to form DNA-binding complexes with Gli proteins (17, 18). Second, the vertebrate Su(fu) can be found in both cytoplasmic and nuclear compartments (22), and the cytoplasmic tethering effect of Su(fu) can be reversed by the action of Fu, which causes nuclear translocation of Su(fu) with Gli (14). Third, genetic experiments in Drosophila indicated that the cytoplasmic sequestration effect of Su(fu) depends on the function of Cos2; in the absence of Cos2, Su(fu) fails to block the nuclear translocation of Ci, but the nuclear Ci is still kept inactive by Su(fu) (21). Our findings here showed that Su(fu) is capable of recruiting the mSin3-HDAC corepressor complex through its interaction with SAP18 to repress transcription. Therefore, Su(fu) exerts its transcriptional repression through two mechanisms. The nuclear repressor role of Su(fu) described here could operate to block the activity of full-length Gli or to augment the truncated Gli repressor that is generated by proteolytic cleavage in the absence of Hh signal. Further experiments using both genetic and biochemical means are required to discern these two possibilities and to address the physiological significance of Su(fu) and SAP18 interaction.
Histone deacetylation is well documented as a general mechanism to repress transcription by forming the closed chromatin structure (27). Usually the histone deacetylase is associated with the mSin3-related scaffold protein in a complex that does not contain DNA-binding affinity. Recruitment of the mSin3-HDAC complex to specific target genes exclusively relies on the interaction with sequence-specific DNA-binding proteins, which could be either direct or indirect through other adapter proteins. SAP18 and another protein, SAP30, were first identified as such adapter proteins in an immunocomplex containing mSin3, and it has been demonstrated that SAP30 is both necessary and sufficient to bridge mSin3-HDAC complex to a subset of repressors that use the nuclear hormone corepressor N-CoR (27). So it is probably not coincidental that Su(fu) specifically binds to the N-terminal domain that is conserved in all members of the Gli family and represses transcription mediated by all Gli proteins. Removal of this N-terminal domain abolishes transcription repression by Gli/Ci (22). Recently, a Drosophila homolog of SAP18 was identified as a binding protein for the GAGA sequence-specific DNA-binding factor (29), lending support to the notion that the same mechanism could operate in Drosophila. Because the transcription-activating activity of Gli is mediated by the C-terminal domain that interacts with the transcription coactivator, CBP/p300, a histone acetylase (30), the multiple controls of activating or attenuating Gli's transcription activity thus converge on modifying the transcriptional competence of chromatin structure through histone acetylation.
Acknowledgments
We thank Drs. K. W. Kinzler and B. Vogelstein for human Gli1 and Gli3 cDNAs, H. Sasaki for Gli reporter constructs, R. Eisenman for mSin3A cDNA, D. Reinberg for FLAG-SAP18 and GST-SAP18, H. Roelink for sonic hedgehog expression plasmid, R. Brent for HeLa cDNA library, R. Derynck for Yeast 2-hybrid vectors. We also thank Drs. Y. Zhang, J. Jiang, D. Robbins, K. Nybaken, and other members of J. M. Bishop's and T. B. Kornberg's laboratories for stimulating discussions. This research was supported by a postdoctoral fellowship from the American Heart Association (to S.Y.C.) and funds from National Institutes of Health Grant CA44338 and the G. W. Hooper Foundation (to J.M.B.).
Abbreviations
- Su(fu)
Suppressor of Fused
- Hh
hedgehog
- Shh
sonic hedgehog
- TSA
trichostatin A
- SAP18
HDAC, histone deacetylase
- Gli-BS
Gli-binding site
- GST
glutathione S-transferase
- HA
hemagglutinin
- Ptc
patched
- Smo
smoothened
References
- 1.Kalderon D. Cell. 2000;103:371–374. doi: 10.1016/s0092-8674(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz i Altaba A. Nat Cell Biol. 1999;1:323–391. doi: 10.1038/14099. [DOI] [PubMed] [Google Scholar]
- 3.Ingham PW. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinzler K W, Vogelstein B. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez M, Brunner M, Hafen E, Basler K. Science. 1996;272:1621–1625. doi: 10.1126/science.272.5268.1621. [DOI] [PubMed] [Google Scholar]
- 6.Aza-Blanc P, Ramirez-Weber F A, Laget M P, Schwartz C, Kornberg T B. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang B, Fallon J F, Beachy P A. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 8.Stone D M, Hynes M, Armanini M, Swanson T A, Gu Q, Johnson R L, Scott M P, Pennica D, Goddard A, Phillips H, et al. Nature (London) 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 9.Marigo V, Davey R A, Zuo Y, Cunningham J M, Tabin C J. Nature (London) 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 10.Robbins D J, Nybakken K E, Kobayashi R, Sisson J C, Bishop J M, Therond P P. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 11.Sisson J C, Ho K S, Suyama K, Scott M P. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 12.Jiang J, Struhl G. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Struhl G. Nature (London) 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 14.Murone M, Luoh S-M, Stone D, Li W, Gurney A, Armanini M, Grey C, Rosenthal A, de Sauvage F J. Nat Cell Biol. 2000;2:310–313. doi: 10.1038/35010610. [DOI] [PubMed] [Google Scholar]
- 15.Aza-blanc P, Lin H Y, Ruiz I, Altaba A, Kornberg T B. Development (Cambridge, UK) 2000;127:4293–4301. doi: 10.1242/dev.127.19.4293. [DOI] [PubMed] [Google Scholar]
- 16.Preat T. Genetics. 1992;132:725–736. doi: 10.1093/genetics/132.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Curr Biol. 1998;8:583–586. doi: 10.1016/s0960-9822(98)70227-1. [DOI] [PubMed] [Google Scholar]
- 18.Pearse II R V, Collier L S, Scott M P, Tabin C J. Dev Biol. 1999;212:323–336. doi: 10.1006/dbio.1999.9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohlmeyer J T, Kalderon D. Nature (London) 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- 20.Methot N, Basler K. Development (Cambridge, UK) 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, Amanai K, Wang B, Jiang J. Genes Dev. 2000;14:2893–2905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kogerman P, Grimm T, Kogerman L, Krause D, Unden A B, Sandstedt B, Toftgard R, Zaphiropoulos P G. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 24.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki H, Hui C C, Nakafuku M, Kondoh H. Development (Cambridge, UK) 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 26.Wang T, Donahoe P K, Zervos A S. Science. 1994;265:674–676. doi: 10.1126/science.7518616. [DOI] [PubMed] [Google Scholar]
- 27.Pazin M, Kadonaga J. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 28.Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sakaki H, Dlugosz A, Nakafuku M, Hui C C. Curr Biol. 1999;9:1119–1122. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- 29.Espinas M L, Canuda S, Fanti L, Pimpinelli S, Casanova J, Azorin F. EMBO Rep. 2000;1:253–259. doi: 10.1093/embo-reports/kvd046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akimaru H, Chen Y, Dai P, Hou D X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Nature (London) 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]