Abstract
Pbx1 is a homeodomain protein that functions in complexes with other homeodomain-containing proteins to regulate gene expression during embryogenesis and oncogenesis. Pbx proteins bind DNA cooperatively as heterodimers or higher order complexes with Meis family members and Hox proteins and are believed to specify cell identity during development. Here, we present evidence that Pbx1, in partnership with Meis1b, can regulate posterior neural markers and neural crest marker genes during Xenopus development. A Xenopus homolog of the Pbx1b homeodomain protein was isolated and shown to be expressed throughout embryogenesis. Xpbx1b expression overlaps with Xmeis1 in several areas, including the lateral neural folds, caudal branchial arch, hindbrain, and optic cup. When ectopically expressed, Xpbx1b can synergize with Xmeis1b to promote posterior neural and neural crest gene expression in ectodermal explants. Further, a physical interaction between these two homeodomain proteins is necessary for induction of these genes in embryonic tissue. In addition, coexpression of Xmeis1b and Xpbx1b leads to a prominent shift in the localization of Xmeis1b from the cytoplasm to the nucleus, suggesting that nuclear transport or retention of Xmeis1b may depend upon Xpbx1b. Finally, expression of a mutant construct in which Xpbx1b protein is fused to the repressor domain from Drosophila Engrailed inhibits posterior neural and neural crest gene expression. These data indicate that Xpbx1b and its partner, Xmeis1b, function in a transcriptional activation complex during hindbrain and neural crest development.
In Xenopus, formation of the anteroposterior axis in the prospective neurectoderm is induced during gastrulation on the dorsal side of the embryo (1). Neural patterning has been proposed to be a two-step process where neurectoderm is first “activated” (anterior state) and subsequently “transformed” or respecified into posterior neurectoderm. Several antagonists of bone morphogenic proteins have been identified which may play a role in the “activation” state (2). Several secreted molecules have “transforming” activity and may be involved in reprogramming this tissue to more posterior cell fates, such as hindbrain and spinal cord (2). Neural crest tissue is induced at the border between the neural plate and epidermis. These cells eventually begin to migrate throughout the embryo and give rise to most of the peripheral nervous system, epidermal pigment cells, and craniofacial cartilage (3). Rhombomeric generation of neural crest cells is observed along the dorsal part of the hindbrain, where they migrate ventrally and give rise to cranial sensory ganglia and populate the pharyngeal arches. The multipotent cells ultimately contribute to the formation of neural, muscular, skeletal, and vascular structures (3). Hox genes are segmentally expressed in the developing vertebrate hindbrain, neural crest cells, and pharyngeal arches, demonstrating an important role in patterning these structures (3).
An array of transcriptional cofactors, such as the homeodomain proteins of the EXD/PBX (PBC; ref. 4) and MEIS/PREP (MEINOX) families regulate the transcriptional activity of HOX proteins during development (5–7). These homeodomain cofactors do not encode any obvious transcriptional activator or repressor domains but they do play important roles during embryonic development. For example, the Drosophila Meis homolog, homothorax (Hth), cooperates with a Drosophila PBX homolog termed extradenticle (Exd). Together, these two homeodomain proteins control antenna determination (8), patterning of the embryonic fly PNS (9, 10), and suppression of eye development (11). The interaction between Hth and Exd triggers the nuclear localization of Exd, thus allowing for proper function of the protein complex (8, 10–14). In vertebrates, aberrant Meis1 gene expression has been shown to be involved in the pathogenesis of murine myeloid tumors and human leukemias (15–17), but new information on the developmental role of these proteins is beginning to emerge. For example, a Xenopus homolog of the mammalian Meis3 gene has been implicated in the caudalization of neural tissue (18, 19), and recent studies in chicken suggest that restriction of Meis1 to proximal regions of the limb is essential for the specification of cell fates along the proximal-distal axis of the limb (20, 21). Also, until recently, little was known about Pbx function in vertebrates.
Pbx1 was identified as a fusion partner with E2A in a translocation breakpoint found in human pre-B cell leukemias (22, 23). Mutations in the Drosophila Exd gene cause homeotic transformations, and Caenorhabditis elegans Pbx mutants display ectodermal and even some mesodermal phenotypes (24, 25). In vertebrates, the expression of Pbx1 and the formation of Pbx/Hox complexes are found in developing neural tissue and in areas of mesenchyme-epithelial interaction (6, 7, 26, 27). Recently, a zebrafish Pbx gene was isolated (28, 29), and null mutants demonstrated that Pbx was critical to segmentation of the hindbrain and pharyngeal pouches (28). Moreover, zebrafish Pbx was shown to function in the same pathway as Meis during hindbrain development (30, 31).
We recently isolated a Xenopus homolog of Meis1b, an alternatively spliced form of Xmeis1. In ectodermal explants, overexpression of Xmeis1b induces expression of neural markers and neural crest marker genes in the absence of mesoderm. Moreover, misexpression of Xmeis1b in developing Xenopus embryos induces ectopic expression of neural markers and neural crest markers along the antero-posterior axis of the neural tube (32). Here, we describe a Xenopus Pbx1b gene, Xpbx1b, which can synergize with Xmeis1b to promote posterior neural markers and neural crest markers in embryonic tissue. We also show that a physical interaction between these two homeodomain proteins is necessary for posterior neural gene-marker induction in ectodermal explants. Moreover, coexpression of Xpbx1b and Xmeis1b leads to a relocation of the Xmeis1b protein from the cytoplasm to the nucleus. Finally, we demonstrate that introduction of the Engrailed repressor domain fused to the Xpbx1b protein inhibits posterior neural and neural crest gene expression during embryogenesis.
Materials and Methods
Isolation and Sequencing of the Xenopus Pbx1b cDNA and Generation of Mutants.
A cDNA-encoding Xenopus Pbx1b was isolated from a Xenopus laevis stage-30 head cDNA library by using human Pbx1a as a probe. From a full-length clone, a Xmeis1b-ΔM1 mutant (lacking amino acids 71–96), ΔM2 (lacking amino acids 148–161), and ΔM1/M2 (lacking amino acids 71–96 and 148–161) mutants were generated by PCR and inserted into pCS2+. Engrailed fusions were made by inserting either the Xmeis1b or Xpbx1b coding region into the 3′ end of the Engrailed repressor domain construct in pCS2+. Flag-tag was added at the C-terminal end of the XPbx1b or Xmeis1b coding regions by PCR and then inserted into pCS2+.
Embryos and Explants.
Wild-type or albino X. laevis embryos were obtained by artificial insemination after induction of female with 300 units of human chorionic gonadotropin and microinjected as described (32). Xenopus Pbx1b and XMeis1b RNA were synthesized and injected, and explants were prepared and cultured, as described (32).
Northern Analysis.
RNA from staged embryos was prepared with Trizol, as suggested by the manufacturer (Life Technologies, Rockville, MD). Seven micrograms of total RNA was separated in an agarose/formaldehyde gel (15). Radiolabeled probes were generated representing a 0.8-kb PstI/Eco0109I restriction fragment of Xpbx1b coding region or a 1.8-kb restriction length fragment from the 3′UTR region of the Xpbx1b cDNA. An 18S ribosomal subunit template (Ambion) was used to generate a control probe.
Whole-Mount in Situ Hybridization.
An N-terminal fragment lacking the homeodomain (277–972) of Xpbx1b was generated by PCR and was subcloned into pCS2+. The digoxygenin-labeled riboprobe was synthesized with T7 RNA polymerase. Plasmids containing Xmeis1a (32), XNrp-1 (33), Krox-20, XAp-2 (34), Xslug (35), Xzic3 (36), and Otx2 were linearized, and digoxygenin-labeled riboprobes were synthesized. In situ hybridization was performed as described (32). Photographs were taken with a dissecting microscope (Nikon SMZ 1500) and a charge-coupled device camera (Sony). For transverse sections, the embryos were embedded in paraffin after whole-mount in situ hybridization and dehydration, and 30-μm sections were cut and mounted on sialynated slides without counterstain.
Reverse Transcriptase (RT)-PCR Assay.
Extraction of total RNA and RT-PCR assay, primer sequences, and conditions were performed as described (32), with the exception of XAG-1 (37). These experiments were repeated three times for consistency.
Immunocytochemistry.
The embryos injected with Flag-tagged Xpbx1b or Flag-tagged Xmeis1b RNA were cultured to stage 9 and fixed with MEMFA (0.1 M Mops, pH 7.4/2 mM EGTA/1 mM MgSO4/4% paraformaldehyde) at 4°C overnight and dehydrated in ethanol. Paraffin-embedded blocks were serially sectioned at 7 μm. Slides were incubated in xylene followed by ethanol dehydration and PBS. FLAG M2 antibody at 1:500 was overlaid on the slides for 1 h at 37°C. After washing in PBS + 0.1% Tween-20 (PTW) for 1 h, peroxidase-conjugated anti-mouse antibody (1:500) was added, and slides were incubated for 1 h at 37°C. After washing in PTW, slides were immunostained with PBS + 0.2 mg/ml diaminobenzidine.
Immunoprecipitation and Western Blot Analysis.
A peptide corresponding to the amino terminal 14 amino acid of Xpbx1b was synthesized, conjugated to keyhole limpet hemocyanin and used to immunize rabbits (Macromolecular Resources, Fort Collins, CO). RNA-injected embryos were cultured until stage 10 and solubilized with lysis buffer [10 μl per embryo: 137 mM NaCl/20 mM Tris·HCl, pH 8.0/2 mM EDTA/1% (vol/vol) Nonidet P-40-containing protease inhibitors (Calbiochem)]. Immunoprecipitation analysis was performed on lysates from 15 embryos per sample, and immune complexes were separated by SDS/10% PAGE. Western analysis was performed as described (38) by using the indicated primary rabbit polyclonal antibodies at 1:1,000 dilution.
Results
Isolation and Expression Pattern of Xpbx1b.
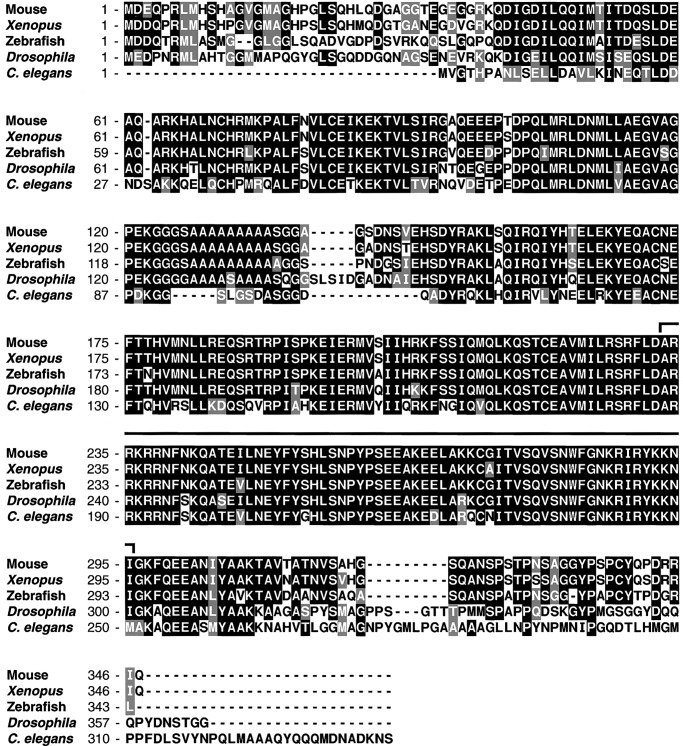
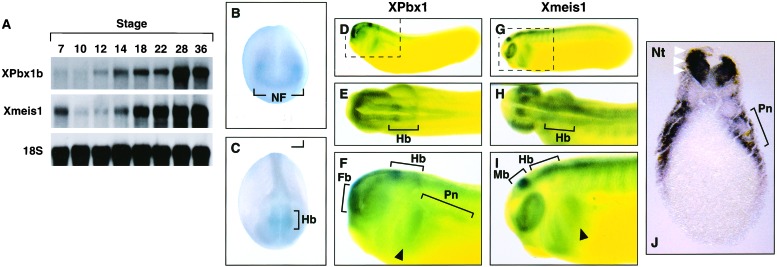
Previous results demonstrated that overexpression of Xmeis1b could induce ectopic expression of hindbrain and neural crest marker genes in ectodermal explants and developing Xenopus embryos. Because these data suggest that Xmeis1b may contribute to the execution of a posterior neural developmental program, we isolated the proposed Xenopus partner gene Xpbx1b. A human Pbx1a cDNA was used to probe an embryonic stage-30 head cDNA library, and sequence analysis revealed that a full-length Xpbx1b cDNA was obtained. The Xpbx1b gene is extremely well conserved, displaying 95% and 80% amino acid identity to two vertebrate proteins, Pbx1b (mouse) and Lazarus (zebrafish Pbx4), respectively. Less homology was evident with regard to the invertebrate proteins Drosophila Exd (70% identity) and C. elegans Ceh-20 (60%). In contrast, the Pbx homeodomains displayed over 90% identity among all of the species (Fig. 1). We next examined the temporal pattern of Xpbx1 mRNA expression during development with Northern blot analysis. A low level of Xpbx1 RNA was observed in unfertilized eggs (data not shown), but subsequent stages revealed an expression pattern similar to Xmeis1, with increased expression during late gastrula through neurula and tailbud stages (Fig. 2A).
Figure 1.
Xpbx1b encodes a TALE family homeodomain-containing protein. Amino acid comparison of the coding sequences of Xenopus Pbx1b, Mouse Pbx1b, Drosophila extradenticle, Zebrafish Pbx 4(Lazarus), and C. elegans Pbx (Ceh-20). Identical residues are shaded in black, conservative differences are shaded in gray, and white represents nonhomology. The bracketed area above the sequence denotes the homeodomain region.
Figure 2.
Temporal and spatial distribution of XPbx1b and Xmeis1 RNA expression during embryogenesis. (A) Northern blot analysis of RNA extracted from embryos at the indicated stages with Xpbx1b and Xmeis1 specific probe. The 18S RNA probe was used as a loading control. Whole mount in situ hybridization analysis of the tissue distribution of XPbx1b (B–F) and Xmeis1 (G–I) transcripts in X. laevis embryos. (B) Stage 15, anterior view. Note expression in lateral neural folds. (C) Stage 21, dorso-anterior view. Note strong staining in the presumptive hindbrain and along neural folds. (D) Lateral view of cleared stage-26 embryo. (E) Stage 26 dorsal view. (F) Enlarged lateral view of enclosure from D. Note the distinct Xpbx1 staining in the forebrain, hindbrain, and caudal branchial arch. (G) Lateral view of cleared stage-25 embryo. (H) Stage 25 dorsal view. (I) Enlarged lateral view of enclosure from G. Note strong expression of Xmeis1 in the midbrain, hindbrain, caudal and rostral branchial arches, and somites. (J) Transverse section through the pronephros of a stage-28 embryo. Note the strong Xpbx1 staining in lateral edge of neural tube (white arrowheads); expression also is observed in the lateral mesoderm surrounding the pronephritic anlage. (B–J) Black arrowheads indicate caudal branchial arch. Black brackets denote the indicated tissues. Fb, forebrain; Hb, hindbrain; Mb, midbrain; NF, neural fold; Nt, neural tube; Pn, pronephros.
To determine the spatial expression of Xpbx1 during development, whole-mount in situ hybridization was performed. Very faint broad expression was detected from the blastula through gastrula stages. At stage 14/15, Xpbx1 expression is diffuse, appearing as a broad arc that will give rise to the forebrain and eyes (Fig. 2B). More intense staining is observed in the lateral neural folds (presumptive neural crest), and expression is also apparent as horizontal stripes in the posterior neural plate that will give rise to the hindbrain (Fig. 2B). Although Xmeis1 expression at this stage has significant overlap with Xpbx1, Xmeis1 is less broad, with more restricted expression in the lateral neural folds and presumptive hindbrain (32). As development proceeds, staining progresses posteriorly along the neural folds. At stage 21, expression is pronounced within the prospective hindbrain (Fig. 2C). A gap in Xpbx1 expression is observed where the caudal portion of the hindbrain meets the rostral portion of the neural fold (Fig. 2C). Above this gap, expression is found in the lateral region of the neural folds, where migratory neural crest resides (Fig. 2C). At later stages (stage 26), Xpbx1 expression becomes intense within the dorsal portion of the forebrain. Staining also is observed within the optic cup, caudal branchial arch, peripheral to the pronephric anlage, and in the dorsal anterior half of the spinal cord (Fig. 2 D–F, J). Xpbx1 expression remains robust throughout the hindbrain but gradually becomes more restricted. At stage 26, two more intense stripes of expression are observed within the hindbrain, where one is more anterior and the other more posterior above the otic vesicle (Fig. 2 D–F). At stage 25, Xmeis1 is found in the optic cup, somites, branchial arches, as well as in the midbrain, hindbrain, and the length of the spinal cord (Fig. 2 G–I). At late stages of development (stage 25/26), both Xmeis1 and Xpbx1 display divergent patterns of expression in the forebrain (Xpbx1), midbrain (Xmeis1), rostral branchial arches and somites (Xmeis1), but also display distinct areas of overlap in the hindbrain, optic cup, caudal branchial arch, and rostral portion of the spinal cord (Fig. 2 D–I). Transverse sections through late stage-28 embryos confirm Xpbx1 expression in the dorsal lateral portion of the neural tube (Fig. 2J). Xpbx1 expression also is observed in the somatic layer of the lateral plate mesoderm that surrounds the pronephritic anlage (Fig. 2J). Collectively, the expression pattern suggests a possible interaction between Xpbx1 and the Xmeis1 binding partner in the patterning of posterior neural and neural crest-derived tissue in embryos.
Xpbx1b and Xmeis1b Cooperate to Induce Neural Markers in Explants.
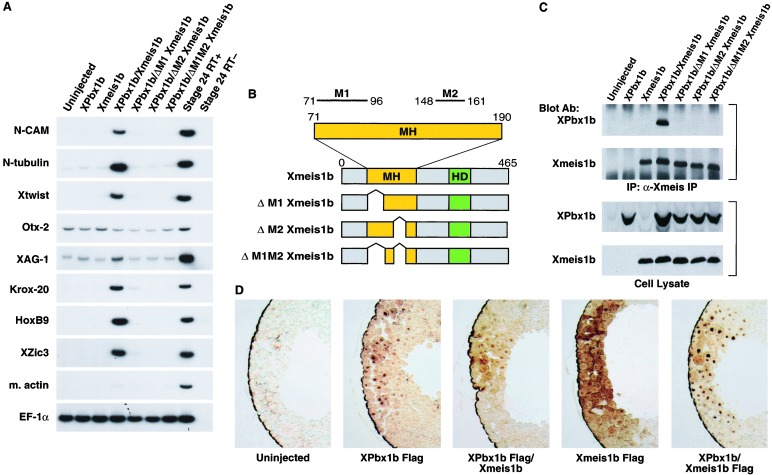
Pbx and Meis have been shown to form dimers, and these interactions are thought to regulate the activity of transcriptional complexes and thus affect developmental programs (4). Therefore, we tested whether Xpbx1b and Xmeis1b could have a functional influence on cell fate in embryonic ectodermal explants. We examined the expression of neural and mesodermal tissue markers by RT-PCR in animal pole explants from embryos injected with Xpbx1b or Xmeis1b RNA or both. Whereas the injection of 2.5 ng of Xmeis1b RNA was previously shown to induce posterior neural and neural crest markers (32), injection at low concentrations of Xmeis1b RNA (0.5 ng) does not induce any of these markers. Expression of Xpbx1b at high or low concentrations (0.5–2.5 ng) also was unable to induce mesodermal or neural markers. In contrast, when Xpbx1b was coexpressed with Xmeis1b, several neural and neural crest markers were induced. These markers included N-CAM (pan-neural marker), N-tubulin (pan-neuron marker), Xtwist (neural crest marker), Krox-20 (hindbrain marker), Hoxb9 (posterior neural marker; Fig. 3A), and Xzic3 (a proneural gene that also promotes the earliest steps in neural crest development; ref. 36). Ectopic Krox-20, Xslug, and N-tubulin expression also was observed in whole embryos (data not shown). In contrast to the prominent induction of posterior neural and neural crest markers, only a very modest effect on XAG-1 (cement gland) and Otx-2 (anterior neural) expression was observed in animal caps (Fig. 3A). Neither Xbrachyury (early mesoderm) nor muscle actin (late mesoderm) transcripts were induced by these products (data not shown, Fig. 3A). These results indicate that the interaction between Xpbx1b and Xmeis1b leads to the induction of posterior neural cell fate markers in the absence of mesoderm.
Figure 3.
Interaction of XPbx1b and Xmeis1b induces posterior neural and neural crest markers in animal cap explants in the absence of mesoderm. (A) The animal pole region of two-cell stage embryos were injected with either XPbx1b RNA (1.0 ng per embryo), Xmeis1b RNA (0.5 ng per embryo), or both RNAs. Animal pole explants were excised at stage 9 and cultured until stage 26. RT-PCR gene analysis was performed for N-CAM (pan-neural), N-tubulin (pan-neuronal), Xtwist (neural crest), Otx-2 (forebrain), XAG-1 (cement gland), Krox-20 (hindbrain), HoxB9 (spinal cord), Xzic3 (proneural and early neural crest marker), muscle actin (dorsal mesoderm), and EF-1a (loading control). Stage-24 embryonic RNA with or without reverse transcriptase (RT+ or RT−) also was used as a positive and negative reaction control. Note that coinjection of XPbx1b and Xmeis1b RNA induced the expression of posterior neural and neural crest cell markers. (B) Schematic representation of the Xmeis1b mutants. Wild-type Xmeis1b consists of a Meis-Homothorax domain (MH, yellow box) and homeodomain (HD, green box). MH domain possesses two subdomains: M1 box (amino acids 71–96) and M2 box (amino acids 148–161) that are important for Pbx binding. Deletions of one or more subdomains are indicated. (C) XPbx1b and Xmeis1b physically interact. The animal pole region of two-cell stage embryos were injected with either XPbx1b (2.5 ng per embryo) or Xmeis1b (2.5 ng per embryo) RNA alone or were coinjected with XPbx1b and Xmeis1b wild-type and mutant RNAs. Embryos were cultured until stage 9, and embryonic extracts were prepared and either directly immunoblotted (Lower) or immunoprecipitated with an anti-Xmeis1 antibody before immunoblotting (Upper). Anti-Meis1 antibody or anti-Pbx1b antibody was used to detect the indicated protein. Note: the Xpbx1b protein was coimmunoprecipitated with the wild-type Xmeis1b but not with any of the Xmeis1b mutants. (D) Nuclear localization of the Xmeis1b protein depends upon the XPbx1b protein in frog embryonic cells. The animal pole region of two-cell stage embryos was injected with either Flag tagged XPbx1b (2.5 ng per embryo) or Flag-tagged Xmeis1b RNA (2.5 ng per embryo) alone or in combination with Xmeis1b or XPbx1b RNA (2.5 ng per embryo). Injected embryos were fixed at stage 9, embedded, sectioned, and immunostained with anti-Flag antibody. Note that Xmeis1b protein was in the cytoplasm when expressed alone but was localized in the nuclei in the presence of XPbx1b. In contrast, XPbx1b was localized in the nuclei regardless of whether exogenous Xmeis1b was present.
Physical Interaction Between Xpbx1b and Xmeis1b Is Critical for Inductive Activity.
To test whether a direct interaction between Xmeis1b and Xpbx1b was necessary for the conversion of ectodermal tissue to a posterior neural cell fate, we generated forms of Xmeis1b with reduced binding activity to Xpbx1b (Fig. 3B). These mutants are termed ΔM1, ΔM2, and ΔM1/M2, depending upon whether the first, second, or both N-terminal Pbx1-binding sites have been removed (Fig. 3B). Coexpression of the mutant RNAs with Xpbx1b RNA in ectodermal explants did not induce any of the neural genes examined. One exception was ΔM1; it displayed a very weak ability to induce the pan-neural gene N-CAM, the posterior neural gene Krox-20, and the neural crest gene Xtwist (Fig. 3A). These data strongly suggest that the physical binding of Xpbx1b and Xmeis1b is necessary for the neural and neural crest inductive activities of these two proteins.
To confirm that the three Xmeis1 mutants were impaired in their ability to physically interact with Xpbx1b, immunoprecipitation and Western analysis was performed on the cell lysates from embryos expressing these proteins (Fig. 3C). Immunoprecipitation with an Xmeis1 polyclonal antibody revealed that Xpbx1b coimmunoprecipitated only with the wild-type Xmeis1b molecule and not with the M1, M2, and M1/M2 mutants (Fig. 3C). These data are consistent with the idea that Xpbx1b and Xmeis1b must be stably associated to affect neural cell fate (Fig. 3C). Further, whereas the interaction between Hth and Exd is essential for the mutual stabilization of both proteins in Drosophila, western analysis shows that this interaction is not necessary for the accumulation of either Xpbx1b or Xmeis1b when expressed in Xenopus embryos (Fig. 3C).
Xpbx1b Affects Localization of Xmeis1b.
In Drosophila, the interaction between Hth and Exd triggers the nuclear localization of Exd, thus allowing for functional activation of the protein complex (8, 10–14). Because the Pbx1 protein is nuclear in proximal cells of the mouse limb and cytoplasmic in distal cells (20, 39), we examined whether a similar mechanism was present in Xenopus embryos (Fig. 3D). Xenopus embryos were injected with RNA encoding a Flag-tagged version of Xpbx1b, either alone or along with Xmeis1b RNA. Immunocytochemistry showed that Xpbx1b localized to the nucleus in the absence or presence of exogenously expressed Xmeis1b. The reciprocal experiment also was performed where Flag-tagged Xmeis1b RNA was injected either alone or with Xpbx1b RNA. Interestingly, when Xmeis1b is expressed alone, it localizes mostly in the cytoplasm, with only a small fraction in the nucleus. In contrast, coexpression of Xmeis1b and Xpbx1b leads to very distinct and prominent nuclear localization of Xmeis1b. These data suggest that nuclear transport or retention of Xmeis1b may depend upon Xpbx1b.
Xpbx1b Fused to the Engrailed Repressor Inhibits Expression of Posterior Neural Markers and Neural Crest Markers.
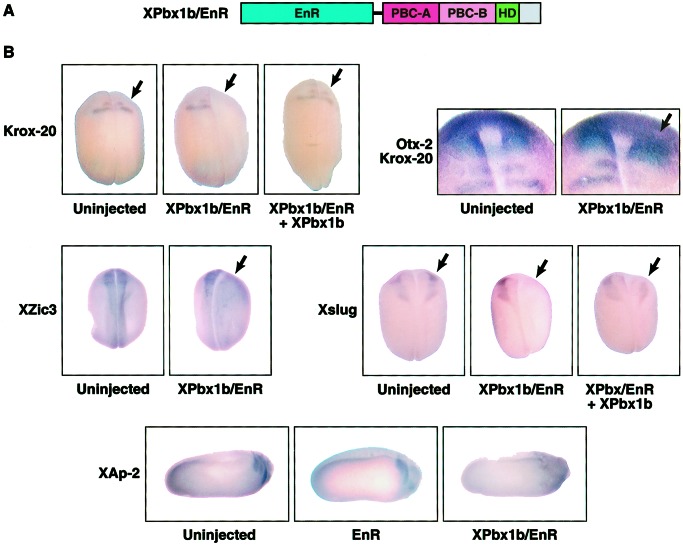
Interactions between Pbx, Meis, and Hox homeodomain-containing proteins have been postulated to play key roles in their activities as transcriptional regulators. Although Hox proteins contain transcriptional activation or repressor domains, Meis (5, 6) and Pbx1 (40) seem to lack such domains. To determine whether the posterior neural and neural crest gene induction observed by the synergistic action of Xpbx1b and Xmeis1b were the result of transcriptional activation or repression, Engrailed–repressor domain fusions were generated (Fig. 4A). One blastomere of two-cell embryos was injected with RNA (2.5 ng) encoding wild-type and/or Engrailed fusion versions (/EnR) of Xpbx1b. In situ hybridization analysis showed that ectopic expression of Xpbx1b induced a modest expansion of Krox-20 and Xslug expression (data not shown) but to a lesser degree than previously reported for Xmeis1b alone (32). In contrast, Xpbx1b/EnR protein resulted in the loss of Krox-20 mRNA expression in the hindbrain (78%; n = 46), whereas the expression of Otx-2 (an anterior neural marker) in the forebrain remained unaffected (Fig. 4B). Various neural markers were examined in early (stages 16 and 18) and late stage (stage 24) embryos expressing Xpbx1b/EnR. Xzic3 (proneural and neural crest) and Xslug (a transcriptional repressor critical for neural crest development; refs. 41 and 42) were both inhibited on the injected side of the embryos [Fig. 4B; 72% (n = 32) and 81% (n = 46), respectively]. The uninjected side displayed a normal Xzic3 or Xslug expression pattern in the neural folds of the embryo and thus acts as a control. In stage-24 embryos, Xpbx1b/EnR markedly reduced XAp-2 (cranial neural crest) expression and showed a low level of XAp-2 staining in the remaining dysmorphic neural crest-derived branchial arches. A similar inhibition of posterior neural and neural crest markers also was observed with an Xmeis1b/EnR construct (data not shown). Embryos expressing only the Engrailed repressor domain showed normal XAp-2 expression and pharyngeal structure, demonstrating the specificity of the effect. Collectively, these data indicate that the inhibitory effects induced by Xpbx1b/EnR show specificity toward posterior neural markers and neural crest markers but not an anterior neural marker.
Figure 4.
XPbx1b/EnR suppresses posterior neural and neural crest markers during embryonic development. (A) Schematic representation of XPbx1b protein fused with an Engrailed repressor domain. The three major conserved domains with vertebrate Pbx1 and C. elegans Ceh-20; PBC-A, PBC-B, and the homeodomain (HD) also are indicated. (B) One blastomere of two-cell stage embryos was either injected with XPbx1b/EnR RNA (0.75 ng per embryo) alone or coinjected with wild-type XPbx1b RNA (2.5 ng per embryo), as indicated. Engrailed control embryos were injected with EnR RNA (2.5 ng per embryo) as noted. Embryos were cultured until stage 16 (Xslug images), stage 18 (Krox-20, Krox-20/Otx-2, and Xzic3 images), or stage 24 (Xap-2 images). Embryos were fixed, and in situ hybridization was performed by using the indicated probe. Arrow indicates injected side of embryo. Note that XPbx1b/EnR suppressed Krox-20 (hindbrain), Xzic3 (proneural and neural crest), Xslug (neural crest), and Xap-2 (neural crest) expression on the injected side, but Otx-2 (anterior neural) expression remained. Also note that coexpression of wild-type Xpbx1b rescued Xslug and Krox-20 expression.
Rescue experiments were undertaken as an additional test of the specificity of the Xpbx1b/EnR repressor activity (Fig. 4B). Embryos injected with only Xpbx/EnR RNA (0.75 ng) displayed a loss of Krox-20 (53%, n = 25) or Xslug expression (64%, n = 25). In contrast, coinjection of wild-type Xpbx1b RNA (2.5 ng) significantly reduced the loss of Krox-20 (18%, n = 28) and Xslug (15%, n = 27) expression induced by Xpbx/EnR. Collectively, these data show that Xpbx1b is a member of a transcriptional activator complex that is important for proper expression of posterior neural and neural crest genes.
Discussion
In this report, we isolate the Xenopus Pbx1b cDNA and show that it has broad expression in neural tissue during embryonic development. Although Xpbx1 expression overlies the reported Xmeis1 pattern in the neural fold (Fig. 2 B and C), there is also overlap at later stages in the hindbrain, optic cup, caudal branchial arch, and dorsal lateral portions of the neural tube (Figs. 2 D–J; ref. 32). The Xpbx1b and Xmeis1b expression patterns along with the coexpression studies reveal a possible link to hindbrain and neural crest development. There are also locations where these two proteins are less likely to interact. For example, although Xmeis1 and Xpbx1 are both expressed quite strongly in the prospective hindbrain region, only Xmeis1 is expressed prominently in the midbrain, and only Xpbx1 is expressed robustly in the forebrain. Thus, a role in hindbrain patterning or posterior neural development is consistent with the opportunity for interaction between these two homeodomain proteins. Lazarus mutants (Pbx4) in zebrafish have hindbrain patterning defects and display defects in cranial neural crest segmentation (28). The Pbx1 knockout mouse also shows severe phenotypic effects in the caudal branchial arches (43).
Although Xpbx1b and Xmeis1b can induce some of the early players in neural crest development (Xslug, Xzic3), it is possible that they play a role later in neural crest development. For example, Xpbx1b and Xmeis1b may exert their influence as the crest cells populate the caudal branchial arches, where strong expression is observed. In addition to presenting evidence that Xpbx1 and Xmeis1 may have the opportunity to interact, we show that the interaction between Xpbx1b and Xmeis1b is essential for the induction of posterior neural and neural crest markers in ectodermal explants. Xmeis1b mutants lacking the M1 (amino acids 71–96) and M2 (amino acids 148–161) boxes were unable to form stable interactions with Xpbx1b in embryos or induce posterior neural markers in explants. The ability of the Xpbx1b and Xmeis1b proteins to form heterodimers allows for complex formation by using various combinations of homeodomain proteins and thus an increased level of complexity in gene regulation of developmental processes. Recent work on the D. rerio Pbx4 gene (lazarus) supports a role for Pbx in neural crest development, where lazarus mutants have defects in the segmentation of cranial neural crest (28). Recent evidence indicates that expression of the Meis protein partially rescues the mutant lazarus phenotype in zebrafish, suggesting Meis functions in the same pathway as Pbx (31). Although it is clear that Xpbx1b and Xmeis1b are collaborative partners in the phenotypic and cell-fate effects described here, it is unclear exactly how their interaction facilitates these events.
In Drosophila (8, 10–14) and mouse (39), Hth or Meis are required to retain Exd or Pbx1 in the nucleus. In our experiments, immunocytochemistry demonstrated that exogenously expressed Xpbx1b localized to the nucleus in the absence of ectopic Xmeis1b. These data suggest that Xenopus Pbx1b may not require interaction with Xmeis1b for nuclear localization. In contrast, coexpression of Xmeis1b and Xpbx1b leads to a dramatic shift in Xmeis1b localization from the cytoplasm to the nucleus. This result is consistent with a similar transport mechanism reported for zebrafish Pbx4 and Meis3 (30). Collectively, these data suggest that Xpbx1b may function as a nuclear transporter of Xmeis1b, and that the synergistic induction of neural markers by the coexpression of Xpbx1b and Xmeis1b in animal caps may depend on this nuclear transport activity. A recent study in zebrafish also reports that coexpression of Meis and Pbx4 causes mutual stabilization of both proteins (31). We have not observed a significant increase in Xmeis1b accumulation when these proteins are coexpressed in Xenopus embryos, but there is a small reproducible increase in Xpbx1b (Fig. 3C). We also have observed an Xmeis1-induced stabilization of Xpbx1b protein in Xenopus oocytes (data not shown), suggesting that such a mechanism may exist in a context-dependent manner.
Although the coexpression of Xpbx1b and Xmeis1b induced posterior neural markers in ectodermal explants and embryos, it was still unclear whether the two proteins were functioning through a transcriptional activator or repressor complex. Therefore, an Engrailed repressor fusion construct was generated for Xpbx1b and then expressed in developing embryos. Xpbx1b/EnR blocks neural crest and posterior neural markers, but not an anterior marker (Otx-2; Fig. 4B) nor a pan-neural marker (Nrp-1; data not shown). These data support the idea that Xpbx1b plays a role in posterior neural development and argues against possible secondary effects caused by inhibiting central nervous system development. The Xpbx1b/EnR repressor activity is rescued by coexpression of wild-type Xpbx1b, demonstrating the specificity of the effect. These data are consistent with the Xpbx1b protein having a role in posterior neural development rather than anterior neurogenesis during early development. Moreover, the Xmeis1b/Xpbx1b complex along with unknown Hox partners may function as a transcriptional activator in this process. A model has been proposed in which the Hox-Pbx complex can act as a repressor or activator of transcription by means of association with corepressors or coactivators (39). These associations are suggested to be a direct determinant of Hox-Pbx function in the patterning of the animal embryo (39). Clarification of how the Xpbx1b/Xmeis1b complex plays a role in the process of hindbrain and neural crest development awaits the isolation of cooperating factors and direct target genes of the Xmeis1b/Xpbx1b complex.
Acknowledgments
We thank Alan Perantoni and Lino Tessarollo for critical reading of the manuscript, Dale Frank, Mitsugu Maeno, Yuichi Watanabe, and Roberto Mayor for helpful discussions. We also thank David Wilkinson (Krox-20), Igor Dawid (N-tubulin), Thomas Sargent (XAP-2), Michael Sargent (Xslug), Jun Aruga (Xzic3), Daniel Kessler (Engrailed pCS2+), and Richard Harland (stage-30 head cDNA library) for reagents. We apologize to many of our colleagues whose work we were unable to cite because of space limitations. R.M. was supported by a Japan Society for the Promotion of Science Research Fellowship.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF480430).
References
- 1.Slack J M, Tannihill D. Development (Cambridge, UK) 1992;114:285–302. doi: 10.1242/dev.114.2.285. [DOI] [PubMed] [Google Scholar]
- 2.Gamse J, Sive H. BioEssays. 2000;22:976–986. doi: 10.1002/1521-1878(200011)22:11<976::AID-BIES4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 3.Trainor P A, Krumlauf R. Nat Rev Neurosci. 2000;1:116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- 4.Mann R S, Affolter M. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 5.Berthelsen J, Zappavigna V, Mavilio F, Blasi F. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs Y, Schnabel C A, Cleary M L. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F. Development (Cambridge, UK) 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 8.Casares F, Mann R S. Nature (London) 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- 9.Salzberg A, D'Evelyn D, Schulze K L, Lee J K, Strumpf D, Tsai L, Bellen H J. Neuron. 1994;13:269–287. doi: 10.1016/0896-6273(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 10.Kurant E, Pai C Y, Sharf R, Halachmi N, Sun Y H, Salzberg A. Development (Cambridge, UK) 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- 11.Pai C Y, Kuo T S, Jaw T J, Kurant E, Chen C T, Bessarab D A, Salzberg A, Sun Y H. Genes Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieckhof G E, Casares F, Ryoo H D, Abu-Shaar M, Mann R S. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 13.Abu-Shaar M, Ryoo H D, Mann R S. Genes Dev. 1999;13:935–945. doi: 10.1101/gad.13.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaw T J, You L R, Knoepfler P S, Yao L C, Pai C Y, Tang C Y, Chang L P, Berthelsen J, Blasi F, Kamps M P, Sun Y H. Mech Dev. 2000;91:279–291. doi: 10.1016/s0925-4773(99)00316-0. [DOI] [PubMed] [Google Scholar]
- 15.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura T, Largaespada D A, Shaughnessy J D, Jr, Jenkins N A, Copeland N G. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence H J, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L, Buchberg A M, Largman C. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 18.Salzberg A, Elias S, Nachaliel N, Bonstein L, Henig C, Frank D. Mech Dev. 1999;80:3–13. doi: 10.1016/s0925-4773(98)00187-7. [DOI] [PubMed] [Google Scholar]
- 19.Dibner C, Elias S, Frank D. Development (Cambridge, UK) 2001;128:3415–3426. doi: 10.1242/dev.128.18.3415. [DOI] [PubMed] [Google Scholar]
- 20.Mercader N, Leonardo E, Azpiazu N, Serrano A, Morata G, Martinez C, Torres M. Nature (London) 1999;402:425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- 21.Mercader N, Leonardo E, Piedra M E, Martinez A C, Ros M A, Torres M. Development (Cambridge, UK) 2000;127:3961–3970. doi: 10.1242/dev.127.18.3961. [DOI] [PubMed] [Google Scholar]
- 22.Kamps M P, Murre C, Sun X H, Baltimore D. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 23.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M L. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 24.Rauskolb C, Smith K M, Peifer M, Wieschaus E. Development (Cambridge, UK) 1995;121:3663–3673. doi: 10.1242/dev.121.11.3663. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Fire A. Development (Cambridge, UK) 2000;127:5179–5190. doi: 10.1242/dev.127.23.5179. [DOI] [PubMed] [Google Scholar]
- 26.Toresson H, Parmar M, Campbell K. Mech Dev. 2000;94:183–187. doi: 10.1016/s0925-4773(00)00324-5. [DOI] [PubMed] [Google Scholar]
- 27.Schnabel C A, Selleri L, Jacobs Y, Warnke R, Cleary M L. Mech Dev. 2001;100:131–135. doi: 10.1016/s0925-4773(00)00516-5. [DOI] [PubMed] [Google Scholar]
- 28.Popperl H, Rikhof H, Chang H, Haffter P, Kimmel C B, Moens C B. Mol Cell. 2000;6:255–267. doi: 10.1016/s1097-2765(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 29.Vlachakis N, Ellstrom D R, Sagerstrom C G. Dev Dyn. 2000;217:109–119. doi: 10.1002/(SICI)1097-0177(200001)217:1<109::AID-DVDY10>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 30.Vlachakis N, Choe S K, Sagerstrom C G. Development (Cambridge, UK) 2001;128:1299–1312. doi: 10.1242/dev.128.8.1299. [DOI] [PubMed] [Google Scholar]
- 31.Waskiewicz A J, Rikhof H A, Hernandez R E, Moens C B. Development (Cambridge, UK) 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- 32.Maeda R, Mood K, Jones T L, Aruga J, Buchberg A M, Daar I O. Oncogene. 2001;20:1329–1342. doi: 10.1038/sj.onc.1204250. [DOI] [PubMed] [Google Scholar]
- 33.Richter K, Good P J, Dawid I B. New Biol. 1990;2:556–565. [PubMed] [Google Scholar]
- 34.Winning R S, Shea L J, Marcus S J, Sargent T D. Nucleic Acids Res. 1991;19:3709–3714. doi: 10.1093/nar/19.13.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayor R, Morgan R, Sargent M G. Development (Cambridge, UK) 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- 36.Nakata K, Nagai T, Aruga J, Mikoshiba K. Proc Natl Acad Sci USA. 1997;94:11980–11985. doi: 10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai C-J, Ekker S C, Beachy P A, Moon R T. Development (Cambridge, UK) 1995;121:2349–2360. doi: 10.1242/dev.121.8.2349. [DOI] [PubMed] [Google Scholar]
- 38.Chong L D, Park E K, Latimer E, Friesel R, Daar I O. Mol Cell Biol. 2000;20:724–734. doi: 10.1128/mcb.20.2.724-734.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saleh M, Huang H, Green N C, Featherstone M S. Exp Cell Res. 2000;260:105–115. doi: 10.1006/excr.2000.5010. [DOI] [PubMed] [Google Scholar]
- 40.Di Rocco G, Mavilio F, Zappavigna V. EMBO J. 1997;16:3644–3654. doi: 10.1093/emboj/16.12.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaBonne C, Bronner-Fraser M. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- 42.Mayor R, Guerrero N, Young R M, Gomez-Skarmeta J L, Cuellar C. Mech Dev. 2000;97:47–56. doi: 10.1016/s0925-4773(00)00412-3. [DOI] [PubMed] [Google Scholar]
- 43.Selleri L, Depew M J, Jacobs Y, Chanda S K, Tsang K Y, Cheah K S E, Rubenstein J L R, O'Gorman S, Cleary M L. Development (Cambridge, UK) 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- 44.Saleh M, Rambaldi I, Yang X J, Featherstone M S. Mol Cell Biol. 2000;20:8623–8633. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]