Abstract
The duplication of Hox clusters and their maintenance in a lineage has a prominent but little understood role in chordate evolution. Here we examined how Hox cluster duplication may influence changes in cluster architecture and patterns of noncoding sequence evolution. We sequenced the entire duplicated HoxAa and HoxAb clusters of zebrafish (Danio rerio) and extended the 5′ (posterior) part of the HoxM (HoxA-like) cluster of horn shark (Heterodontus francisci) containing the hoxa11 and hoxa13 orthologs as well as intergenic and flanking noncoding sequences. The duplicated HoxA clusters in zebrafish each house considerably fewer genes and are dramatically shorter than the single HoxA clusters of human and horn shark. We compared the intergenic sequences of the HoxA clusters of human, horn shark, zebrafish (Aa, Ab), and striped bass and found extensive conservation of noncoding sequence motifs, i.e., phylogenetic footprints, between the human and horn shark, representing two of the three gnathostome lineages. These are putative cis-regulatory elements that may play a role in the regulation of the ancestral HoxA cluster. In contrast, homologous regions of the duplicated HoxAa and HoxAb clusters of zebrafish and the HoxA cluster of striped bass revealed a striking loss of conservation of these putative cis-regulatory sequences in the 3′ (anterior) segment of the cluster, where zebrafish only retains single representatives of group 1, 3, 4, and 5 (HoxAa) and group 2 (HoxAb) genes and in the 5′ part of the clusters, where zebrafish retains two copies of the group 13, 11, and 9 genes, i.e., AbdB-like genes. In analyzing patterns of cis-sequence evolution in the 5′ part of the clusters, we explicitly looked for evidence of complementary loss of conserved noncoding sequences, as predicted by the duplication-degeneration-complementation model in which genetic redundancy after gene duplication is resolved because of the fixation of complementary degenerative mutations. Our data did not yield evidence supporting this prediction. We conclude that changes in the pattern of cis-sequence conservation after Hox cluster duplication are more consistent with being the outcome of adaptive modification rather than passive mechanisms that erode redundancy created by the duplication event. These results support the view that genome duplications may provide a mechanism whereby master control genes undergo radical modifications conducive to major alterations in body plan. Such genomic revolutions may contribute significantly to the evolutionary process.
Hox genes encode transcription factors that control pattern formation along the anterior-posterior axis (1). Hox genes are organized in clusters produced by a series of tandem duplications of progenitor gene(s) long before the protostome-deuterostome split (2). Although cluster organization has been maintained in both protostome (e.g., Drosophila) and deuterostome (e.g., human) lineages, the number of Hox clusters varies. To date, all protostome lineages have a single Hox cluster (3–8). The number of clusters in deuterostomes ranges from one (e.g., amphioxus; ref. 5) to several [e.g., four in mammals (9) and seven in zebrafish (10)]. Hox clusters have been shaped by different molecular evolutionary processes including tandem gene duplications (2, 11), cluster duplication and subsequent gene loss (10, 12), gene sequence divergence, functional divergence, and regulatory element evolution. Hox clusters also display the phenomenon of colinearity, in which the position of a gene in the cluster is related to its spatiotemporal pattern of expression along the A–P axis. The study of Hox clusters, therefore, provides the opportunity to understand the relationship between molecular evolution, genome organization, and gene expression. Here we report on the molecular evolution of the HoxA cluster in representatives of the three major gnathostome lineages, horn shark (cartilaginous fish), common zebrafish (ray-finned fish), and human (lobe-finned fish).
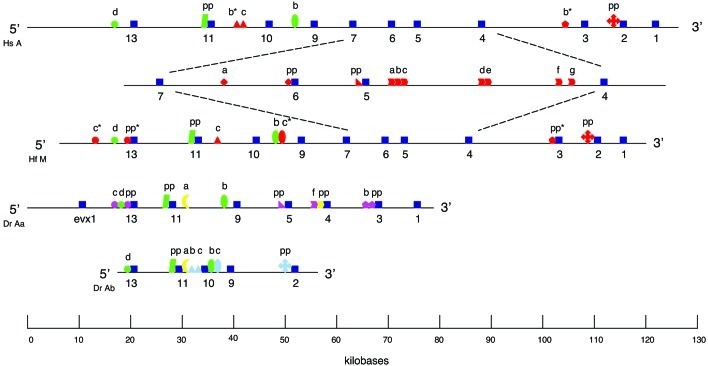
HoxA cluster organization and gene complement is highly conserved between horn shark and human (13). Here we first asked whether this conservation extends to noncoding DNA sequences, i.e., can the criterion of evolutionary conservation identify candidate cis-regulatory sequences that may be responsible for HoxA cluster and gene regulation? To address this question, we sequenced the 5′ (posterior) part of the HoxM (HoxA-like) cluster of the horn shark (Heterodontus francisci), which includes the previously undiscovered hoxa11 and unsequenced hoxa13 orthologs, as well as considerable stretches of noncoding DNA and spliced these sequences to the partial HoxA-like cluster of horn shark reported in ref. 13, generating a contig that spans the entire cluster. We aligned this sequence to a contig that contains the entire HoxA cluster sequence of human. We identified several putative cis-regulatory sequences that are conserved, some to a striking degree, between these two distantly related taxa (Fig. 1; Table 1). Our findings suggest that human and horn shark each may retain and share in common features of the “archetypal” HoxA cluster in the gnathostome common ancestor.
Figure 1.
HoxA clusters for human (Hs A), horn shark (Hf M), zebrafish (Dr Aa), and zebrafish (Dr Ab) are drawn schematically. Coding regions of each gene on a cluster are represented by dark blue squares. For human and horn shark only, the region spanning from hoxa7 to hoxa4 is enlarged. Phylogenetic footprint clusters (PFCs) within each intergenic region share the same geometric shape; letters above shape are as found in Table 1. Note that except for the region between hoxa7 and hoxa4, the only PFCs shown are those that are present in human and/or horn shark as well as at least one of the zebrafish clusters. PFCs shared between human and horn shark are red shapes. A red shape with an asterisk designates a PFC that is found only in human or horn shark. PFCs on the zebrafish HoxAa cluster that are homologous to horn shark and/or human are magenta shapes. PFCs on the zebrafish HoxAb cluster that are homologous to horn shark and/or human are light blue shapes. PFCs shared only by zebrafish HoxAa and HoxAb clusters or by at least one zebrafish cluster and the HoxA cluster of striped bass (Table 1) are yellow shapes. PFCs shared by all four clusters (taxa) are green shapes. See text and Table 1 for description. The line indicates distance in kilobases (kb).
Table 1.
Summary of PFCs detected in HoxA clusters
| PFC 5′–3′ | Number of PF, N | Homo sapiens A | Heterodontus francisci M | Danio rerio Aa | Danio rerio Ab | Morone saxatilis A |
|---|---|---|---|---|---|---|
| Upstream of 13-a* | 2 | 45,156–45,210 (10,752 nt) | 6,519–6,573 (9,775 nt) | |||
| Upstream of 13-b | 1 (n = 16 nt) | 45,454–45,469 (10,454 nt) | 6,799–6,814 (9,496 nt) | |||
| Upstream of 13-c | 4 | 8,558–8,898 (7,737 nt) | 21,743–21,961 (1,768 nt) | |||
| Upstream of 13-d | 3 | 53,837–53,897 (2,071 nt) | 13,232–13,292 (3,063 nt) | 22,665–22,689 (905 nt)† | 58,337–58,378 (827 nt)† | |
| 13pp | 4 | 16,136–16,285 (159 nt) | 23,322–23,560 (248 nt) | |||
| 13-11-a | 3 | 59,511–59,575 (11,330 nt) | 19,139–19,201 (10,846 nt) | |||
| 13-11pp | 6 | 70,667–70,784 (174 nt) | 29,795–29,916 (190 nt) | 31,067–31,250 (197 nt) | 67,995–68,103 (180 nt) | |
| 11-10-a | 3 | 75,351–75,404 (6,312 nt) | 34,437–34,489 (9,023 nt) | |||
| 11-10-b | 1 (n = 9 nt) | 76,034–76,042 (5,629 nt) | 71,322–71,330 (2,244 nt) | |||
| 11-10-c | 1 (n = 18 nt) | 76,089–76,106 (5,574 nt) | 35,064–35,081 (8,396 nt) | 71,450–71,473 (2,116 nt) | ||
| 11-9-a*‡ | 10 | 29,667–29,816 (10,185 nt) | 71,146–71,280 (7,641 nt) | |||
| 10-9-a* | 8 | 85,317–85,661 (4,867 nt) | 46,403–46,773 (6,926 nt) | 3,010–3,044 (3,462 nt)† | ||
| 10-9-b | 2 | 86,431–86,491 (4,097 nt) | 47,562–47,662 (5,767 nt) | 37,297–37,324 (2,555 nt) | 76,764–76,822 (2,025 nt) | 3,394–3,475 (3,078 nt) |
| 10-9-c | 1 (n = 21 nt) | 48,332–48,352 (4,997 nt) | 77,065–77,085 (1,724 nt) | |||
| 10-9-d | 2 | 90,066–90,146 (461 nt) | 52,913–52,992 (416 nt) | |||
| 10-9pp | 10 | 90,272–90,449 (257 nt) | 53,088–53,301 (242 nt) | 6,219–6,238 (253 nt) | ||
| 9-7-a | 2 | 94,195–94,235 (5,425 nt) | 56,999–57,039 (5,339 nt) | |||
| 9-7-b* | 8 | 94,470–94,680 (4,970 nt) | 57,233–57,438 (5,105 nt) | |||
| 9-7-pp* | 9 | 99,257–99,340 (183 nt) | 62,153–62,338 (185 nt) | 11,537–11,630 (104 nt) | ||
| 7-6-a | 9 | 103,244–103,410 (4,992 nt) | 66,475–66,640 (5,462 nt) | 14,808–14,945 (3,892 nt)§ | ||
| 7-6pp | 10 | 108,024–108,213 (212 nt) | 71,722–71,914 (215 nt) | |||
| 6-5pp* | 9 | 112,162–112,359 (216 nt) | 74,586–74,780 (219 nt) | 53,287–53,475 (265 nt)¶ | 18,425–18,613 (275 nt)§ | |
| 5-4-a* | 4 | 114,053–114,615 (10,749 nt) | 76,427–76,535 (11,435 nt) | |||
| 5-4-b | 5 | 114,717–114,793 (10,535 nt) | 76,648–76,710 (11,214 nt) | |||
| 5-4-c* | 13 | 115,547–115,810 (9,705 nt) | 77,569–77,832 (10,293 nt) | 21,576–21,750 (7,532 nt) | ||
| 5-4-d | 4 | 119,381–119,436 (5,871 nt) | 81,957–82,012 (5,905 nt) | 23,523–23,578 (5,585 nt) | ||
| 5-4-e | 13 | 119,820–120,077 (5,432 nt) | 82,457–82,716 (5,404 nt) | 24,230–24,303 (4,878 nt) | ||
| 5-4-f* | 12 | 121,994–122,212 (3,258 nt) | 84,830–85,048 (3,032 nt) | 59,806–59,946 (1,813 nt) | ||
| 5-4-g | 5 | 122,792–122,860 (2,460 nt) | 85,618–85,689 (2,244 nt) | |||
| 5-4 pp | 12 | 61,457–61,619 (162 nt) | 28,945–29,108 (163 nt) | |||
| 4-3-a | 5 | 128,868–128,945 (16,478 nt) | 91,111–91,190 (15,251 nt) | |||
| 4-3-b* | 5 | 142,345–142,462 (3,000 nt) | 70,156–70,268 (1,576 nt) | |||
| 4-3pp* | 9 | 106,192–106,356 (170 nt) | 71,561–71,726 (171 nt) | |||
| 3-2-a* | 5 | 152,808–152,911 (677 nt) | 113,693–113,795 (558 nt) | |||
| 3-2pp* | 11 | 153,150–153,375 (335 nt) | 113,960–114,181 (291 nt) | 90,567–90,778 (271 nt)‖ | ||
| 2-1-a | 6 | 156,894–157,044 (3,179 nt) | 117,382–117,518 (2,791 nt) |
Column 1, PFCs detected in HoxA clusters. For each region, the distal most (5′-most) element is designated a. The proximal promoter is designated pp. Column 2, N = number of phylogenetic footprints in each PFC. When N = 1, the number of nucleotides (n) is given. Columns 3–7, nucleotide positions of each PFC in HoxA cluster sequence files (nucleotides 181, 288–344, 666) and their GenBank accession nos.: human, AC004079, AC004080, and AC010990; horn shark, AF479755; zebrafish (Aa), AC107364; zebrafish (Ab), AC107365; striped bass, AF089743. Numbers in parentheses indicate the distance, in nucleotides, from the first nucleotide of the PFC to the nucleotide immediately 5′ to the ATG of the anterior gene for a given intergenic region. See text for description.
Indicates that this PFC has at least one TAAT or ATTA motif.
Indicates that only part of the PF or PFC is shared with horn shark or human.
This intergenic region reflects the absence of the group 10 gene on the zebrafish HoxAa cluster (Fig. 1). Nucleotides in parentheses are the position from hoxa9a.
This PFC was detected in the region between hoxa7 and hoxa5 of the striped bass HoxA cluster. Nucleotides in parentheses are the position from hoxa5.
Detected in the intergenic region between hoxa9a and hoxa5a of the zebrafish HoxAa cluster. Nucleotides in parentheses are the position from hoxa5a.
Detected in the intergenic region between hoxa9b and hoxa2b of the zebrafish HoxAb cluster. Nucleotides in parentheses are the position from hoxa2b.
Working in this framework, we next asked how HoxA cluster architecture and patterns of putative cis-regulatory sequences evolve subsequent to cluster duplication. The common zebrafish (Danio rerio) possesses two HoxA clusters (HoxAa and HoxAb) as a result of duplication of either the entire genome or local regions (10). At present, evidence suggests that other teleosts including pufferfish (Fugu rubripes; ref. 14), cichlid (Oreochromis niloticus; ref. 12), and likely the striped bass (Morone saxatilis; refs. 12 and 15 and this study) also possess more than one HoxA cluster. To address the second question, we sequenced the entire HoxAa and HoxAb clusters of the common zebrafish and present a detailed description of their structural and putative cis-regulatory features. Differences in cluster architecture and patterns of putative cis-regulatory sequences between the duplicated HoxA clusters of zebrafish and the single HoxA clusters of horn shark and human (Fig. 1; Table 1) have yielded significant insights and raised interesting questions on the molecular evolutionary processes that have shaped the HoxA cluster in gnathostomes.
Materials and Methods
Genomic DNA Sequencing.
Two P1 artificial chromosome (PAC) genomic clones containing the entire HoxAa [241-I7 (10, 16) and HoxAb (10-O19); ref. 17] clusters of zebrafish were shotgun-sequenced and contiged at the Whitehead Institute Center for Genome Research (Massachusetts Institute of Technology, Cambridge, MA). Clones 241-I7 and 10-O19 have the GenBank accession nos. AC107364 and AC107365, respectively. In addition, we sequenced an isolated PAC clone (45E) of the horn shark (GenBank accession no. AF479755) that extends the HoxM (HoxA-like) cluster 5′ of the group 10 gene and spliced the sequence to nucleotide position 1 of clone Het1 (GenBank accession no. AF224262; ref. 13), generating a 124,487-bp contig that contains the entire HoxA cluster (Fig. 1). The human HoxA cluster sequence spans nucleotide positions 181,288–344,666 of a larger contig (GenBank accession nos. AC004079, AC004080, and AC010990).
Cluster Alignment and Sequence Comparisons.
We assembled, aligned, and analyzed nucleotide sequences of the entire HoxA clusters of horn shark, zebrafish (Aa, Ab), and human (Fig. 1). Some analyses included partial HoxA cluster sequences of the striped bass (GenBank accession no. AF089743; ref. 15). HoxA clusters were confirmed as orthologous by using two methods: (i) whole cluster alignments, drawn as percentage identity plots, using the PIPMAKER program (http://bio.cse.psu.edu/pipmaker/; ref. 18) and (ii) evolutionary analyses of coding regions (data not shown). Individual sequence files of each HoxA intergenic region also were constructed for each taxon. For example, four intergenic sequence files were created for the DrHoxAb sequence (hox13-11b, hox11-10b, hox10-9b, and hox9-2b; see Fig. 1).
Phylogenetic Footprints (PFs) and PFCs.
PFs are defined as short blocks of noncoding DNA sequence, typically 6 bp or more, that are 100% conserved in taxa that have an additive evolutionary time of at least 250 million years (19). So-identified PFs are putatively functional cis elements and in several cases have been shown to regulate gene expression (e.g., ref. 20). We identified PFs in intergenic regions using CLUSTAL W (21), PIP (18), and BAYESIAN (22) alignment methods; the latter method does not require preset gap penalties. Most PFs were found to be (i) in close proximity to one another, i.e., at least two and as many as 13 PFs are located within 200 bp, and (ii) located at comparable distances, in nucleotides, from the 3′ gene of each intergenic region (Table 1). We designated these PFCs, organized and named according to the specific intergenic region, starting with the letter “a” and moving in a 5′-to-3′ direction. A PFC located within 350 bp upstream of the start codon is a “proximal promoter.” The results are summarized in Table 1. The full data set of PF and PFC sequences is available at the website of C.-h.C. (http://lifesci.rutgers.edu/∼molbiosci/Professors/chiu.html).
Results
HoxA Cluster Architecture and Evolution by Duplication.
Here we sequenced an isolated PAC clone of the horn shark that extends the HoxA-like cluster (13) at the 5′ end (Fig. 1). Although clone 45E extends nearly 60 kb upstream of hoxa13, it does not contain an evx1 gene. The exact number of Hox clusters in the horn shark is yet to be determined; however, the results illustrated in Fig. 1 are consistent with the possibility that cartilaginous (e.g., horn shark) and lobe-finned [e.g., lungfish (23) and human (9)] fishes each possess a single HoxA-like cluster with a diagnostic architecture and size of ≈110 kb.
We also sequenced PAC clones containing the entire HoxAa and HoxAb clusters of zebrafish. In striking contrast to the HoxA clusters of human and horn shark, the duplicated zebrafish clusters are highly modified in structure, most notably in reduced gene complement (10) and overall size (Fig. 1). Counting from the first nucleotide of the hoxa13a gene to the last nucleotide of the hoxa1a gene, the zebrafish HoxAa cluster is 57,871 bp. The evx1 gene is only ≈10 kb upstream from the hoxa13a gene, much shorter than the 44-kb distance in human. Counting from the first nucleotide of the Hoxa13b gene to the last nucleotide of the Hoxa2b gene, the zebrafish HoxAb cluster is only 33,347 bp, approximately one-third the size of the HoxA clusters of human or horn shark. The genes in paralog groups 7 and 6 are lost in both zebrafish clusters. The genes in paralog groups 5–1 are maintained in complementary sets in the zebrafish clusters (Fig. 1; ref. 10). The segment 3′ of the group 9 paralog in the HoxAb cluster is much shorter than the same segment in the HoxAa cluster; both are shorter than the orthologous segments of the horn shark and human HoxA clusters (Fig. 1). Interestingly, the posterior segments of each zebrafish HoxA cluster that contain the Abd-B-like genes (paralog groups 9, 10, 11, and 13), have remained similar in size to one another. Unlike the situation in the 3′ end of the clusters, however, only one gene, paralog group 10 on the zebrafish Aa cluster, has been lost after the cluster duplication (Fig. 1). Note that the intergenic distance between the group 11 and 9 paralogs on the HoxAa cluster is not reduced by the loss of the group 10 gene. These observations highlight different patterns of evolution of the 3′ and 5′ parts of the HoxA clusters after duplication (see below).
The architecture of the striped bass partial HoxA cluster (15) resembles the zebrafish HoxAa cluster with the presence of group 9, 5, and 4 genes and the absence of a group 6 gene and resembles the HoxAb cluster with the presence of a group 10 gene. Differential partitioning of genes after HoxA cluster duplication has been documented also for Fugu (14); hence, it is possible that the striped bass has more than one HoxA cluster. Accelerated rates of hoxa4 and hoxa9 coding sequence evolution in the striped bass (12) is consistent with cluster duplication, as was found for the Hoxa11a and Hoxa11b genes of zebrafish (16).
HoxA cis-Sequence Patterns Detected in Human–Horn Shark Comparisons.
Noncoding sequences between genes, i.e., intergenic regions, can contain two types of functional sequences: proximal promoter elements, defined here as the first 350 bp immediately 5′ of the first codon, and “distal” elements, which act to enhance or suppress gene expression. We identified putatively functional, intergenic cis sequences using two criteria: evolutionary conservation and distance, in nucleotides, of cis-sequence elements to each other and to the anterior-most (3′) gene of each pair. As illustrated in Table 1, we found that the HoxA clusters of human and horn shark share a striking degree of cis-sequence conservation. PFCs are distributed throughout homologous regions of the human and horn shark HoxA clusters, with the highest concentration of PFCs near the 4, 5, and 6 orthologs (Fig. 1). The 2, 5, 6, 7, 9, and 11 orthologs of human and horn shark share conserved, putative proximal promoters. The 9-7-pp sequence (Table 1), which is a proximal promoter of hoxa7, also is conserved in the hoxa7 proximal region of mouse and chick (24). A number of PFCs detected upstream (6-5-a, 6-5-pp) and downstream (5-4-a,b,c,d,e) of the hoxa5 gene (Table 1; Fig. 1) coincide with an 11.1-kb genomic fragment of mouse hoxa5 (from −3.8 kb to + 7.3 kb) that reproduces, to a large extent, the endogenous expression of hoxa5 in transgenic mouse embryos (25). Our findings are consistent with the observations that for vertebrate taxa, (i) functional cis sequences in one species are evolutionarily conserved (26, 27) and (ii) cis sequences identified using evolution-based methods are functional when tested (16, 20, 28, 29). It will be important to determine whether the additional PFCs identified in this study are functional.
Patterns of cis-Sequence Evolution in the Duplicated HoxAa and HoxAb Clusters of Zebrafish and the HoxA Cluster of Striped Bass.
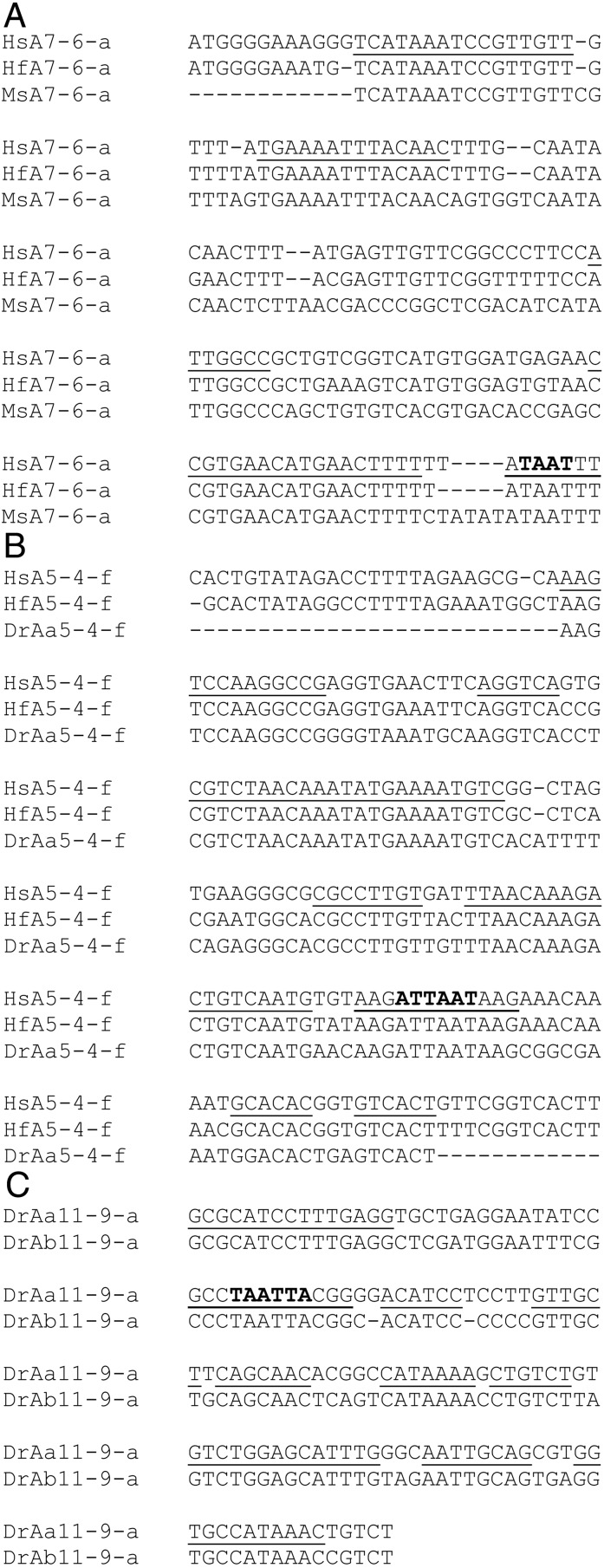
A major objective of this study was to investigate the patterns of cis-sequence evolution in the duplicated HoxAa and HoxAb clusters of zebrafish. We carried out this objective using two approaches, and each yielded interesting patterns. First, we used the PFCs identified in the orthologous HoxA clusters of human and horn shark (Fig. 1; Table 1) as a guide to search for homologous PFCs in the zebrafish HoxAa and HoxAb clusters. We also included the sequences of a partial HoxA cluster of the striped bass (15). The first, most frequent pattern is that the teleost HoxA clusters do not have homologues, as judged by sequence conservation (see Discussion), of several PFCs shared by human and shark (Table 1). The second pattern shows that there are very few PFCs that are shared in all clusters and all taxa, i.e., are symplesiomorphic for the archetypal HoxA cluster (Fig. 1; Table 1). Indeed, we detected only five PFCs: upstream of 13-c, shared in human, horn shark, and zebrafish (Aa and Ab); 13-11-pp (hoxa11 proximal promoter), shared in human, horn shark, and zebrafish (Aa and Ab); 10-9-b, shared in human, horn shark, zebrafish (Aa and Ab), and striped bass; 9-7-pp (hoxa7 proximal promoter), shared in human, horn shark, and striped bass; and 7-6-a, shared in human, horn shark, and striped bass (Fig. 2A). Note, the latter two examples are interesting, because striped bass, human, and horn shark possess hoxa7 orthologs, whereas the gene has been lost in both zebrafish clusters (Fig. 1; ref. 15). The third major pattern is that, for some HoxA genes, zebrafish and/or striped bass intergenic sequences retain a subset of the full spectrum of PFCs identified in the orthologous HoxA region of human and horn shark (Fig. 1; Table 1). A good example is PFC 5-4-f, which is one of seven PFCs located in the hoxa5–hoxa4 intergenic region of the human and horn shark HoxA clusters and is the only PFC detected in the homologous segment of the zebrafish HoxAa cluster, which has these two genes (Figs. 1 and 2B; Table 1). In the homologous segment of the striped bass HoxA cluster, only PFCs 5-4-c, 5-4-d, and 5-4-e are found (Fig. 1; Table 1).
Figure 2.
PFCs detected in orthologous HoxA clusters. Abbreviations are as described in the Fig. 1 and Table 1 legends. PFs conserved among all taxa in the alignment are underlined. TAAT or ATTA motifs are in bold. (A) PFC 7-6-a. (B) PFC5-4-f. (C) PFC 11-9-a.
In the second phase of this analysis, we carried out different combinations of intergenic sequence alignments to identify PFCs shared by teleosts, i.e., autapomorphies that may or may not be associated with duplication, and differential partitioning of elements conserved with human or horn shark, but not both. The example for a PFC that is shared only by the zebrafish HoxAa and HoxAb clusters is 11-9-a, which consists of 10 closely spaced PFs (Figs. 1 and 2C). By using parsimony, this element was acquired in the ray-finned fish lineage before the duplication of the HoxA clusters in zebrafish. We note, however, that the alternative scenario, i.e., independent loss of this element in the separate lineages leading to both human and horn shark, cannot be excluded entirely. Most significant to this study, however, is this signature of extensive cis-sequence conservation in the paralogous zebrafish HoxAa and Ab clusters (see Discussion).
We found six examples of PFCs that are shared between one zebrafish HoxA cluster and a HoxA cluster of either human or shark, but not both (Fig. 1; Table 1). The parsimonious interpretation of this pattern is that the PFC is ancestral (plesiomorphic) but that it has been lost in (i) the HoxA clusters of either horn shark or human and (ii) one of the zebrafish HoxA (Aa or Ab) clusters after duplication.
Discussion
Our investigations on the molecular evolution of the HoxA cluster in taxa that represent the three major gnathostome lineages have produced two significant findings. First, the single HoxA clusters of two distantly related taxa, the horn shark and human, are remarkably conserved in architecture and putatively functional cis-regulatory sequences. Second, the HoxAa and HoxAb clusters of zebrafish exhibit striking modifications of these conserved structural and putative regulatory features, most likely the outcome of HoxA cluster duplication. In fact, Hox cluster duplication and retention of duplicates in a lineage have played a prominent, although relatively little understood, role in the evolution of chordates. Major architectural changes associated with cluster duplication include compression and gene loss (refs. 10, 14, and 15 and this study). Indeed, compression of Hox clusters during evolution of the chordates is a definite trend if one considers that the single Hox cluster of the protochordate Amphioxus is ≈400 kb (11), each of the four Hox clusters (A, B, C, and D) in human is ≈110 kb, and the additional, i.e., greater than four, Hox clusters of euteleosts are considerably shorter than 110 kb (Fig. 1; ref. 15). The duplicated HoxA clusters of zebrafish also exhibit a trend of gene loss, particularly in the 3′ cluster segment where hoxa7 and hoxa6 are lost in both clusters and the other 3′ genes are retained in complementary sets: hoxa2 in the HoxAb cluster and hoxa5, hoxa4, hoxa3, and hoxa1 in the HoxAa cluster (Fig. 1; ref. 10). Interestingly, we found cluster compression to be correlated only loosely with gene loss. Our data on the exact intergenic distances of genes housed in the zebrafish HoxAa and HoxAb clusters (Fig. 1) show that the distance between hoxa11a and hoxa9a in the HoxAa cluster is slightly larger than the same intergenic region in the HoxAb cluster, even though the HoxAa cluster does not have a hoxa10a. Hence, our results on the zebrafish provide further confirmation that cluster compression is caused primarily by a decrease in intergenic distances, as found previously for pufferfish (14) and striped bass (15).
Cluster and/or gene(ome) duplications create functional redundancy that can be resolved by different molecular evolutionary mechanisms including the loss of one of the duplicate daughters, and the remaining daughter carries out the ancestral function(s) (e.g., semiconservative; ref. 30) that could account for the trend of gene loss and shortening of intergenic sequences or by retention of both daughter duplicates via subfunctionalization (31, 32). A highly feasible mechanism by which subfunctionalization occurs is the so-called duplication-deletion-complementation model (32) whereby the two duplicate genes retain complementary subsets of the cis-regulatory sequences and functional repertoire of the progenitor gene. Such a mechanism, acting on duplicated Hox clusters, could produce shortening of intergenic, noncoding sequences, which suggests that changes in the zebrafish HoxAa and HoxAb cluster architecture and patterns of conservation of cis-regulatory sequences could be a passive consequence of the processes outlined above that erode genetic redundancy created by the duplication event. However, we found patterns of noncoding sequence evolution that are largely inconsistent with the expectations that follow from these processes.
Conservation of noncoding sequences is a powerful method of identifying functional cis-regulatory sequences (20, 27). We found the single HoxA clusters of horn shark and human to share a striking degree of putatively functional cis-sequences, particularly among the “anterior” (3′) HoxA genes (Fig. 1; Table 1). These results imply that there is strong selection for maintaining cis-regulatory sequence motifs that most likely are necessary for essential (plesiomorphic) Hox gene functions ancestral for gnathostomes. Hence, it is reasonable to expect that a cis sequence that is conserved in the ancestral lineage before duplication will be retained in one of the duplicated daughter genes, especially if its sister duplicate is lost, and the remaining gene has to perform the function(s) of the ancestral gene. Our results on the duplicated HoxA clusters of zebrafish are inconsistent with this expectation. Although we did find a small number of conserved cis sequences in the 3′ segment of the zebrafish HoxAa or HoxAb clusters that retain at most a single copy of any 3′ gene (Fig. 1), the majority have been lost. We infer from these results that the essential Hox gene functions in zebrafish are performed with different cis-regulatory elements (e.g., phenogenetic drift; ref. 33) than of the ancestral gene with cis elements highly conserved in horn shark and human. It is important to note that although the majority of noncoding sequences of the zebrafish HoxAa and HoxAb clusters show little or no homology at the sequence level to HoxA cluster sequences of horn shark and human (Table 1), it is possible that they are homologous in function. This phenomenon has been reported elsewhere for nonvertebrate metazoans (e.g., even-skipped stripe 2 element of Drosophila; refs. 34 and 35).
Nevertheless, our results are intriguing, because despite the fact that human and horn shark diverged from one another more than 400 million years ago and have considerable differences in morphology, selection acting on the HoxA clusters has been strong enough to maintain their remarkable conservation in putatively functional cis-regulatory sequences in the absence of a cluster duplication. Yet, after HoxA cluster duplication in the zebrafish, radical remodeling of cis sequences occurred even when only one gene is retained to perform these functions. Interestingly, some of the cis elements conserved in human and horn shark and absent in zebrafish are retained in the striped bass and vice versa (Table 1). If modification of cis-regulatory elements is caused only by relaxed selection, then the homologous, ancestral cis elements in all teleost lineages should be affected. The retention of different ancestral elements in zebrafish and striped bass suggests that cis-regulatory elements in these different lineages have been shaped by distinct and different selection pressures.
As noted above, the 5′ segment of the duplicated zebrafish clusters retains genes from paralog groups 9, 11, and 13 (Fig. 1). If two genes are maintained after gene duplication, this can be caused either by subfunctionalization (32) or the acquisition of a new function by one or both genes (36). In the former case, each duplicated gene retains a subset of the ancestral functions, and thus both are maintained by stabilizing selection. The duplication-degeneration-complementation model of this process (32) proposes that if the various functions of the ancestral genes are regulated by modular enhancers, subfunctionalization simply may be the result of the passive loss of complementary enhancer sequences. Such a process is entirely nonadaptive, i.e., driven only by the chance fixation of degenerative mutations. This model makes strong predictions about the pattern of sequence conservation after duplication. Cis-regulatory sequences conserved in the ancestral lineage are expected to be retained in complementary sets among the two duplicates. Here we examined patterns of putatively functional cis-sequence evolution to test these predictions.
If each PFC is indicative of a modular enhancer, one possibility is that the duplicated zebrafish paralogs retain two, complementary subsets of the PFCs detected in the HoxA clusters of human and horn shark. Of the nine PFCs described in the 5′ region of human and horn shark HoxA clusters (excluding proximal elements), four have no counterpart in the zebrafish, one is maintained in both paralogs, and only four are found in either the HoxAa or HoxAb clusters (Fig. 1; Table 1). In addition, one extensive PFC 11-9-a (Table 1; Figs. 1 and 2C), which comprises 10 individual PFs, is found only in the zebrafish HoxAa and HoxAb clusters (Table 1). Thus, only 4 of 10 PFCs exhibit patterns that are consistent with the predictions of the duplication-degeneration-complementation model. It is possible that individual PFs in a PFC are parts of different modular enhancers. If so, one would expect that for an individual PFC identified in human and horn shark, each of the two zebrafish paralogs retain complementary sets of PFs. We found no example of this pattern in our data.
Although enhancer modularity has been shown for a number of genes (37), it may not be the rule for Hox genes. A possible reason for the conservation of Hox clusters as clusters may be that extensive sharing of enhancers among genes in the same Hox cluster prevents the dislocation of Hox genes from each other. Enhancer sharing also may explain the extent of noncoding sequence conservation between horn shark and human. In the absence of enhancer modularity, complementary degeneration of enhancers may not be an option for Hox genes.
In interpreting our findings, we think it likely that after Hox cluster duplication the lineage benefits from an increased evolvability in the developmental genetic system. If this increased evolvability further coincides with an adaptive radiation, the duplicated Hox clusters may experience adaptive remodeling by directional selection rather than just the erosion of genetic redundancy. Duplicated Hox paralogs exhibit increased rates of coding sequence evolution (12, 16), experience differential purifying selection (38), and tests for directional selection on duplicated genes are significant for all zebrafish Hox gene paralogs tested (hoxb5, hoxb6, and hoxc6) but only for 2 of 22 other duplicated genes (39). It is possible, therefore, that Hox cis-regulatory elements also can be subject to directional selection after duplication. Such an adaptive remodeling may be an important reason for why duplicated Hox clusters are maintained. In fact, there are only four clusters described for amniotes, and possibly no more exist in all sarcopterygians. Yet there are many vertebrate lineages known to have undergone tetraploidization; hence passive erosion of genetic redundancy rarely if ever leads to the retention of new Hox clusters. Thus we think that further efforts in understanding the evolution of Hox clusters should be directed toward testing the possibility that Hox cluster duplication and retention is an adaptive phenomenon. To do this task it will be necessary to obtain a much more fine-grained taxon sampling, in particular among basal actinopterygians (associated with the teleost duplications) and the cartilaginous fishes (associated with the duplication event leading to the four gnathostome clusters).
Acknowledgments
We thank Kenta Sumiyama, Steven Irvine, Tom Powers, and Ashley Carter for insightful comments on this work. We also thank Eric S. Lander and the Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research sequencing teams for the sequencing of the zebrafish clones. This research was supported by a National Science Foundation/Sloan Postdoctoral Fellowship in Molecular Evolution (to C.-h.C.), National Science Foundation Grants IBN-9905403 (to F.H.R. and G.P.W.) and IBN-9905408 (to C.A.), and National Institutes of Health Grant R24-RR14085-2 (to C.A.).
Abbreviations
- PF
phylogenetic footprint
- PFC
phylogenetic footprint cluster
- PAC
P1 artificial chromosome
Footnotes
References
- 1.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 2.Kappen C, Schughart K, Ruddle F H. Proc Natl Acad Sci USA. 1989;86:5459–5463. doi: 10.1073/pnas.86.14.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akam, M., Averof, M., Castelli-Gair, J., Dawes, R., Falciani, F. & Ferrier, D. (1994) Dev. Suppl. 209–215. [PubMed]
- 4.Wang B B, Muller-Immergluck M M, Austin J, Robinson N T, Chisholm A, Kenyon C. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Fernandez J, Holland P W. Nature (London) 1994;370:563–566. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- 6.Martinez P, Rast J P, Arenas-Mena C, Davidson E H. Proc Natl Acad Sci USA. 1999;96:1469–1474. doi: 10.1073/pnas.96.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powers T P, Hogan J, Ke Z, Dymbrowski K, Wang X, Collins F H, Kaufman T C. Evol Dev. 2000;6:311–325. doi: 10.1046/j.1525-142x.2000.00072.x. [DOI] [PubMed] [Google Scholar]
- 8.Devenport M P, Blass C, Eggleston P. Evol Dev. 2000;6:326–339. doi: 10.1046/j.1525-142x.2000.00074.x. [DOI] [PubMed] [Google Scholar]
- 9.Krumlauf R. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 10.Amores A, Force A, Yan Y, Joly L, Amemiya C, Fritz A, Ho R K, Langeland J, Prince V, Wang Y L, et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 11.Ferrier D E, Minguillon C, Holland P W, Garcia-Fernandez J. Evol Dev. 2000;5:284–293. doi: 10.1046/j.1525-142x.2000.00070.x. [DOI] [PubMed] [Google Scholar]
- 12.Malaga-Trillo E, Meyer A. Am Zool. 2001;41:676–686. [Google Scholar]
- 13.Kim C-B, Amemiya C, Bailey W, Kawasaki K, Mezey J, Miller W, Minoshima S, Shimizu N, Wagner G, Ruddle F H. Proc Natl Acad Sci USA. 2000;97:1655–1660. doi: 10.1073/pnas.030539697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aparicio S, Hawker K, Cottage A, Mikawa Y, Zuo L, Venkatesh B, Chen E, Krumlauf R, Brenner S. Nat Genet. 1997;16:79–83. doi: 10.1038/ng0597-79. [DOI] [PubMed] [Google Scholar]
- 15.Snell E A, Scemama J L, Stellwag E J. J Exp Zool. 1999;285:41–49. [PubMed] [Google Scholar]
- 16.Chiu C-h, Nonaka D, Xue L, Amemiya C T, Wagner G P. Mol Phylogenet Evol. 2000;17:305–316. doi: 10.1006/mpev.2000.0837. [DOI] [PubMed] [Google Scholar]
- 17.Chiu C-h, Amemiya C T, Carr J L, Bhargava J, Hwang J K, Shashikant C S, Ruddle F H, Wagner G P. Dev Genes Evol. 2000;210:105–109. doi: 10.1007/s004270050016. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tagle D A, Koop B F, Goodman M, Slightom J L, Hess D L, Jones R T. J Mol Biol. 1988;203:439–455. doi: 10.1016/0022-2836(88)90011-3. [DOI] [PubMed] [Google Scholar]
- 20.Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, Liu J S, Lawrence C E. Bioinformatics. 1998;14:25–39. doi: 10.1093/bioinformatics/14.1.25. [DOI] [PubMed] [Google Scholar]
- 23.Longhurst T J, Joss J M. J Exp Zool. 1999;285:140–145. doi: 10.1002/(sici)1097-010x(19990815)285:2<140::aid-jez6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 24.Gaunt S J. Dev Dyn. 2001;221:26–36. doi: 10.1002/dvdy.1122. [DOI] [PubMed] [Google Scholar]
- 25.Larochelle C, Tremblay M, Bernier D, Aubin J, Jeannotte L. Dev Dyn. 1999;214:127–140. doi: 10.1002/(SICI)1097-0177(199902)214:2<127::AID-AJA3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 26.Nonchev S, Maconochie M, Vesque C, Aparicio S, Ariza-McNaughton L, Manzanares M, Maruthainar K, Kuroiwa A, Brenner S, Charnay P, Krumlauf R. Proc Natl Acad Sci USA. 1996;93:9339–9345. doi: 10.1073/pnas.93.18.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shashikant C S, Kim C-B, Borbely M A, Wang W C, Ruddle F H. Proc Natl Acad Sci USA. 1998;95:15446–15451. doi: 10.1073/pnas.95.26.15446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sumiyama, K., Kim, C.-B. & Ruddle, F. H. (2001) 71, 260–262. [DOI] [PubMed]
- 29. Chiu, C.-h. & Hamrick, M. W. (2002) Evol. Anthropol., in press.
- 30.Ruddle F H, Amemiya C T, Carr J L, Kim C-B, Ledje C, Shashikant C S, Wagner G P. Ann NY Acad Sci. 1999;870:238–248. doi: 10.1111/j.1749-6632.1999.tb08884.x. [DOI] [PubMed] [Google Scholar]
- 31.Hughes A L. Proc R Soc London Ser B. 1994;256:119–124. [Google Scholar]
- 32.Force A, Lynch M, Pickett F B, Amores A, Yan Y L, Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss K M, Fullerton S M. Theor Popul Biol. 2000;57:187–195. doi: 10.1006/tpbi.2000.1460. [DOI] [PubMed] [Google Scholar]
- 34.True J R, Haag E S. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig M Z, Bergman C, Patel N H, Kreitman M. Nature (London) 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 36.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 37.Kirchhamer C V, Yuh C H, Davidson E H. Proc Natl Acad Sci USA. 1996;93:9322–9328. doi: 10.1073/pnas.93.18.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dermitzakis E T, Clark A G. Mol Biol Evol. 2001;18:557–562. doi: 10.1093/oxfordjournals.molbev.a003835. [DOI] [PubMed] [Google Scholar]
- 39.Van De Peer Y, Taylor J S, Braasch I, Meyer A. J Mol Evol. 2001;53:436–446. doi: 10.1007/s002390010233. [DOI] [PubMed] [Google Scholar]