Abstract
Double-stranded RNA-mediated interference (RNAi) has recently emerged as a powerful reverse genetic tool to silence gene expression in multiple organisms including plants, Caenorhabditis elegans, and Drosophila. The discovery that synthetic double-stranded, 21-nt small interfering RNA triggers gene-specific silencing in mammalian cells has further expanded the utility of RNAi into mammalian systems. Here we report a technology that allows synthesis of small interfering RNAs from DNA templates in vivo to efficiently inhibit endogenous gene expression. Significantly, we were able to use this approach to demonstrate, in multiple cell lines, robust inhibition of several endogenous genes of diverse functions. These findings highlight the general utility of this DNA vector-based RNAi technology in suppressing gene expression in mammalian cells.
Double-stranded RNA (dsRNA) can trigger silencing of homologous gene expression by a mechanism termed RNAi (for RNA-mediated interference) (1). RNAi is an evolutionarily conserved phenomenon and a multistep process that involves generation of active small interfering RNA (siRNA) in vivo through the action of an RNase III endonuclease, Dicer. The resulting 21- to 23-nt siRNA mediates degradation of the complementary homologous RNA (reviewed in refs. 2–4). RNAi has been used as a reverse genetic tool to study gene function in multiple model organisms, including plants, Caenorhabditis elegans, and Drosophila where large dsRNAs efficiently induce gene-specific silencing (1, 5–7).
One obstacle to achieving RNAi in mammals is that dsRNAs longer than 30 nt will activate an antiviral response, leading to the nonspecific degradation of RNA transcripts and a general shutdown of host cell protein translation (8, 9). As a result, the long dsRNA, with a few exceptions (10, 11), does not produce RNAi activity, and RNAi therefore is not a general method for silencing specific genes in mammalian cells. This obstacle has been recently overcome by Tuschl and colleagues (12) who found that gene-specific suppression in mammalian cells can be achieved by vitro-synthesized siRNA that are 21 nt in length, long enough to induce gene-specific suppression, but short enough to evade the host interferon response.
In this article, we describe a DNA vector-based approach to achieve RNAi in mammalian cells. With this approach, small RNAs are predicted to be synthesized from a DNA template under the control of an RNA polymerase III (Pol III) promoter in transfected cells. Pol III has the advantage of directing the synthesis of small, noncoding transcripts whose 3′ ends are defined by termination within a stretch of 4–5 thymidines (Ts) (13). These properties make it possible to use DNA templates to synthesize, in vivo, small RNAs with structural features close to what has been found to be required for active siRNAs synthesized in vitro (14).
Using this DNA vector-based RNAi approach, we show that transfected as well as endogenous genes can be efficiently inhibited. We have examined the effects of the in vivo-synthesized siRNAs on a transfected reporter gene, a housekeeping gene, and genes involved in cell cycle control and DNA methylation. In each and every case, we find that these small RNAs efficiently and specifically inhibit the synthesis of proteins encoded by the corresponding genes. Taken together, we have developed an RNAi approach that uses a DNA template to synthesize small RNA in vivo. This technique can be broadly used for analysis of gene functions and thus will significantly facilitate the use of the RNAi technology in cell culture and vertebrate animals.
Materials and Methods
Construction of Plasmids That Contain DNA Templates for the Synthesis of siRNAs Under the Control of the U6 Promoter.
Plasmid pmU6 (−315/+1) (gift of S. Altman, Yale University, New Haven, CT) was used as a template for PCR isolation of the U6 promoter (−315 to +1) with an added ApaI cloning site at the transcriptional initiation site, which was cloned into Bluescript (BS) to generate the parent plasmid BS/U6. A general strategy for constructing an RNAi plasmid involved subcloning an inverted repeat into BS/U6 at the ApaI site. The selection of the coding sequences for siRNA was empirically determined but they started with GG and were analyzed by blast research to ensure that they did not have significant sequence homology with other genes. Insertion of the individual repeat motifs into BS/U6 was achieved in two separate steps. For example, to generate the BS/U6/gfp RNAi plasmid, a 22-nt oligo (oligo 1) corresponding to nucleotides 106–127 of the green fluorescent protein (GFP) coding region was first inserted into the BS/U6 vector digested with ApaI (blunted) and XhoI. The inverted motif that contains the 6-nt spacer and five Ts (oligo 2) was then subcloned into the XhoI and EcoRI sites of the intermediate plasmid to generate BS/U6/gfp. Oligo 1 is 5′-GGCGATGCCACCTACGGCAAGC-3′ (forward) and 5′-TCGAGCTTGCCGTAGGTGGCATCGCC-3′ (reverse). Oligo 2 is 5′-TCGAGCTTGCCGTAGGTGGCATCGCCCTTTTTG-3′ (forward) and 5′-AATTCAAAAAGGGCGATGCCACCTACGGCAAGC-3′ (reverse).
Construction of RNAi plasmids for the endogenous genes [lamin A/C, cyclin-dependent kinase-2 (cdk-2), and DNA methyltransferase-1 (dnmt-1)] was essentially the same as above. The sequences for the bodies of the siRNAs for lamin A/C, cdk-2, and dnmt-1 were taken from GenBank accession nos. XM-086566 (nucleotides 1627–1647), XM-049150 (nucleotides 652–672), and NM-001379 (nucleotides 598–617), respectively.
Cell Culture and Transfections.
HeLa, U-2 OS, H1299, and C-33A (American Type Culture Collection) cells were cultured in DMEM (GIBCO) supplemented with 10% of heat-inactivated FBS. Cells grown on coverslips in 6-well plates were transfected by using a calcium phosphate method and harvested 2–3 days after the transfection. Separate plasmids encoding GFP and siRNAs were generally used at a ratio of 1:10–1:30.
Immunofluorescence Microscopy.
Cells were harvested 3 days posttransfection for analysis. They were washed once with PBS and fixed with 3% paraformaldehyde in PBS for 20 min at room temperature. The cells were permeabilized with PBS containing 0.5% of Igepal CA-630 nonionic detergent (Sigma) for 10 min and washed twice in PBS containing 0.1% of Igepal (washing buffer). After blocking with washing buffer containing 10% FBS, cells were incubated with the appropriate primary antibodies for 2–4 h at room temperature. The anti-DNMT-1, anti-CDK-2, and anti-lamin B antibodies (Santa Cruz Biotechnology) were used at the dilutions of 1/50, 1/150, and 1/100 in blocking buffer, respectively. The monoclonal anti-hemagglutinin (HA) antibody was from Babco (Richmond, CA), and anti-lamin A/C antibody was from Cell Signaling (Beverly, MA). They were used at the dilutions of 1/300 and 1/100, respectively. After three washes, the cells were incubated with the corresponding secondary antibodies for 30 min at room temperature and washed three times with the washing buffer and once with PBS. The coverslips were then rapidly rinsed in water before being mounted in Vectashield medium (Vector Laboratories). The coverslips were analyzed by fluorescence microscopy (Leica, Deerfield, IL) using objective ×60, and the data were acquired with a Sony digital charge-coupled device camera and processed by Adobe photoshop software.
Western Blotting.
Two days after transfection, cells were washed with PBS and collected by scraping. They were lysed in ice-cold Tris buffer (50 mM, pH 7.5) containing 5 mM EDTA, 300 mM NaCl, 0.1% Igepal, 0.5 mM NaF, 0.5 mM Na3VO4, 0.5 mM PMSF, and antiprotease mixture (Roche Molecular Biochemicals), sonicated, and centrifuged at 13,000 × g for 10 min. The supernatant was used for protein determination by the Bradford procedure (Bio-Rad) and Western blotting. The proteins were resolved on 12% SDS-polyacrylamide gels, transferred onto nitrocellulose membranes, and incubated with the appropriate antibodies. The anti-GFP antibody was obtained from Santa Cruz Biotechnology and used at a 1/100 dilution. The anti-HA mAb was used at a 1/2,000 dilution. The peroxidase-based detection was performed with Chemiluminescence Reagent (NEN Life Science) according to the manufacturer's instructions.
Results and Discussion
siRNA Synthesized from DNA Templates in Vivo Efficiently Inhibited a Transfected Gene in Mammalian Cells.
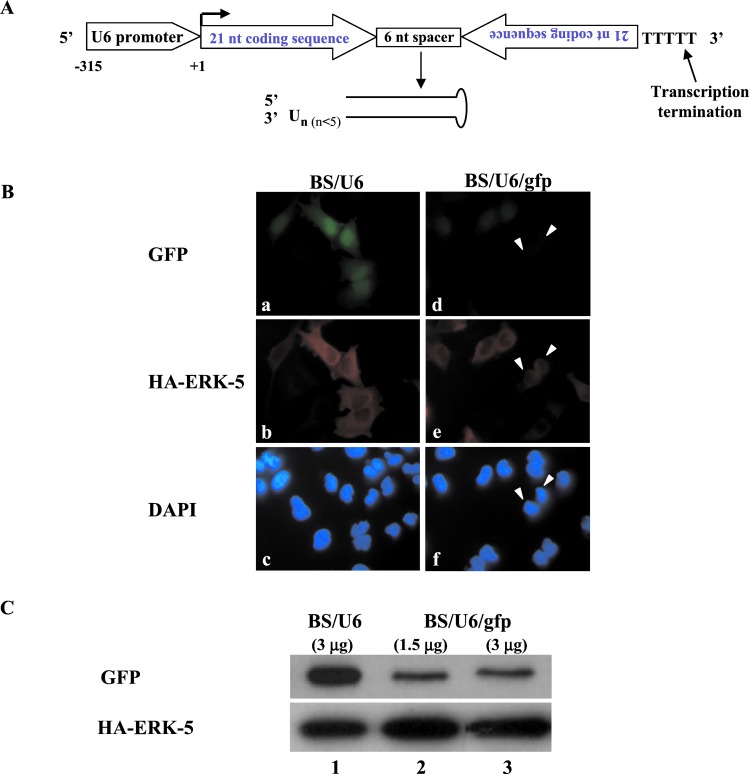
Tuschl and colleagues (14) defined the active, in vitro synthesized siRNA as a 21-nt-long dsRNA with symmetrical 2- to 3-nt 3′ overhangs (14). In other organisms such as C. elegans and Drosophila, the input RNA can be either in the form of a long dsRNA or a hairpin dsRNA (15, 16). Presumably, both forms of RNA are further cleaved by Dicer, a RNase III enzyme, to generate 21- to 23-nt-long siRNA (17–19). To synthesize, from a DNA template, a small RNA displaying features close to these requirements, we used RNA Pol III, which directs transcription that terminates at a run of 4–5 Ts, making it possible to design RNA with defined ends. The strategy we adopted is shown in Fig. 1A. Briefly, we inserted DNA fragments that acted as templates for the synthesis of small RNAs under the control of the mouse U6 promoter that directs the synthesis of a Pol III-specific RNA transcript (20). The resulting RNA is composed of two identical 21-nt sequence motifs in an inverted orientation, separated by a 6-bp spacer of nonhomologous sequences. Five Ts that function as a termination signal for Pol III (13) were added at the 3′ end of the repeat (Fig. 1A). This RNA is predicted to fold back to form a hairpin dsRNA with a 3′ overhang of several Ts (Fig. 1A). Although the exact structure of this small RNA is unknown, it robustly inhibited gene expression in vivo as described below. We therefore use the term siRNA to refer to these molecules.
Figure 1.
A siRNA synthesized from a DNA template in vivo inhibited expression of a transfected gene. (A) Strategy for generating siRNA from DNA template in vivo. An inverted repeat is inserted at the +1 position of the U6 promoter (−315 to + 1). The individual motif is 21 nt long and corresponds to the coding region of the gene of interest. The two motifs that form the inverted repeat are separated by a spacer of 6 nt. The transcriptional termination signal of five Ts is added at the 3′ end of the inverted repeat. The resulting siRNA is predicted to fold back to form a hairpin dsRNA as shown (drawing not to scale). The selection of the nucleotide sequence to be included in the siRNA vector is empirical and it is unknown whether certain regions of a given mRNA would be more or less susceptible to RNAi. (B) Analysis of gfp siRNA on GFP expression in HeLa cells. Vector BS/U6 or the plasmid carrying DNA templates that direct synthesis of gfp siRNA (BS/U6/gfp) were cotransfected with CMV-GFP and CMV-HA-ERK-5 into HeLa cells with a ratio of 20:1 (effector versus target plasmids) to ensure that cells that received GFP and HA-ERK-5 also received the RNAi plasmid. (a–c) Cells transfected with BS/U6 vector together with CMV-GFP and CMV-HA-ERK-5. (a) GFP-positive cells; (b) same field of cells stained with the anti-HA antibody to detect HA-ERK-5 expression; (c) same cells stained with 4′,6-diamidino-2-phenylindole (DAPI) to indicate all cells in the field. (d–f) Cells transfected with BS/U6/gfp and the GFP and HA-ERK-5 plasmids. Solid arrows indicate cells that are positive for HA-ERK-5 but display nearly undetectable GFP. All corresponding images were taken at the same exposure. (Magnification: ×60.) (C) Western blot analysis of GFP expression in cells cotransfected with either BS/U6 or BS/U6/gfp. (Upper) GFP proteins. (Lower) HA-ERK-5 detected with the anti-HA monoclonal antibody. Lane 1: BS/U6 vector control. Lanes 2 and 3: Cells transfected with 1.5 and 3.0 μg of the BS/U6/gfp plasmid. In all transfections, 100 ng of CMV-GFP and 0.5 μg of HA-ERK-5 plasmids were used.
Previous studies showed that the length of the 3′ overhangs plays a role in determining the activity of siRNA synthesized in vitro (14). Although the exact number of Ts has not been determined, we expect the 3′ overhang of the siRNA transcribed by Pol III in our system not to exceed five Ts (Fig. 1A). This prediction is based on the fact that human Pol III stops within or immediately after the five Ts (13). This structure preserves some of the features of the siRNA defined by Tuschl and colleagues (12) but is nevertheless distinct. Notably, only one end of the siRNA is exposed and its 3′ overhang is predicted to be slightly longer than 2–3 nt. Despite these differences, these siRNAs functioned effectively to inhibit gene expression in mammalian cells. We used this strategy to generate DNA templates for synthesis of siRNAs corresponding to the gfp (BS/U6/gfp), human lamin A/C (BS/U6/lamin A/C), cdk-2 (BS/U6/cdk-2), and dnmt-1 (BS/U6/dmnt-1) genes.
To determine whether this DNA vector-based approach can be used to inhibit gene expression in mammalian cells, we first tested DNA template-derived siRNA on a transfected plasmid encoding the GFP. We constructed a plasmid, BS/U6/gfp, that carries the U6 promoter linked to an inverted repeat matching a 21-nt coding region within the gfp gene. We transfected either the BS/U6 or BS/U6/gfp vector together with the target cytomegalovirus (CMV)-GFP plasmid and an unrelated HA-ERK-5 plasmid into HeLa cells. We then assayed for GFP and HA-ERK-5 expression within the same transfected cells by immunostaining (Fig. 1B). Whereas vector BS/U6 had no effect on GFP expression, BS/U6/gfp greatly diminished its expression. In most transfected cells (i.e., displaying HA-ERK-5 expression), GFP expression was reduced to near background levels as shown in Fig. 1B (two such cells are indicated by solid arrows in d). Significantly this effect is gene-specific because BS/U6/gfp did not inhibit the expression of cotransfected HA-ERK-5 (Fig. 1B, compare b and e). The presence of HA-ERK-5 did not affect RNAi because in its absence we observed similar level of inhibition of GFP by the GFP siRNA (data not shown). The immunostaining results were further confirmed by Western blotting, as shown in Fig. 1C. We used two different doses of the BS/U6/gfp plasmid in the experiment (1.5 and 3.0 μg) and observed similar reduction of GFP levels (Fig. 1C, compare lanes 2 and 3). Further experiments will be necessary to determine whether there is a dose–response in the inhibition mediated by siRNA synthesized from DNA templates. Compared with the BS/U6 vector control, the inhibition of GFP expression by BS/U6/gfp was estimated to be more than 80% (Fig. 1C, compare lanes 2 and 3 with lane 1). The lack of complete inhibition may in part be caused by high level of GFP expression that was directed by a strong CMV promoter. As shown below, RNAi inhibition of endogenous genes appeared to be more robust. Taken together, these findings strongly suggest that the DNA vector-based RNAi approach functions in mammalian cells.
Efficient Inhibition of Three Endogenous Genes by siRNAs Synthesized from DNA Templates in Vivo.
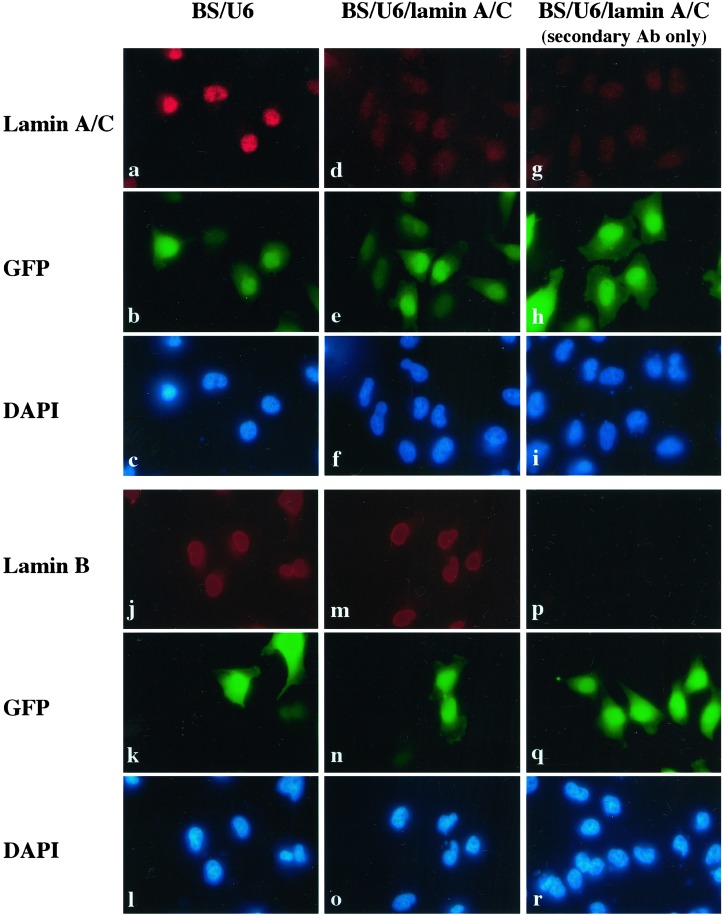
We next wanted to determine whether this approach functions to inhibit expression of endogenous genes. We analyzed three endogenous genes with diverse functions. The first gene we targeted for repression was the human lamin A/C gene, which has been shown to be effectively inhibited in cell culture by in vitro-synthesized siRNAs (12). We transfected HeLa cells with either BS/U6 vector or BS/U6/lamin A/C, which directs synthesis of a lamin A/C siRNA in vivo, together with CMV-GFP to mark the transfected cells. As shown in Fig. 2, whereas BS/U6 vector had no significant effect on lamin A/C expression (a), the plasmid BS/U6/lamin A/C reduced lamin A/C expression in transfected cells to levels comparable to those seen with the secondary antibody alone (compare d with a and g). Significantly, BS/U6/lamin A/C siRNA had no effect on the expression level of the related lamin B gene (Fig. 2, compare m with j), suggesting that the observed RNAi effect is gene-specific.
Figure 2.
Human lamin A/C siRNA derived from DNA template in vivo specifically inhibited expression of endogenous lamin A/C. HeLa cells were transfected with either BS/U6 vector or the RNAi plasmid BS/U6/lamin A/C together with CMV-GFP to mark transfected cells. (a and d) Cells stained with the anti-lamin A/C antibody. (g) BS/U6/lamin A/C-transfected cells stained with the secondary antibody alone to indicate background signals. b, e, and h, corresponding to a, d, and g, respectively show the same sets of cells positive for GFP, as a marker of transfected cells. c, f, and i are images of the same fields (as in a, d, and g, respectively) of cells stained with 4′,6-diamidino-2-phenylindole (DAPI) to identify all cells. Cells transfected with either the vector BS/U6 or the RNAi plasmid BS/U6/lamin A/C were also stained with antibodies that recognized the related lamin B protein (j and m). Corresponding images were taken at the same exposure. (Magnifications: ×60.)
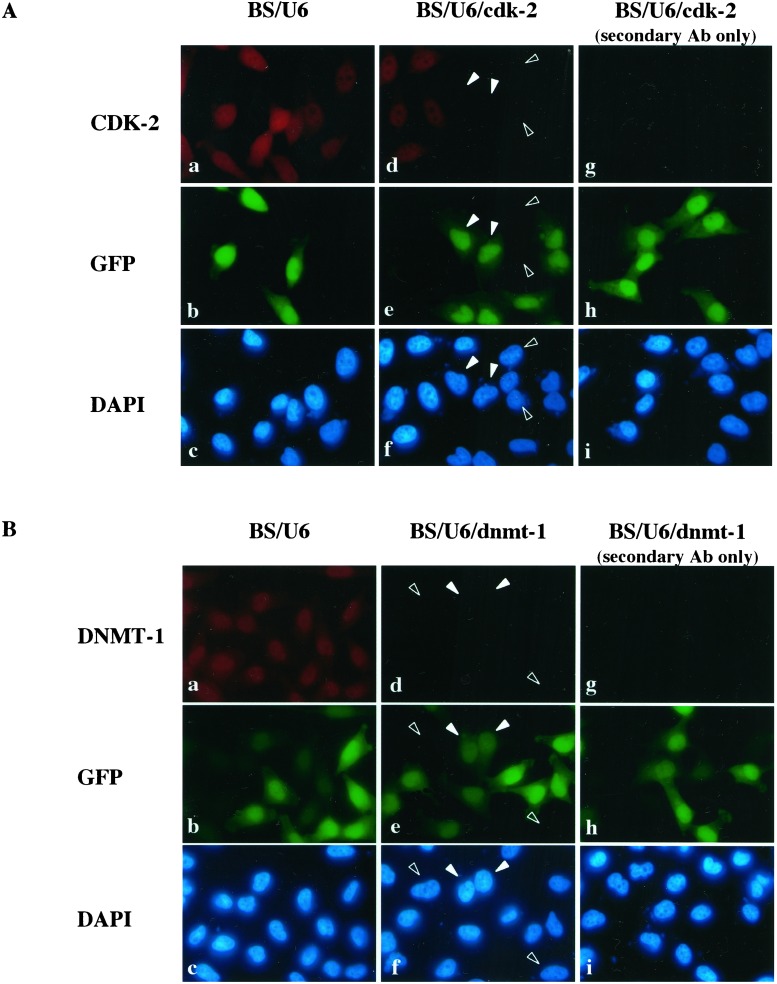
Having demonstrated that the DNA vector-based RNAi worked well to inhibit the expression of the lamin A/C gene, we asked whether this strategy functioned broadly to inhibit genes of interest by examining two additional genes involved in different aspects of cell biology. The second gene we targeted for repression was the human cdk-2 gene, which plays an important role in cell cycle control (reviewed in ref. 21). We constructed a plasmid carrying a DNA template that directs the synthesis of a cdk-2 siRNA and carried out transient transfection in HeLa cells as described above. GFP was cotransfected to mark the transfected cells. As shown in Fig. 3A, cells that have been transfected with the BS/U6/cdk-2 plasmid had significantly reduced, close to background, level of CDK-2 protein compared with BS/U6 vector-transfected cells (compare panel d with a and g, two transfected cells were indicated by solid arrows). In contrast, cdk-2 expression in nontransfected cells was comparable to that observed in cells transfected with the vector control (Fig. 3Aa). Because we transfected significantly more BS/U6/cdk-2 plasmid than GFP plasmid (20:1 ratio), some of the cells may have received only the RNAi plasmid and not the GFP plasmid. As expected, some GFP-negative cells (indicated by open arrows in Fig. 3A) also had reduced CDK-2 expression near to background level (Fig. 3A, compare d with a and g). Taken together, these data indicate that cdk-2 siRNA worked efficiently to reduce CDK-2 expression in vivo.
Figure 3.
Inhibition of expression of a cell cycle control gene and a DNA methyltransferase. (A) Inhibition of CDK-2 expression. HeLa cells were transfected with either BS/U6 vector or BS/U6/cdk-2 and CMV-GFP to mark transfected cells as above. Solid arrows indicate two of the GFP-positive cells (transfected cells) in which CDK-2 expression was below the level of detection. Open arrows indicate two GFP-negative cells in which CDK-2 expression is also undetectable. (B) Inhibition of a DNA methyltransferase (DNMT-1) expression. HeLa cells were transfected with either BS/U6 vector or BS/U6/dnmt-1 and CMV-GFP to mark transfected cells as above. Solid arrows indicate two GFP-positive cells (transfected cells) in which DNMT-1 expression was barely detectable. Open arrows indicate two GFP-negative cells in which DNMT-1 expression is also undetectable. The latter are likely to have received the RNAi plasmid only, because of the excess ratio of the RNAi vector versus GFP-encoding plasmid used in transfection.
We next targeted DNMT-1, which plays an important role in maintaining patterns of CpG methylation and the epigenetic control of gene expression in mammals (22). As shown in Fig. 3B, similar to what has been observed for lamin A/C and cdk-2, dnmt-1 expression can be efficiently inhibited by BS/U6/dnmt-1 siRNA in vivo (compare d with a). Once again, we observed GFP-negative cells (indicated by open arrows in Fig. 3B) that also had reduced DNMT-1 expression. As mentioned above, these cells may have received only BS/U6/dnmt-1 vector but not the GFP plasmid, because of the high RNAi-to-GFP plasmid ratio used for the cotransfections.
Lastly, we asked whether siRNAs synthesized from DNA templates in vivo can inhibit gene expression in different cell lines. In addition to HeLa cells, we analyzed the activity of the cdk-2 and lamin A/C siRNA plasmids in three additional cell lines: H1299 (nonsmall cell lung carcinoma), C-33A (human papilloma virus negative cervical carcinoma), and U-2 OS (osteosarcoma). Cells were transfected with either BS/U6 or siRNA plasmid together with CMV-GFP to mark the transfected cells. For each data point shown in Table 1, 200 GFP-positive cells were counted for the expression of either CDK-2 or lamin A/C. In the presence of the BS/U6 vector, the CDK-2-negative/GFP-positive cells ranged from 0.2% to 5.2%, whereas the lamin A/C-negative/GFP-positive cells ranged from 1.2% to 4.3%. In contrast, in the presence of the cdk-2 or lamin A/C siRNA plasmid, we observed dramatic increases in the number of CDK-2 or lamin A/C-negative/GFP-positive cells (86.9% to 97.7%; and 93.5% to 95.2%, respectively, Table 1). Taken together, our data demonstrate that siRNAs synthesized from DNA templates in vivo cause robust, near complete inhibition of endogenous gene expression in a variety of cells, suggesting that this technique is generally applicable to studying gene function in mammalian cells.
Table 1.
Inhibition of cdk-2 and lamin A/C gene expression in different human cell lines
| Cells | %
of CDK-2-negative/GFP-positive cells
|
% of Lamin
A/C-negative/GFP-positive cells
|
||
|---|---|---|---|---|
| BS/U6 | BS/U6/cdk-2 | BS/U6 | BS/U6/lamin A/C | |
| HeLa | 0.2 ± 0.4 (n = 6) | 97.7 ± 1.0 (n = 6) | 2.0 ± 0.8 (n = 4) | 95.2 ± 2.1 (n = 4) |
| H1299 | 4.0 ± 0.5 (n = 3) | 86.9 ± 1.5 (n = 3) | 4.3 ± 1.7 (n = 3) | 93.5 ± 1.8 (n = 3) |
| U-2 OS | 5.2 (n = 2) | 92.4 (n = 2) | 1.2 (n = 2) | 93.8 (n = 2) |
| C-33A | 3.7 (n = 2) | 92.3 (n = 2) | n.d. | n.d. |
Cells were cotransfected with CMV-GFP and either BS/U6, BS/U6/cdk-2, or BS/U6/lamin A/C and were analyzed by immunofluorescence for endogenous CDK-2 or lamin A/C expression. For each data point, 200 GFP-positive cells were scored for the presence of CDK-2 or lamin A/C signals. The values correspond to the percentage of GFP-positive cells that display a reduction of the CDK-2 or lamin A/C immunostaining near the background level and are presented as the average ± SD. n, number of transfection experiments. n.d., not determined.
We have provided strong evidence that a DNA vector-based RNAi approach functions effectively to silence endogenous gene expression in mammalian cells. This process is expected to greatly facilitate the use of the RNAi technology for gene function studies in mammalian cells and perhaps in vertebrate animals as well. The technology can be adapted to analyzing gene function over a long period through stable inhibition. It also can be adapted to establish an inducible siRNA system that can knock down gene expression in a regulated fashion. This latter feature is necessary for studying genes whose products are required for cell viability. Finally, the vector-based RNAi technology makes it possible to consider RNAi as a reverse genetic tool for genome level analysis of mammalian gene functions.
Acknowledgments
We thank Laising Yen for helpful discussion and Grace Gill for critical reading of the manuscript. We thank Sidney Altman for the U6 promoter plasmid pmU6 (−315/+1) and Matt Meyerson for human lamin A/C antibodies. We thank Margaret Po-Shan Luke for technical assistance. E.B.A. is supported by a fellowship from the Taplin Foundation, and Yujiang Shi is supported by a Program in Cancer Biology Training Grant from the National Cancer Institute (T32CA72320). This work is supported by a grant from the National Institutes of Health to Yang Shi (GM53874).
Abbreviations
- Pol III
RNA polymerase III
- RNAi
RNA-mediated interference
- dsRNA
double-stranded RNA
- siRNA
small interfering RNA
- GFP
green fluorescent protein
- BS
Bluescript
- Ts
thymidines
- HA
hemagglutinin
- CMV
cytomegalovirus
- CDK-2
cyclin-dependent kinase-2
- DNMT-1
DNA methyltransferase-1
References
- 1.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein E, Denli A M, Hannon G J. RNA. 2001;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 4.Zamore P D. Nat Struct Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- 5.Chuang C F, Meyerowitz E M. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misquitta L, Paterson B M. Proc Natl Acad Sci USA. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuschl T, Zamore P D, Lehmann R, Bartel D P, Sharp P A. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baglioni C, Nilsen T W. Interferon. 1983;5:23–42. [PubMed] [Google Scholar]
- 9.Williams B R. Biochem Soc Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- 10.Paddison P J, Caudy A A, Hannon G J. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Proc Natl Acad Sci USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 13.Bogenhagen D F, Sakonju S, Brown D D. Cell. 1980;19:27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tavernarakis N, Wang S L, Dorovkov M, Ryazanov A, Driscoll M. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 16.Kennerdell J R, Carthew R W. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 18.Ketting R F, Fischer S E, Bernstein E, Sijen T, Hannon G J, Plasterk R H. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight S W, Bass B L. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel G R, Maser R, Calvet J, Pederson T. Proc Natl Acad Sci USA. 1986;83:8575–8579. doi: 10.1073/pnas.83.22.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper J W, Adams P D. Chem Rev. 2001;101:2511–2562. doi: 10.1021/cr0001030. [DOI] [PubMed] [Google Scholar]
- 22.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]