Abstract
Nematode parasites show a characteristic aggregated distribution among hosts. This observation has important implications for pathogenesis, immunology, and control of these infections, but the relative roles of environment and genetics in determining these patterns have remained uncertain. This paper presents the results of the first genome scan for susceptibility to infection with roundworm (Ascaris lumbricoides). Data on 375 genetic markers were generated for each of 444 members of a genetically isolated Nepalese population, the Jirels. Ascaris worm burden as assessed by egg counts was measured in these same individuals by using the Kato Katz thick smear method. The extensive genealogical data available for the population allowed assignment of all 444 individuals to a single pedigree that contained 6,209 pairs of relatives that were informative for genetic analysis. A variance components linkage analysis resulted in the unequivocal localization of two genes (one on chromosome 1 and another on chromosome 13) with clear, significant effects on susceptibility to Ascaris infection. This is the first evidence that individual quantitative trait loci influence variation in Ascaris burden in humans.
Ascaris lumbricoides is the most common helminthic infection in developing nations, affecting over a quarter of the world's population (1, 2). Heavy worm loads can result in serious morbidity and even mortality as a consequence of intestinal blockage (3–7). Long-term infection with roundworm has been implicated in the development and persistence of childhood malnutrition and may have lasting effects on anthropometric measures of growth (5, 8–12).
In addition to the importance of the disease because of the worldwide scale of infection, the significance of helminthic infections has dramatically increased with the advent of HIV infections (13). Bentwich and colleagues (14, 15) have proposed that helminthic infections have deleterious effects on the ability to mount an effective immune response to other infections, particularly HIV and tuberculosis. Furthermore, they have suggested that the rapid progression to AIDS observed in developing countries may be attributable in part to helminthic coinfections (14, 15). Helminthic coinfections may also diminish the efficacy of future vaccines (13, 15). Clearly, helminthic infections can have a significant impact on epidemiological patterns of other diseases that have devastating health consequences (13).
Helminthic infections are characteristically overdispersed in human populations, with a small proportion of people harboring the majority of parasitic worms present (16, 17). Although this aggregation has been attributed most commonly to variability in exposure patterns, some have suggested the possible role of genetic factors (17, 18). A familial patterning to Ascaris infection frequently has been noted (19–21).
Our previous studies have demonstrated that there is a substantial genetic component to susceptibility to Ascaris infection in humans (22). The focal population for these studies was the Jirel population of eastern Nepal, a Tibeto–Burman language speaking hybrid group derived from Sherpas and Sunwars approximately 10–11 generations ago (23, 24). We used data on Ascaris egg counts in a single large pedigree comprising 1,261 members of the Jirel population to determine that genetic factors accounted for approximately 30% (h2 = 0.291, P < 0.0001) of the variation in eggs per gram of feces (EPG) as assessed by the Kato–Katz thick smear method (22). Systematic environmental factors accounted for an additional 6% of the variation. This strong evidence for the role of genetic factors in determining susceptibility to Ascaris infection provided impetus for a genome scan in the same population to find the locations of specific genes involved in determining susceptibility and resistance.
Although these previous studies are compelling in their general support of the possibility of genetic factors influencing Ascaris infection, classical genetic approaches that look only at the phenotypic variability among classes of relatives are not able to unequivocally disentangle genetic factors from shared environmental components of disease susceptibility. However, modern genomic approaches to the dissection of the determinants of infectious disease susceptibility do not confound genetic and environmental effects because the genetic effects are assessed from the cosegregation of a disease trait and a specific marker locus throughout an extended pedigree. It is impossible for an environmental effect on a disease trait to mimic this segregation pattern throughout an extended pedigree. Thus, linkage analysis approaches can be used to demonstrate the roles of genes in determining patterns of variation in parasitic burden among individuals.
Ultimately, identifying the specific genes involved in susceptibility to roundworm infection may suggest new biological pathways to be targeted by pharmacological treatment and intervention mechanisms. Localization of the genes by linkage analysis is the first important step toward identifying genes influencing Ascaris infection. Additionally, this easily quantifiable infection may serve as a model disease for understanding the genetic mechanisms involved in other more complex infectious diseases.
Methods
The trait we focused on for this analysis was Ascaris EPG as determined by the Kato–Katz thick smear method (measured in six repeated samples over a 2-day period) in 444 members of the single Jirel pedigree used in our previous quantitative genetic analyses of roundworm infection. The 444 individuals represented a highly informative branch of the pedigree. They were selected for the genome scan based only on pedigree informativeness, and were not selected in anyway with respect to Ascaris phenotype.
The 444 individuals were recruited between 1995 and 1996 from 98 households through house-to-house surveys of the Jiri region, which is located 190 kilometers east of Kathmandu, the capital city of Nepal. The sample included 218 males and 226 females, and the average age was 27 years with a range between 3 years and 85 years. Subsequent to recruitment, two small fecal samples were obtained for assessment of egg counts and a blood sample was drawn to obtain DNA. The collection protocol was approved by the Institutional Review Board of the University of Texas Health Sciences Center at San Antonio and the Nepal Health Research Council in Kathmandu, Nepal.
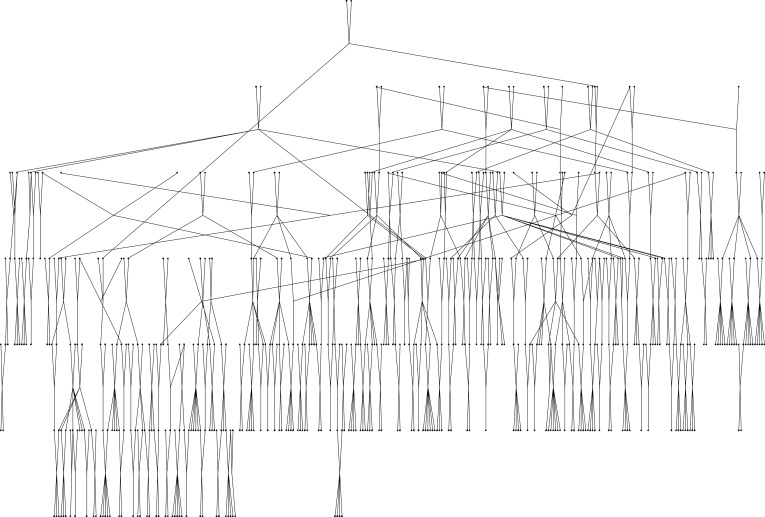
The population has been the subject of detailed genetic study since 1985 (23, 24), which has resulted in a wealth of pedigree information. These extensive genealogical records allowed assignment of all 444 individuals to a single pedigree that was very powerful for genetic analysis. The sample includes a total of 6,209 pairs of relationships that are informative linkage analysis, including 472 parent–offspring pairs, 345 sibling pairs, 805 second-degree relative pairs, 955 third-degree relative pairs, 853 fourth-degree relative pairs, and 2,779 higher-degree relative pairs. Table 1 presents the number of pairs for each type of relationship for which there are 20 or more pairs in the pedigree. The total pedigree used in the analysis is illustrated in Fig. 1. This pedigree is one of the largest family configurations ever analyzed without resorting to pedigree simplification in a human linkage study (25).
Table 1.
Pairwise relationships among genotyped individuals
| Number of pairs | Relationship | Coefficient of relationship |
|---|---|---|
| 472 | Parent–offspring | 1/2 |
| 345 | Siblings | 1/2 |
| 534 | Avuncular | 1/4 |
| 239 | Grandparent–grandchild | 1/4 |
| 24 | Half siblings | 1/4 |
| 644 | First cousins | 1/8 |
| 219 | Grand avuncular | 1/8 |
| 68 | Half avuncular | 1/8 |
| 799 | First cousins once removed | 1/16 |
| 117 | Half first cousins | 1/16 |
| 28 | Great grand avuncular | 1/16 |
| 799 | Second cousins | 1/32 |
| 240 | Half first cousins once removed | 1/32 |
| 161 | First cousins twice removed | 1/32 |
| 452 | Second cousins once removed | 1/64 |
| 253 | Half second cousins | 1/64 |
| 24 | Second cousins and third cousins | 5/256 |
| 278 | Half second cousins once removed | 1/128 |
| 112 | Third cousins | 1/128 |
| 67 | Half first cousins twice removed | 1/128 |
| 187 | Half third cousins | 1/256 |
| 35 | Third cousins once removed | 1/256 |
Only relationships with more than 20 pairs are shown.
Figure 1.
Pedigree of the Jirel population used for the analysis of susceptibility to Ascaris.
A 10-centimorgan (cM) genome scan involving 375 dinucleotide short tandem repeat markers evenly distributed across all 22 autosomes was conducted by using the ABI Prism Linkage Mapping Set, version 2 (Perkin–Elmer). All genotyping was performed on an automated ABI DNA Sequencer (model 377). The average heterozygosity was 0.74 per marker. The average marker density per autosome was one marker per every 9.1 cM
We performed quantitative trait linkage analyses using the variance component method as implemented in the computer program solar. Multipoint identity-by-descent probability matrices were calculated by using the method of Almasy and Blangero (26). This approach allowed us to analyze the Jirel pedigree in its entirety, thereby maintaining all of the power inherent to the single extended pedigree of 444 individuals. Additionally, covariate effects, including age, sex, and their interactions, were simultaneously estimated in all analyses to minimize the effects of these potential confounding variables.
Because of this extreme kurtosis of Ascaris EPG, we used a log transformation [ln(EPG + 1)]. Even after transformation, Ascaris EPG is markedly non-normal (with a kurtosis of 2.11). Because leptokurtic distributions can lead to excessive Type 1 errors when variance components are calculated assuming underlying multivariate normality, we used a robust method to calculate multipoint logarithm of odds (LOD) scores. We used a method that utilizes a scalar correction to the LOD score and yields the correct Type I error distribution regardless of the traits distribution (27, 28). All P values were obtained analytically and then validated empirically by using intensive computer simulation of the exact pedigree and data structure.
Results and Discussion
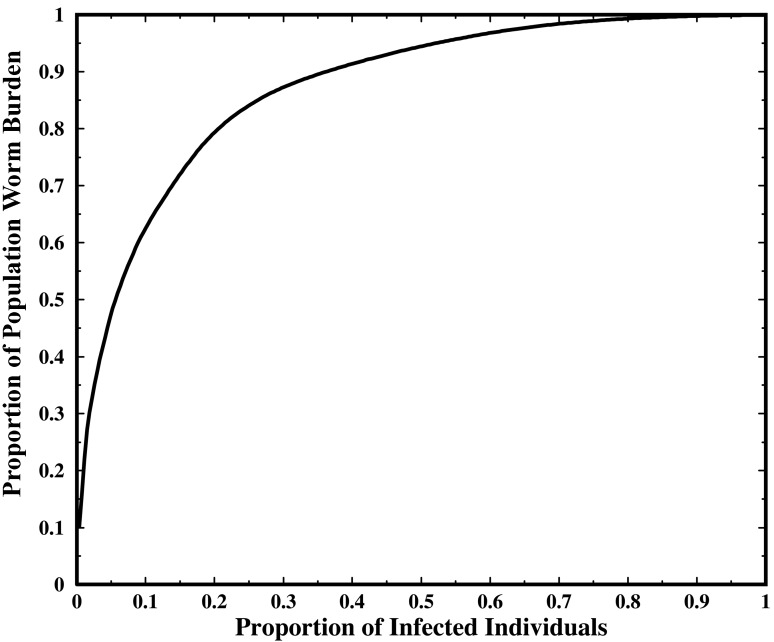
The prevalence of infection was 27.2% and the geometric mean EPG was 1,453, indicating a moderate average burden. As is typical of helminthic infections, worm burden is highly overdispersed. Fig. 2 shows the relative distribution of total worm burden (as measured by EPG) in the population. From the graph it is apparent that 20% of the infected individuals account for over 80% of the total infection burden.
Figure 2.
Relative distribution of total worm burden (as measured by EPG) in the population.
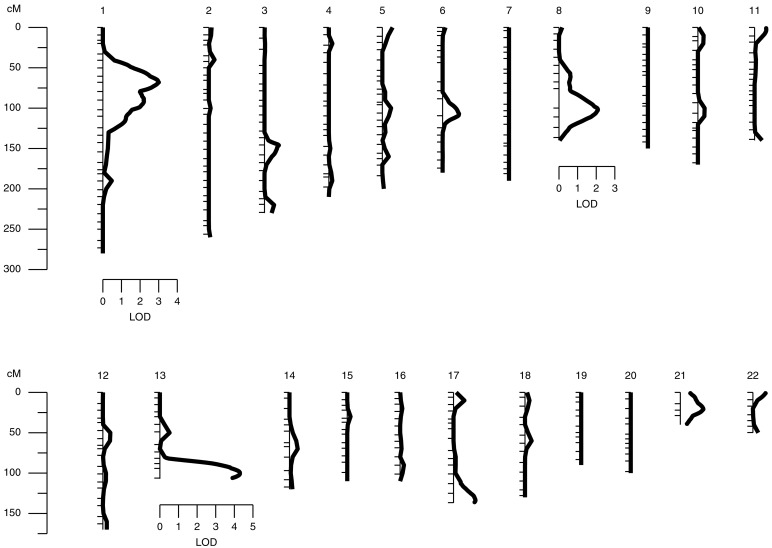
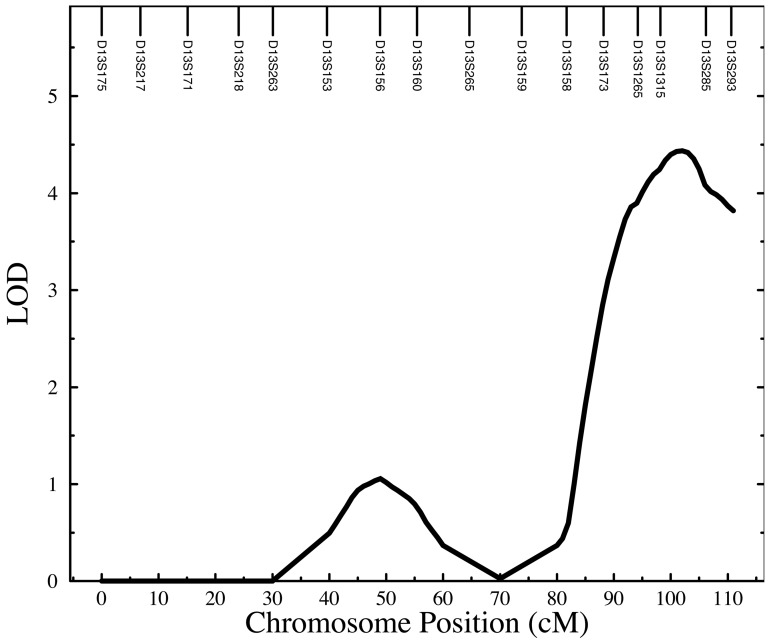
We found strong evidence for two distinct loci influencing susceptibility to Ascaris infection. Fig. 3 provides a string plot of the linkage results for all chromosomes. The strongest signal is found near the q terminus (13q32–q34) of chromosome 13 as is shown in detail in Fig. 4. Two adjacent markers also exhibit two-point LOD scores greater than 3 (D13S1265 LOD = 3.61; D13S285 LOD = 3.26). The maximum multipoint robust LOD score occurring at 101 cM was 4.30 corresponding to a nominal P value of 4 × 10−6 and a genome wide P value of 0.0013. We typed an additional three markers to reduce gaps in this area and the subsequent multipoint robust LOD score rose to 4.43 (nominal P value = 3.1 × 10−6; genome-wide P value = 0.00091) peaking at 102 cM in the region of 13q33. Fig. 4 provides a graphical summary of the evidence for this quantitative trait locus (QTL) on chromosome 13. The observed LOD score represents strong evidence that a gene in this area of chromosome 13 harbors an important quantitative trait locus influencing human host Ascaris burden.
Figure 3.
Genome scan results for Ascaris egg counts Ln(EPG + 1) in 444 Jirels: robust LOD results.
Figure 4.
Genome scan results for Ascaris egg counts Ln(EPG + 1) in 444 Jirels: chromosome 13.
The 1-LOD support interval encompasses a 20-cM region starting at 90 cM and extending to the q terminus of chromosome 13. This relatively gene-poor region includes 7 megabases (Mb) of DNA sequence and contains only 19 known genes and an additional 10 predicted genes. However, bioinformatic analysis of this region has revealed a single major candidate locus in this region. This gene, TNFSF13B (also known as BlyS), is a member of the tumor necrosis factor (TNF) superfamily of cytokines and is a major regulator of B cell activation and Ig secretion (29–31). This 37-kb gene is located within 1.5 Mb of marker D13S1315, which exhibited the single highest two-point LOD score.
Because TNSF13B is a major regulator of B cells, and the immune response to Ascaris infection is primarily B cell regulated, we examined the effect of this QTL on total serum IgE levels. Total serum IgE was measured in a single batch by using a commercial enzyme immunoassay (Kallestad Total IgE Microplate kit, San Ofi Diagnostics, Pasteur, Chaska, MN) based on the sandwich technique using solid phase coated microplate wells for separation. The intra-assay coefficient of variation was ≈5%. The heritability of serum IgE levels was high in this population (h2 = 0.53, P < 0.0000001). IgE levels were associated with Ascaris EPG (P = 0.002) and another important helminthic infection, hookworm, as assessed by EPG (P = 5 × 10−5). To obtain a measure of IgE more specific to Ascaris infection, we removed the effect of hookworm infection intensity by regression simultaneously in our genetic analyses. Additional covariates for IgE included sex, age, and sex by age interactions. A test of pleiotropy revealed that the chromosome 13 QTL also significantly influenced serum IgE levels (LOD = 1.73, P = 0.0024). This finding strongly supports our contention that TNFSF13B should be considered the major positional candidate gene for the QTL.
As seen in Fig. 3, there is also clear evidence for a second quantitative trait locus influencing susceptibility to Ascaris infection on chromosome 1. On this chromosome, the LOD score exhibits a sharp peak at 68 cM (1p32) with a maximum value of 3.01 (nominal P value = 0.0001; genome-wide P value = 0.033). The peak is significant and provides clear evidence for a second locus influencing the Ascaris worm burden trait. However, unlike the chromosome 13 QTL, the chromosome 1 QTL does not appear to influence serum IgE levels (LOD = 0.0).
Finally, there is also suggestive evidence for a third locus influencing Ascaris worm burden on chromosome 8. In this case, the LOD achieves a maximum at 101 cM (8q12–q13) with a maximum value of 2.09 (nominal P value = 9.0 × 10−3, genome-wide P value = 0.328). Although the evidence for this possible quantitative trait locus is substantially less than that for the other two, it may significantly improve with an increase in sample size.
There are two interesting possible candidate genes in this area of chromosome 8. The first is IL7, whose product is produced in intestinal epithelial cells and regulates mucosal lymphocytes (32) and has been shown to block helminthic development (33). The other major positional candidate gene is the activated B-cell factor 1 (ABF1) which is a transcription factor of the form basic helix-loop-helix that regulates B cell differentiation. Interestingly, this putative QTL may also influence serum IgE levels (LOD = 0.91, P = 0.02), which is consistent with the locus influencing B cell function in some way.
The clear localization of two genes influencing susceptibility to Ascaris burden in this single human pedigree also affirms the superiority of extended human pedigrees over simpler family structures such as sibling pairs for mapping quantitative traits. This superiority can easily be seen by comparing the expected LOD scores per individual in our Jirel pedigree versus that for sibling pairs, the most commonly used family design in human genetics. By using the theory developed by Williams and Blangero (34), we calculate that the Jirel pedigree structure is 22.8 times more efficient (in terms of the expected LOD score) for detecting a quantitative trait locus that accounts for 5% of the trait's variance than a classical sibling pair design. Thus, although there has been considerable speculation that linkage-based designs may not be sufficient for finding genes influencing common diseases (35), this supposition is based on the assumption of suboptimal study designs such as sibling pair studies.
These results are the first linkages reported for genes influencing susceptibility to infection with A. lumbricoides. The quantitative trait loci on chromosomes 1 and 13 provide strong evidence for the influence of at least two discrete genes on Ascaris burden. Although the linkage reported for chromosome 8 is not significant, the involvement of the candidate genes in the area is biologically plausible. The success of the study in localizing two genes influencing such a complex disease process in only 444 individuals is caused by the power of large extended human pedigrees for mapping loci influencing quantitative traits (see the discussion by Blangero and colleagues; ref. 27). With the recent availability of statistical genetic methods that can handle pedigrees of arbitrary size and complexity in quantitative trait linkage analyses, we anticipate that a refocusing of research designs on large complex families will be particularly useful for searches for genes influencing infectious diseases. Because of the lack of confounding by environmental factors, which often play important roles in determining exposure patterns for parasitic infections and other transmissible diseases, the genomic approach ultimately will lead to the identification of additional human QTLs for this important class of diseases.
Acknowledgments
The generous cooperation of the Jirel people is gratefully acknowledged. Robin S. Shrestha, Suresh Bijukchhe, Suman Jirel, Dev Jirel, Mithe Jirel, Chatra Jirel, Chandra Jirel, Mari Hui, and Cheryl Raindl provided much appreciated technical assistance. This work was supported by National Institutes of Health Grants AI44406, AI37091, and MH59490. The solar software used to analyze the Jirel pedigree is available on our web site, www.sfbr.org.
Abbreviations
- EPG
eggs per gram of feces
- LOD
logarithm of odds
- QTL
quantitative trait locus
- cM
centimorgan
- Mb
megabase
References
- 1.Pawlowski Z S. In: Tropical and Geographic Medicine. Warren K S, Mahmoud A A F, editors. New York: McGraw–Hill; 1990. pp. 369–378. [Google Scholar]
- 2.Tanowitz H B, Weiss L M, Wittner M. Gastroenterologist. 1994;2:39–49. [PubMed] [Google Scholar]
- 3.Mpairwe J B. J Helminthol. 1991;65:286–288. doi: 10.1017/s0022149x00010877. [DOI] [PubMed] [Google Scholar]
- 4.Crompton D W T, Neisham M, Pawlowski Z S. Ascariasis and Its Public Health Significance. London: Taylor and Francis; 1985. [Google Scholar]
- 5.de Silva N R, Chan M S, Bundy D A P. Trop Med Int Health. 1997;2:519–528. doi: 10.1046/j.1365-3156.1997.d01-320.x. [DOI] [PubMed] [Google Scholar]
- 6.de Silva N R, Guyatt H L, Bundy D. Trop Med Int Health. 1997;2:189–190. doi: 10.1046/j.1365-3156.1997.d01-241.x. [DOI] [PubMed] [Google Scholar]
- 7.Chrungoo R K, Hangloo V K, Faroqui M M, Khan M. J Indian Med Assoc. 1992;90:171–174. [PubMed] [Google Scholar]
- 8.Crompton D W T. Trans R Soc Trop Med Hyg. 1992;86:577–579. doi: 10.1016/0035-9203(92)90133-w. [DOI] [PubMed] [Google Scholar]
- 9.Gupta M C. J Trop Pediatr. 1990;36:189–191. doi: 10.1093/tropej/36.4.189. [DOI] [PubMed] [Google Scholar]
- 10.Stephenson L S. Impact of Helminth Infections on Human Nutrition. London: Taylor and Francis; 1987. [Google Scholar]
- 11.Thein-Hlaing, Than-Saw, Myat-Lay-Kyin Trans R Soc Trop Med Hyg. 1991;85:519–522. doi: 10.1016/0035-9203(91)90241-p. [DOI] [PubMed] [Google Scholar]
- 12.Thein-Hlaing, Thane-Toe, Than-Saw, Myat-Lay-Kyin, Myint-Lin Trans R Soc Trop Med Hyg. 1991;85:523–528. doi: 10.1016/0035-9203(91)90242-q. [DOI] [PubMed] [Google Scholar]
- 13.Bundy D A, Sher A, Michael E. Parasitol Today. 2000;16:273–274. doi: 10.1016/s0169-4758(00)01689-6. [DOI] [PubMed] [Google Scholar]
- 14.Bentwich Z, Kalinkovich A, Weisman Z. Immunol Today. 1995;16:187–191. doi: 10.1016/0167-5699(95)80119-7. [DOI] [PubMed] [Google Scholar]
- 15.Bentwich Z, Kalinkovich A, Weisman Z, Borkow G, Beyers N, Beyers A D. Immunol Today. 1999;20:485–487. doi: 10.1016/s0167-5699(99)01499-1. [DOI] [PubMed] [Google Scholar]
- 16.Anderson R M, May R M. Adv Parasitol. 1985;24:1–101. doi: 10.1016/s0065-308x(08)60561-8. [DOI] [PubMed] [Google Scholar]
- 17.Anderson R M, Medley G F. Parasitology. 1985;90:629–660. doi: 10.1017/s0031182000052288. [DOI] [PubMed] [Google Scholar]
- 18.Schad G A, Anderson R M. Science. 1985;228:1537–1540. doi: 10.1126/science.4012307. [DOI] [PubMed] [Google Scholar]
- 19.Chai J-Y, Seo B-S, Lee S-H. Korean J Parasitol. 1983;21:142–149. [Google Scholar]
- 20.Williams D, Burke G, Hendley J O. J Pediatr. 1974;84:853–854. doi: 10.1016/s0022-3476(74)80762-6. [DOI] [PubMed] [Google Scholar]
- 21.Forrester J E, Scott M E, Bundy D A P, Golden M H B. Trans R Soc Trop Med Hyg. 1988;82:282–288. doi: 10.1016/0035-9203(88)90448-8. [DOI] [PubMed] [Google Scholar]
- 22.Williams-Blangero S, Subedi J, Upadhayay R P, Manral D B, Rai D R, Jha B, Robinson E S. Am J Trop Med Hyg. 1999;60:921–926. doi: 10.4269/ajtmh.1999.60.921. [DOI] [PubMed] [Google Scholar]
- 23.Blangero J. Int J Anthropol. 1988;3:289–299. [Google Scholar]
- 24.Williams-Blangero S. Am J Phys Anthropol. 1990;82:61–72. doi: 10.1002/ajpa.1330820108. [DOI] [PubMed] [Google Scholar]
- 25.Dyer T D, Blangero J, Williams J T, Goring H H H, Mahaney M C. Genet Epidemiol. 2001;21 Suppl. 1:S236–S243. doi: 10.1002/gepi.2001.21.s1.s236. [DOI] [PubMed] [Google Scholar]
- 26.Almasy J, Blangero J. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blangero J, Williams J T, Almasy L. Genet Epidemiol. 2000;19:S8–14. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Blangero J, Williams J T, Almasy L. Adv Genet. 2001;42:151–181. doi: 10.1016/s0065-2660(01)42021-9. [DOI] [PubMed] [Google Scholar]
- 29.Moore P A, Belvedere O, Orr A, Pieri K, LaFleur D W, Feng P, Soppet D, Charters M, Gentz R, Parmelee D, et al. Science. 1999;285:260–263. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 30.Schneider P, MacKay F, Steiner V, Hofmann K, Bodmer J L, Holler N, Ambrose C, Lawton P, Bixler S, Acha-Orbea H, et al. J Exp Med. 1999;7:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan M, Marsters S A, Grewal I S, Wang H, Ashkenazi A, Dixit V M. Nat Immunol. 2000;1:37–41. doi: 10.1038/76889. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe Y, Uneo Y, Yajima T, Iwao Y, Tsuchiya M, Ishikawa H, Aiso S, Hibi T, Ishii H. J Clin Invest. 1995;95:2945–2953. doi: 10.1172/JCI118002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolowczuk I, Nutten S, Roye O, Delacre M, Capron M, Murray R M, Trottein F, Auriault C. Infect Immun. 1999;67:4183–4190. doi: 10.1128/iai.67.8.4183-4190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams J T, Blangero J. Ann Hum Genet. 1999;63:545–563. doi: 10.1017/S0003480099007848. [DOI] [PubMed] [Google Scholar]
- 35.Risch N, Merikangas K. Science. 1996;273:545–563. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]