Abstract
Cervical cancer is one of the most common malignant tumors among women worldwide. Its primary etiology is closely associated with human papillomavirus infection, which poses a serious threat to the health of women. N6-methyladenosine (m6A) modifications notably affect the biological characteristics of tumor cells, such as their proliferation, metastasis and chemoresistance, by regulating the stability, translation and degradation of RNA. It also serves an important regulatory role in the pathogenesis of cervical cancer. The present review details the mechanisms underlying m6A modification in cervical cancer and analyzes its impact on tumor progression. Moreover, it explores the potential clinical applications of m6A modification as a biomarker and therapeutic target to provide new insights and evidence regarding the early diagnosis and individualized treatment of patients with cervical cancer.
Keywords: cervical cancer, N6-methyladenosine, signaling pathway, biomarker
1. Introduction
Cervical cancer remains a threat to the health of women worldwide, particularly in developing nations where elevated incidence and mortality rates persist owing to inadequate healthcare infrastructure. As reported by the World Health Organization, 604,000 new cases of cervical cancer were diagnosed globally in 2020, with over 300,000 fatalities (1,2). The pathogenesis of cervical cancer is inextricably linked to persistent infection with high-risk (HR)-human papillomavirus (HPV), notably genotypes 16 and 18, which are recognized as the principal oncogenic drivers (3,4). Epidemiological advancements have demonstrated that early screening programs and HPV vaccination notably reduce the disease burden (3,5). However, suboptimal vaccination coverage persists in several countries, particularly in low-income regions, despite validated vaccine efficacy (3,4). Stagnation in the decline in cervical cancer incidence underscores the importance of elucidating its molecular pathogenesis to develop improved preventive and therapeutic strategies.
Progression from HPV infection to invasive carcinoma involves intricate interactions between viral oncoproteins, host genetic susceptibility and microenvironmental factors (5). HR-HPVs integrate into host genomes, inducing genomic instability via dysregulation of cell cycle checkpoints. The viral oncoproteins, E6 and E7, orchestrate malignant transformation by degrading the tumor suppressor, p53 and retinoblastoma protein, respectively, thereby promoting uncontrolled proliferation and evasion of apoptosis (6). Persistent viral infection coupled with compromised host immunosurveillance facilitates the progression of cervical intraepithelial neoplasia to invasive carcinoma (7). Co-carcinogenic factors, including prolonged oral contraceptive use, tobacco smoking and immunosuppressive states (such as human immunodeficiency virus co-infection), synergistically enhance cervical cancer risk. Cigarette smoke constituents exacerbate carcinogenesis by inducing chronic inflammation and impairing local immune responses (8). Nutritional deficiencies (such as vitamins A and C) and environmental carcinogens may modulate cancer susceptibility via oxidative stress pathways (9). A comprehensive understanding of these multifactorial interactions is crucial for the development of targeted prevention strategies.
N6-methyladenosine (m6A), the most prevalent internal RNA modification in eukaryotes, precisely controls RNA metabolism, gene expression and cellular homeostasis (10). Emerging evidence implicates m6A dysregulation in several malignancies, including cervical cancer. As shown in Fig. 1, the m6A regulatory machinery comprises writers [such as methyltransferase 3 (METTL3)], erasers [such as fat mass and obesity-associated protein (FTO)] and readers [such as YTH domain-containing family protein (YTHDF)1 and insulin-like growth factor 2 mRNA-binding protein (IGF2BP)2] that collectively determine the fate of RNA. Clinical analyses have reported that METTL3 overexpression in cervical cancer tissues is associated with an advanced disease stage and poor prognosis. Mechanistically, METTL3 stabilizes the cathepsin L mRNA to enhance tumor cell migration and invasion. Conversely, FTO-mediated demethylation regulates oncogenic transcripts, including zinc finger E-box binding homeobox 1 (ZEB1) and MYC, suggesting its potential as a therapeutic target (11). m6A also governs the stability and functionality of long non-coding (lnc)RNAs, thereby modulating tumor proliferation and metastatic potential (12,13). Notably, m6A reprogramming influences tumor metabolism by regulating aerobic glycolysis, whereas distinct m6A modification patterns are associated with immune cell infiltration characteristics in the tumor microenvironment, potentially affecting responsiveness to immunotherapy (14,15). These findings suggest that m6A is a promising diagnostic biomarker and novel therapeutic target (Table I). The present review systematically examines the molecular mechanisms underlying m6A-mediated cervical carcinogenesis and evaluates its translational potential. By delineating how m6A modifiers orchestrate oncogenic signaling networks, innovative strategies for the early detection of cervical cancer and development of personalized therapy are proposed. Further investigation into the immunomodulatory roles of m6A may unlock combinatorial treatment approaches that integrate epigenetic targeting with immune checkpoint inhibitors.
Figure 1.
Dynamic regulatory mechanism of m6A modification. Writers are responsible for adding m6A modifications to mRNA, erasers for removing these modifications, and readers for recognizing and responding to m6A modifications. These collectively regulate gene expression and cellular functions. m6A, N6-methyladenosine. WTAP, Wilms tumor 1-associated protein; KIAA1429/VIRMA, Vir-like m6A methyltransferase associated; RBM15/RBM15B, RNA binding motif protein 15/15B; METTL14, methyltransferase like 14; ZC3H13, zinc finger CCCH-type containing 13; FTO, fat mass and obesity-associated protein; ALKBH5, AlkB homolog 5; YTHDF1, YTH N6-methyladenosine RNA binding protein 1; YTHDC1, YTH domain containing 1; HNRNPC, heterogeneous nuclear ribonucleoprotein C; IGF2BP1, insulin-like growth factor 2 mRNA-binding protein 1.
Table I.
m6A regulators and the potential targets in cervical cancer.
| m6A regulators | Targets in cervical cancer | (Refs.) |
|---|---|---|
| m6A writers | ||
| METTL3 | DLG2, HDAC6, MYC, PDE3A, IRF5, PPP6C, NEK2, KRAS, HSPA9, | (21–44) |
| circRNF13, circSTX6, Linc00426, lncMETTL4-2, LncFOXD2, miR-193b, CTSL, DARS, CTNNB1, CDC25B, TXNDC5, ZFAS1 and HK2 | ||
| METTL14 | DIRAS1, HOXB13, FTH1, TAPBP, TRIM11, CYP1B1 and DARS | (45–50) |
| RBM15 | EZH2, DCN, lncHEIH, OTUB2 and C-MYC | (51–54) |
| KIAA1429 | BTG2 and LARP1 | (55,56) |
| ZC3H13 | Circ_0081723, CKAP2 and CENPK | (57–61) |
| m6A erasers | ||
| FTO | LncHOXC13-AS, BMP4, E2F1, MYC, PIK3R3 and ZEB1 | (62–68) |
| ALKBH5 | SIRT3, circCCDC134, MMP2, MMP9, PAK5 and GAS5 | (69–73) |
| m6A readers | ||
| YTHDF1 | HK2, MCT1 and PDK4 | (26,38) |
| YTHDF2 | TAPBP, DDX6, CTNNB1, CARMN, circCCDC134, PAK5 and GAS5 | (69,70,77,78) |
| YTHDF3 | PDE3A, LRP6 and RAD51D | (57–61) |
| HNRNPA2B1 | LDHA | (10) |
| IGF2BP1 | TRIM11, SYVN and SIRT3 | (48,72,80) |
| IGF2BP2 | CTSL, MYC and FOXM1 | (82,83) |
| IGF2BP3 | lncKCNMB2-AS1, SCD and PDK4 | (76,84,85) |
| EIF3A | HYOU1 | (61) |
| HNRNPC | FOXM1 | (87) |
DLG2, discs large MAGUK scaffold protein 2; HDAC6, histone deacetylase 6; MYC, MYC proto-oncogene BHLH transcription factor; PDE3A, phosphodiesterase 3A; IRF5, interferon regulatory factor 5; PPP6C, protein phosphatase 6 catalytic subunit; NEK2, NIMA related kinase 2; KRAS, KRAS proto-oncogene; HSPA9, heat shock protein family A member 9; circRNF13, circ ring finger protein 13; circSTX6, circ syntaxin 6; lncFOXD2, lnc forkhead box D2; CTSL, cathepsin L; DARS, aspartyl-tRNA synthetase; CTNNB1, catenin beta 1; CDC25B, cell division cycle 25B; TXNDC5, thioredoxin domain containing 5; ZFAS1, ZNFX1 antisense RNA 1; HK2, hexokinase 2; PIK3R3, phosphoinositide-3-kinase regulatory subunit 3; ZEB1, zinc finger E-box binding homeobox 1; MMP2, matrix metallopeptidase 2; PAK5, P21 activated kinase 5; GAS5, growth arrest specific 5; TAPBP, TAP binding protein; CARMN, cardiac mesoderm enhancer-associated non-coding RNA; TRIM11, tripartite motif containing 11; FOXM1, forkhead box M1; SCD, stearoyl-CoA desaturase; HYOU1, hypoxia up-regulated 1.
2. Targets and signaling pathway regulatory networks of m6A writers in cervical cancer
Key m6A regulators as biomarkers and therapeutic targets in cervical cancer
Dynamic imbalance of the m6A methyltransferase system in cervical cancer is a core epigenetic feature that drives disease progression. In cervical cancer, dysregulated m6A modification enzymes (writers, erasers and readers) act as potential prognostic biomarkers and therapeutic targets, of which the high expression levels of certain ones are associated with reduced overall survival (16). However, reduced m6A mRNA methylation levels have been notably associated with advanced cervical cancer stages, larger tumor size, lower differentiation, lymph node invasion and recurrence. Reduced m6A mRNA methylation has also been associated with the progression and poor prognosis of cervical cancer, suggesting that it may act as a potential therapeutic target (17). For example, WT1 associated protein, RNA binding motif protein 15 (RBM15), Cbl proto-oncogene like 1 and YTHDC2 act as hub genes and potential biomarkers for cervical cancer, with RBM15 being markedly overexpressed and the others downregulated. RBM15 gene knockdown inhibits cancer progression and the JAK-STAT pathway, highlighting its therapeutic potential (18).
METTL3 in the proliferation and metastasis of cervical cancer
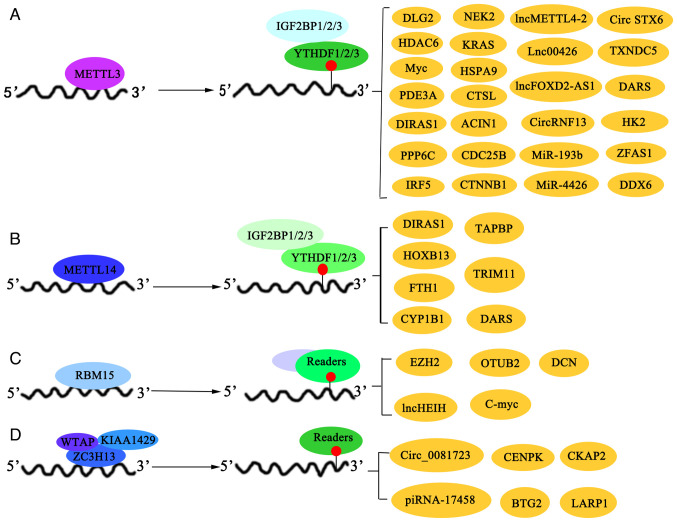
METTL3, the core regulatory hub, cooperates with HPV to enhance genome stability. Integrative analyses of HPV-related cancers have revealed that METTL3, an m6A regulator, is associated with poor prognosis, HPV status and an immunosuppressive microenvironment, suggesting that it has potential as a therapeutic target. High METTL3 expression is associated with reduced immune cell infiltration and increased levels of immunosuppressive checkpoint molecules, and its inhibition in combination with anti-programmed cell death protein 1 (PD-1) therapy shows promise for the treatment of cervical squamous cell carcinoma (CSSC) (19). Moreover, a marked association between m6A RNA methylation regulators, the tumor microenvironment, programmed death-ligand 1 (PD-L1) expression and immune infiltration was reported in cervical cancer, identifying a prognostic signature that could mediate PD-L1 expression and immune cell dynamics in the tumor immune microenvironment (20). Furthermore, METTL3 is involved in regulating key signaling pathways involved in the progression of cervical cancer. Disks large homolog 2 (DLG2) expression is downregulated in cervical cancer. Its upregulation inactivates the Hippo/YAP signaling pathway and inhibits the malignant behavior of cancer cells. As shown in Fig. 2A, METTL3 destabilized the DLG2 mRNA by increasing m6A modification levels, thereby negatively regulating DLG2 expression. Moreover, low DLG2 expression counteracted the inhibitory effects of METTL3 silencing in cancer cells. This indicates that DLG2 acts as a tumor suppressor in cervical cancer. Moreover, METTL3 promotes the progression of cervical cancer by downregulating DLG2 expression and activating the Hippo/YAP signaling pathway (21).
Figure 2.
In cervical cancer, writers and their interacting readers jointly regulate target mRNA to promote or inhibit the progression of cervical cancer. The readers include (A) METTL3, (B) METTL14, (C) RBM15, (D) WTAP, KIAA1429 and ZC3H13. METTL, methyltransferase; RBM15, RNA binding motif protein 15; WTAP, WT1-associated protein; KIAA1429, Vir-like m6A methyltransferase associated; ZC3H13, zinc finger CCCH-type containing 13; TXNDC5, thioredoxin domain containing 5; PDE3A, phosphodiesterase 3A; CTSL, cathepsin L; DDX6, DEAD-box helicase 6; TAPBP, TAP binding protein; FTH1, ferritin heavy chain 1; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; CENPK, centromere protein K; CKAP2, cytoskeleton associated protein 2; LARP1, La ribonucleoprotein 1.
The transcription factor, interferon regulatory factor 5 (IRF5), induced by m6A modification, promotes cervical cancer progression by upregulating protein phosphatase 6 catalytic subunit; its activity is regulated by METTL3, suggesting that it could be another potential therapeutic target (22). METTL3 gene knockdown inhibits cervical cancer by disrupting the AKT/mTOR pathway, and its upregulation drives tumor growth through m6 modification of the phosphodiesterase 3A (PDE3A) mRNA, highlighting the METTL3/YTHDF3/PDE3A axis as a potential therapeutic target (23). METTL3 promotes cervical cancer progression via m6A modification-mediated upregulation of NIMA-related kinase 2 (NEK2), which together activate the Wnt/β-catenin pathway and inhibit apoptosis, suggesting that the METTL3-NEK2 axis could be a potential therapeutic target for cervical cancer (24). Previous research indicated that individuals with elevated METTL3 levels exhibited reduced disease-free survival, whereas those with increased MYC expression showed decreased overall survival rates. Thus, METTL3 may promote the occurrence and development of cervical cancer by regulating the translation of the oncogene, MYC, through m6A modification, leading to enhanced cell viability and malignant behavior (25). The study also demonstrated that RNA m6A modification, mediated by upregulated METTL3, downregulated nuclear receptor subfamily 4 group A member 1 (NR4A1) in cervical cancer, facilitating malignancy. This process involves the YTHDF2-DDX6 pathway and the recruitment of the lysine demethylase 1A (LSD1)/histone deacetylase (HDAC)1/CoREST complex to the AKT serine/threonine kinase 1 (AKT1) promoter, highlighting the role of m6A modification in cervical cancer progression (26).
METTL3-driven m6A modification promotes cancer cell dissemination via β-catenin expression and membrane trafficking. The METTL3/β-catenin axis acts as a potential target for the inhibition of cancer metastasis (27). Additionally, METTL3-mediated m6A modification of the heat shock protein family A9 mRNA in cervical cancer cells led to increased stability and translation efficiency of mortalin, an exosomal protein. Overexpression of exosomal mortalin promotes cancer cell proliferation, migration and invasion, whilst inhibiting cellular senescence by blocking the nuclear transport of p53 and inactivating it. These findings highlight the notable role of METTL3 and exosomal mortalin in cervical cancer progression and suggest their potential as early diagnostic biomarkers and therapeutic targets (28). METTL3 enhances HDAC6 translation by facilitating its m6A modification and subsequent interaction with YTHDF3, thereby modulating cilia dynamics and alpha-tubulin acetylation, which affect cervical cancer cell growth and tumor progression (29). METTL3 upregulates cell cycle progression and proliferation in cervical cancer by enhancing cell division cycle 25B translation via m6A/YTHDF1-dependent mechanisms, revealing a novel role of m6 methylation in regulating the cell cycle (30). Finally, METTL3 participates in metabolic reprogramming of cervical cancer cells. For instance, METTL3 targets the 3′-UTR of the hexokinase 2 (HK2) mRNA and enhances its stability by recruiting YTHDF1 for m6A modification, contributing to cervical cancer progression and the Warburg effect; its upregulation in cancer tissues is associated with poor prognosis, highlighting a potential therapeutic target (31). METTL3-induced m6A methylation of solute carrier 38A1 promotes cervical cancer growth by stabilizing its mRNA, suggesting that targeting this pathway could be a novel therapeutic strategy for cervical cancer treatment (32). Moreover, METTL3 was reported to target thioredoxin domain containing 5 for m6A modification, which promoted cervical cancer progression by alleviating endoplasmic reticulum stress, suggesting the role of METTL3 as a biomarker and therapeutic target (33).
METTL3 regulates the expression of non-coding (nc)RNA in cervical cancer
METTL3 regulates ncRNA expression. For example, m6A modification, involving writers METTL3 and METTL14, as well as erasers, FTO and AlkB homolog 5, RNA demethylase (ALKBH5), serves a pivotal role in regulating the stability and translation of the mRNA of the tumor suppressor, DIRAS family GTPase 1, which is markedly downregulated in cervical cancer cells, and contributes to its anti-oncogenic effects (34). LINC00426, an m6A-modulated lncRNA, induces epithelial-mesenchymal transition (EMT) in cervical cancer by binding to ZEB1, which confers resistance to certain chemotherapies and sensitizes cells to imatinib, suggesting its potential as a therapeutic target (35). Aspartyl-tRNA synthetase (DARS)-antisense RNA 1 (DARS-AS1), an oncogenic lncRNA, promotes cytoprotective autophagy in cervical cancer by recruiting METTL3/METTL14 to enhance DARS mRNA m6A modification and translation, forming a hypoxia-induced pathway that could be a potential therapeutic target (36). Circ0000069, upregulated in cervical cancer due to m6A modification, promotes cell proliferation and migration by inhibiting microRNA (miRNA/miR)-4426, indicating a role for this circular (circ)RNA in cervical cancer progression (37). CircRNF13, an m6A-modified circRNA, confers radioresistance to cervical cancer by stabilizing the C-X-C motif chemokine ligand 1 (CXCL1) mRNA, suggesting that the METTL3/YTHDF2/circRNF13/CXCL1 axis is a potential therapeutic target for countering radiotherapy resistance (38). circSTX6, overexpressed in cervical cancer, is associated with poor prognosis and promotes cell survival, proliferation, invasion and migration via its interaction with Spi-1 proto-oncogene (SPI1), leading to IL-6 upregulation and JAK2/STAT3 pathway activation. METTL3-induced m6A modification of circSTX6, recognized by YTHDC1, enhances circSTX6 stability by forming a positive regulatory loop with SPI1. This insight deepens the understanding of cervical cancer pathogenesis and suggests that circSTX6 may be a novel therapeutic target (39). Moreover, METTL3 fosters cervical cancer progression by stabilizing the lncRNA, forkhead box D2-AS1, which recruits LSD1 to the P21 promoter, facilitating H3K4 demethylation and transcriptional silencing of the tumor suppressor, p21, thereby representing a novel therapeutic axis in cervical cancer (40). Another study revealed a pivotal role of METTL3 in CSCC, where it upregulates and stabilizes the m6A-modified lncRNA, METTL4-2, which in turn promotes cell migration, proliferation and tumor growth, suggesting that the METTL3-METTL4-2 axis could be a novel therapeutic target for CSCC (41). ZNFX1 antisense RNA 1 serves an oncogenic role in cervical cancer by suppressing miR-647 via a METLL3-mediated m6A-dependent mechanism, and its upregulation is associated with poor prognosis and aggressive tumor behavior, suggesting its potential as a therapeutic target (42). MiR-30c-5p promotes ferroptosis and inhibits the growth and metastasis of cervical cancer by targeting the METTL3/KRAS axis, revealing a novel molecular mechanism for cervical cancer progression (43). Downregulation of miR-193b by m6A methylation, mediated by METTL3 (Fig. 2A), promotes cervical cancer aggressiveness by targeting Cyclin D1, indicating a novel mechanism for tumor progression (44).
METTL14-mediated m6A remodeling and oncogenic signaling in cervical cancer
Emerging evidence has highlighted context-dependent regulation of the m6A methylation machinery under apoptotic stress. A comparative analysis of chemotherapy-induced apoptosis revealed that cisplatin treatment selectively reduces METTL14 and FTO transcript levels in cervical cancer cells, unlike that observed in TNF-α-mediated apoptosis pathways. Notably, cisplatin exposure also markedly diminishes the levels of m6A reader proteins, IGF2BP2 and IGF2BP3, suggesting stress-specific rewiring of the m6A epitranscriptome that may underlie heterogeneous therapeutic responses across cell death inducers (45). Owing to this regulatory plasticity, METTL14 demonstrates multifaceted oncogenic roles. As shown in Fig. 2B, sorafenib promotes ferroptosis by destabilizing the ferritin heavy chain 1 mRNA via m6A methylation, indicating its role in regulating cancer progression via the PI3K/AKT signaling pathway (46). Utilizing m6A-dependent upregulation of homeobox B13, METTL14 activates NF-κB signaling and drives tumor invasion. It is associated with advanced International Federation of Gynecology and Obstetrics (FIGO) stages and is emerging as a potential therapeutic target (47). In collaboration with insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1), METTL14 stabilizes tripartite motif containing 11 transcripts to enhance PH domain and leucine rich repeat protein phosphatase 1 ubiquitination and proteasomal degradation, thereby activating the AKT pathway and enhancing cell proliferation, migration and invasion, indicating that it is a potential therapeutic target for cervical cancer (48). Consistent with these pro-tumorigenic functions, a previous study reported that silencing of METTL14 in cervical cancer cells led to cell cycle arrest and suppression of the PI3K/AKT/mTOR pathway. METTL14 knockdown inhibited the growth, migration and invasion of cervical cancer cells, including both HPV-positive and -negative cell lines, suggesting that METTL14 serves an oncogenic role in cervical cancer progression. This is supported by bioinformatics analysis indicating that METTL14 upregulation is a poor prognostic factor in patients with cervical cancer (49). Notably, the oncogenic activity of METTL14 is modulated by ncRNA crosstalk. piRNA-14633 is highly expressed in cervical cancer tissues and cells, where it promotes cell viability, proliferation, migration and invasion via the METTL14/cytochrome P450 1B1 signaling axis, suggesting a notable role for piRNA-14633 in cervical cancer progression. In vitro and in vivo experiments demonstrated that piRNA-14633 affects m6A RNA methylation and METTL14 mRNA stability, with implications for potential therapeutic targeting in cervical cancer (50).
RBM15 orchestrates multi-layered epitranscriptomic networks in cervical cancer
Analogous to METTL14, RBM15 drives cervical carcinogenesis through a multi-layered epitranscriptomic network, demonstrating remarkable pathway heterogeneity. For example, as shown in Fig. 2C, RBM15-mediated m6A modification of enhancer of zeste 2 polycomb repressive complex 2 (EZH2) promotes EMT and the proliferation of cervical cancer cells, suggesting that the RBM15/EZH2/FN1 signaling cascade is a potential therapeutic target (51). During stromal remodeling in cervical cancer tissues, there is an association between elevated RBM15 expression and increased malignancy, whereas RBM15 knockdown reduces the malignancy of cervical cancer by mediating the m6A modification of decorin, leading to increased decorin expression and inhibition of tumor growth both in vitro and in vivo (52). RBM15 promotes cervical cancer cell stemness and progression by stabilizing the oncogenic lncRNA, ‘High Expression In Hepatocellular carcinoma, which competitively adsorbs miR-802, upregulates epithelial growth factor receptor (EGFR) and enhances tumor growth and metastasis (53). Research has also reported that RBM15-mediated m6A modification upregulated OTU deubiquitinase, ubiquitin aldehyde binding 2 (OTUB2), which in turn promoted cervical cancer progression via the AKT/mTOR signaling pathway. Enhanced OTUB2 expression is associated with poor outcomes in patients with CSCC and endocervical adenocarcinoma, and its silencing inhibits cancer cell proliferation and metastasis whilst promoting apoptosis (54). Unlike the role of METTL14 in chemoresponse and ferroptosis regulation, RBM15 predominantly orchestrates tumor plasticity and stromal crosstalk, although both proteins synergistically amplify oncogenic signaling via AKT/mTOR activation. This hierarchical regulatory architecture underscores the therapeutic potential of co-targeting m6A writers and downstream effectors (such as OTUB2 inhibitors combined with EGFR blockade) (53,54).
Vir like M6A methyltransferase associated (KIAA1429) and zinc finger CCCH-type containing 13 (ZC3H13): Viral mimicry and metabolic networks in cervical cancer
The methyltransferase, KIAA1429, which complements the roles of METTL3, METTL14 and RBM1, has emerged as a dual-function modulator of viral mimicry and plasticity. As shown in Fig. 2D, KIAA1429 enhances cervical cancer tumorigenesis by modulating the m6A modification and stability of the La ribonucleoprotein 1, translational regulator mRNA in HPV+ models (55). KIAA1429 facilitates EMT in cervical cancer by mediating the m6A modification of BTG anti-proliferation factor 2, thereby promoting cancer cell malignancy. Silencing of KIAA1429 inhibits cervical cancer cell proliferation, migration, invasion and EMT, whilst enhancing apoptosis, highlighting its role in tumor progression (56).
Moreover, ZC3H13 cells exemplify metabolic stemness-related crosstalk. Cervical cancer samples exhibited marked upregulation of hsa_circ_0081723, with elevated levels associated with advanced histological grade, increased FIGO stage, reduced cellular differentiation and a worse prognosis. ZC3H13-mediated m6A modification of hsa_circ_0081723 facilitates cervical cancer progression via the AMPK/p53 signaling pathway. These findings suggest that the ZC3H13-hsa_circ_0081723 axis is a novel and potentially tractable therapeutic target for the clinical management of cervical cancer (57). Overexpression of ZC3H13 and cytoskeleton associated protein 2 (CKAP2) in cervical cancer is associated with an unfavorable prognosis, with ZC3H13 contributing to malignancy through the m6A modification of CKAP2, which stimulates cell proliferation, invasion and migration (58). In cervical cancer, copy number variations predominantly result in gene amplification in HR groups and deletions in low-risk groups; ZC3H13 downregulation is associated with increased proliferation and invasion of cancer cells, along with reduced RNA methylation levels. By contrast, rapamycin treatment inversely affects these cellular behaviors by increasing m6A levels, suggesting that m6A mRNA methylation is integral to cervical cancer progression and may act as a prognostic indicator and therapeutic target (59). ZC3H13-mediated m6A modification of the centromere protein K mRNA promotes cervical cancer stemness and chemoresistance (60). Additionally, rs1059288, a genetic variant of TAP binding protein (TAPBP), is associated with m6A-associated hereditary risk: It enhances TAPBP expression to promote lymph node invasion and carboplatin resistance, bridging epigenetic dysregulation with genomic instability (61). These findings highlight the complex interplay between m6A writers and cervical cancer pathogenesis and offer multiple avenues for targeted interventions.
3. Targets and signaling pathways of m6A erasers in cervical cancers
The m6A erasers FTO and ALKBH5: Bifunctional regulators of cervical cancer progression
The m6A erasers, FTO and ALKBH5, dynamically regulate cervical cancer progression by modulating distinct molecular targets and signaling networks. Their roles vary depending on the downstream pathways and cellular context, yielding dual oncogenic or tumor-suppressive effects (62–78) (Fig. 3).
Figure 3.
Targets and signaling pathways of m6A erasers in CC. The m6A erasers FTO and ALKBH5 dynamically regulate CC progression by modulating distinct molecular targets and signaling networks. m6A, N6-methyladenosine; FTO, fat mass and obesity-associated protein; ALKBH5, AlkB homolog 5, RNA demethylase; CC, cervical cancer. HIF1A, hypoxia-inducible factor 1 subunit alpha; CCDC134, Coiled-coil domain containing 134; GSK3β, Glycogen synthase kinase 3β; E2F1, E2F transcription factor 1; PAK5, P21 (RAC1) activated kinase 5.
FTO drives cervical cancer progression through metabolic and epigenetic reprogramming
FTO, the most extensively studied m6A eraser in cervical cancer, is commonly upregulated in cervical cancer tissues and is strongly associated with cervical carcinoma (62). Moreover, FTO expression is elevated in patients with HPV-positive cervical cancer, is associated with advanced FIGO stages, and is higher in cancerous tissues than in paracancerous tissues, with FTO knockdown increasing PIK3R3 m6A levels and affecting FoxO pathway activation (63). FTO is a multidimensional oncogenic driver that promotes tumorigenesis via several mechanisms. As shown in Fig. 3, the E6 and E7 oncogenes activate the transcription of GSK3B for metabolic reprogramming; glycogen synthase kinase 3β (GSK3β) can promote the ubiquitination-mediated proteasomal degradation of FTO and reduce the level of FTO protein; and FTO inhibits the maturation and translation of the HK2 mRNA by retaining the HK2 pre-mRNA in the nucleus. Therefore, E6/E7 regulates HK2 expression in cervical cancer via GSK3β/FTO signaling (64). FTO facilitates adipogenesis by regulating adipogenic pathways and inducing pre-adipocyte differentiation (65). FTO knockdown markedly suppressed xenograft tumor growth and reduced the bone morphogenetic protein 4 (BMP4) level in vivo. Collectively, the results demonstrate that FTO promotes cervical cancer progression in vitro and in vivo by regulating the BMP4/Hippo/Yes1 associated transcriptional regulator (YAP1)/WW domain containing transcription regulator 1 (TAZ) pathway, suggesting that FTO acts as an oncogenic molecule and that the FTO/BMP4 Hippo/YAP1/TAZ axis may act as a valuable target for cervical cancer treatment (66).
Furthermore, in transcriptional and post-transcriptional networks, FTO stabilizes the lncRNA, HOXC13 antisense RNA (HOXC13-AS), which recruits CBP/p300 to acetylate the frizzled class receptor 6 (FZD6) promoter, thereby epigenetically upregulating FZD6 and activating the Wnt/β-catenin signaling to drive cervical cancer proliferation, invasion and EMT. This suggests that HOXC13-AS is a potential target for cervical cancer treatment (67). Moreover, FTO demethylates E2F1, MYC and ZEB1 mRNAs, promoting the cell cycle and invasion (62,68).
Context-dependent roles of ALKBH5 in cervical cancer
ALKBH5 exhibits dual regulatory functions depending on the HPV status and target specificity in cervical carcinogenesis. Emerging evidence has revealed an oncogenic role in distinct m6A-dependent networks. In HPV-positive tumors, ALKBH5 is upregulated by E7-mediated E2F1 activation and histone H3K27Ac/H3K4Me3 modifications, leading to erasure of the m6A mark on the P21 (RAC1) activated kinase 5 (PAK5) mRNA. This demethylation stabilizes PAK5 transcripts via YTHDF2 recognition and activates RAC1/CDC42 signaling to potentiate metastasis (69). ALKBH5 further stabilizes oncogenic circCCDC134 by removing m6A marks, enabling dual pro-metastatic functions: Nuclear recruitment of p65 to enhance transcription and cytoplasmic sponging of miR-503-5p to derepress MYB expression (70). Silencing of metastasis associated lung adenocarcinoma transcript 1 decreases ALKBH5 expression via miR-141-3p, leading to reduced matrix metallopeptidase (MMP)2 and MMP9 expression and suppression of cell migration and invasion, suggesting that the MALAT-ALKBH5 axis may promote HPV-positive cervical cancer cell proliferation and metastasis (71). These findings suggest that ALKBH5 can act as a molecular rheostat tuned to viral status, with its downstream effectors (PAK5 and hypoxia inducible factor-1α) being targets for precise therapeutic intervention.
Emerging evidence have also delineated the context-dependent tumor-suppressive functions of ALKBH5 through metabolic and epigenetic regulation. In CSSC, ALKBH5 destabilizes sirtuin 3 mRNA via m6A-IGF2BP1 recognition and suppresses acetyl-CoA carboxylase α deacetylation and lipid accumulation to inhibit tumor growth, a mechanism that is inversely associated with clinical prognosis (72). Concurrently, the growth arrest specific 5 (GAS5)-AS1 lncRNA inhibits cervical cancer growth and metastasis by stabilizing GAS5 via interaction with ALKBH5 and reduction of GAS5 m6A modification, which is crucial for epigenetic regulation in cervical carcinogenesis (73). These mechanisms, including metabolic suppression and RNA stabilization, exemplify the pivotal role of ALKBH5 as a safeguard against the advancement of carcinogenesis.
4. m6A readers in cervical cancer as orchestrators of tumor heterogeneity and therapeutic resistance
YTHDF readers orchestrate immune-metastatic crosstalk in cervical cancer
The YTHDF family of proteins (YTHDF1, YTHDF2 and YTHDF3) regulate mRNA stability and translation by recognizing the m6A mark. Moreover, YTHDF1 was reported to predict unfavorable clinical outcomes in cervical cancer, which were negatively associated with CD8+ T-cell infiltration (74). As shown in Fig. 4, YTHDF1 drives immune evasion by stabilizing monocarboxylate transporter 1 (MCT1) mRNA to enhance lactate accumulation and upregulates RAN binding protein 2 translation to accelerate tumor growth and invasion (74,75). Its synergy with IGF2BP3 via pyruvate dehydrogenase kinase 4 (PDK4) stabilization amplifies glycolysis, establishing a metabolic vulnerability targeted by dual YTHDF1/PDK4 inhibition (76). Additionally, YTHDF2 collaborates with METTL3 to degrade the tumor suppressor NR4A1 mRNA, relieving its repression of AKT1 transcription, thereby driving malignant progression (26). YTHDF2 also mediates the METTL3-dependent degradation of circRNF13, indirectly stabilizing the pro-metastatic CXCL1 mRNA, revealing its dual role in radioresistance (38). YTHDF3 promotes lymph node metastasis via the LDL receptor related protein 6-YAP-VEGF-C axis and confers radioresistance by enhancing RAD51 paralog D translation, and its overexpression was reported to predict a poor radiotherapy response (77,78). YTHDC2 has emerged as a pan-cancer immune modulator, and its hypomethylation is associated with cytotoxic T cell exhaustion in CSSC (79). These findings suggest that reader-specific inhibitors (such as YTHDF3 antisense oligonucleotides) and reader-immune checkpoint combinations (such as the IGF2BP3/PD-L1 blockade) are precise strategies for dismantling the m6A-regulated oncogenic network.
Figure 4.
m6A readers and related signaling pathways in the development, metastasis and radioresistance of CC. m6A readers affect tumor development, metastasis and radioresistance via regulating downstream signaling pathways. The Hippo/YAP, Wnt/β-catenin and PI3K/MAPK signaling pathways are all highly activated and serve a role in the tumor cell proliferation, differentiation and metastasis of CC cells. m6A, N6-methyladenosine; CC, cervical cancer; NR4A1, nuclear receptor subfamily 4 group A member 1; AKT1, AKT serine/threonine kinase 1; SYVN, synoviolin; NLRP3, NLR family pyrin domain containing 3; FOXM1, forkhead box M1; MAPK, mitogen-activated protein kinase; FASN, fatty acid synthase.
Context-dependent oncogenicity of the IGF2BP family: Metabolic reprogramming and immune crosstalk
The IGF2BP family exhibits context-dependent oncogenicity. The lncRNA interactome diversifies reader functions, in which IGF2BP1 partners with leucine rich repeat containing 75A-AS to destabilize synoviolin 1, activating NLR family pyrin domain containing 3/IL-1β/SMAD2/3 signaling. By contrast, YTHDF2 degrades the tumor suppressor lncRNA, cardiac mesoderm enhancer-associated non-coding RNA, in concert with miR-21-5p (80,81). IGF2BP2 stabilizes MYC and forkhead box M1 (FOXM1) mRNAs to drive HPV-mediated aerobic glycolysis and Rho GTPase activating protein 12-dependent metastasis (82,83), whilst IGF2BP3 hijacks lipid metabolism via stearoyl-CoA desaturase upregulation and activates glutaminolysis through glutaminase/glutamate dehydrogenase 1 stabilization, fostering the Treg-mediated immune escape (84,85). Paradoxically, Parkin-mediated ubiquitination of IGF2BP3 at K213 suppresses PI3K/MAPK signaling, underscoring its regulatory complexity (86).
Metabolic, metastatic and drug resistance dynamics of heterogeneous nuclear ribonucleoprotein C (HNRNP) and eukaryotic translation initiation factor 3 (EIF3) family proteins in cervical cancer
HNRNP readers sculpt tumor spatial heterogeneity. HNRNPA2B1 accelerates glycolysis via lactate dehydrogenase A stabilization, whilst HNRNPC induces FOXM1 exon skipping to promote lymphatic metastasis (10,87). Clinically, EIF3A-mediated stabilization of hypoxia up-regulated 1 has been reported to be associated with cisplatin resistance, whereas the TAPBP rs1059288 risk allele (G) enhances m6A modification via METTL14/YTHDF2, which drives TAPBP overexpression and activates the JAK/STAT/major histocompatibility complex class I polypeptide-related sequence B pathway to confer multidrug resistance (61).
5. Multidimensional biomarker network in cervical cancer
Within epigenetic regulatory hubs, METTL14-driven aberrant m6A modifications shape the immunosuppressive microenvironment via two mechanisms: i) Upregulation of glycolytic activity to induce PD-1 expression in tumor-associated macrophages, impairing phagocytic function; and ii) reinforcing the Warburg effect through m6A-dependent transcriptome remodeling, thereby fueling pro-metastatic metabolic loops (88). RCAN family member 3 (RCAN3), a pan-cancer epigenome-immune modulator, serves a pivotal role in CSSC: High RCAN3 expression is associated with poor overall survival and m6A modifier genes (such as FTO and YTHDF2), driving immune evasion via CD8+ T cell exhaustion (89).
In the present review, the association between m6A modification-related regulators and immune microenvironment characteristics was systematically evaluated using a comprehensive analysis of 24 distinct immune cell subtypes in patients with CSSC using the bioinformatics toolkit available at the Xiantao website (http://www.xiantao.love). In particular, RNA-sequencing data from The Cancer Genome Atlas-CSSC project STAR pipeline were downloaded in TPM format, as well as clinical data (https://portal.gdc.cancer.gov). Using the single-sample gene set enrichment analysis algorithm provided in the R package GSVA (1.46.0) (90), immune infiltration profiles for the corresponding cloud-based data were calculated using markers of 24 immune cell types (91). A correlation analysis was performed between several m6A regulators and the immune infiltration matrix. The results were visualized using a Laplace plot using the ggplot2 [3.4.4] package (Fig. 5) (90,91). Spearman's correlation analysis revealed a significant inverse relationship between the expression levels of m6A regulators and the prevalence of Th1 cells (P<0.001), cytotoxic cells (P<0.001), neutrophils (P<0.01) and dendritic cells (P<0.001). Conversely, a positive correlation was observed for the number of T helper cells (P<0.001).
Figure 5.
Lollipop plot of the immune infiltration profiles of the m6A regulators. A correlation analysis was performed using the ggplot2 package between several m6A regulators in the data and the immune infiltration matrix. m6A, N6-methyladenosine. The immune cell types included T helper cells, Th2 cells, Tcm, NK cells, Tem, eosinophils, mast cells, NK CD56 bright cells, CD8 T cells, Tgd, Tfh, macrophages, Th17 cells, neutrophils, iDC, pDC, T cells, DC, aDC, NK CD56dim cells, B cells, Treg, Th1 cells and cytotoxic cells. Treg, T-regulatory cells; Th2, type 2 T-helper; NK, natural killer; DC, dendritic cells; aDC, activated DC; iDC, interdigitating DC; pDC, plasmacytoid DC; Tcm, central memory T-cells; Tem, effector memory T-cells; Tgd, gamma delta T cells; Tfh, T follicular helper cells. *P<0.05, **P<0.01 and ***P<0.001.
Additionally, for clinical translation, the aforementioned biomarkers have been reported to form a cross-pathway regulatory network. GLTP and RCAN3 jointly mediate m6A-immune checkpoint interactions, whereas the Egl-9 family hypoxia inducible factor 1-METTL14 axis synergistically suppresses macrophage antigen presentation via lactate metabolism. Multidimensional biomarker panels stratify immunotherapy responsiveness, and targeting of the METTL14-glycolysis pathway combined with PD-1 inhibitors was reported to rescue macrophage dysfunction (88,89,92).
6. Conclusions and future perspectives
Despite significant advancements in the understanding of the roles of m6A modifications in cervical cancer, several critical challenges remain.
Mechanistic complexity and contradictory evidence
The m6A regulatory system exhibits context-dependent duality in function cervical cancer. For instance, YTHDF1 presents a particularly challenging paradox, highlighting the need for nuanced interpretation of the evidence. Evidence supporting a pro-tumorigenic role comes from preclinical studies, such as cell line and xenograft models, where YTHDF1 promotes tumor growth by enhancing the translational efficiency of cyclins, and its deficiency markedly suppresses tumor progression (93). However, contradictory evidence arises from clinical data analyses, which paradoxically associate elevated YTHDF1 expression with improved patient prognosis. Furthermore, whilst the preclinical evidence suggests YTHDF1 depletion attenuates tumor development (93), it unexpectedly increases cancer cell resistance to cisplatin-based chemotherapy in these models, adding another layer of complexity. This contradictory ‘pro-tumorigenic yet therapy-sensitizing’ duality observed across different experimental systems (cell lines compared with clinical cohorts) suggests phase-dependent regulatory roles of YTHDF1 across distinct stages of tumor evolution; however, further robust clinical evidence is needed to fully resolve this paradox and understand the contextual factors driving these opposing effects.
Translational bottlenecks
Most studies on m6A in cervical cancer are confined to preclinical models (such as cell lines and mice), representing a marked evidence gap for clinical translation. The strength of evidence supporting the predictive efficacy of m6A biomarkers is currently limited, as large-scale, multicenter cohort validations are largely lacking. Whilst several m6A regulators show promise as potential biomarkers in preliminary studies, the evidence often comes from single-center analyses or small patient cohorts, which may not be generalizable. Existing m6A-targeted inhibitors (such as STM2457 and FB23-2) have low selectivity, off-target effects and pharmacokinetic limitations, as evidenced by preclinical and early-phase studies (94). This highlights the urgent need for further preclinical development and, critically, robust clinical evidence from well-designed trials to support tissue-specific or functionally reversible intervention strategies.
Sample heterogeneity
The high molecular heterogeneity of cervical cancer (such as existence of HPV subtypes and immune microenvironment variations) presents a challenge for interpreting existing evidence and may dilute the predictive power of m6A biomarkers for specific subgroups. Evidence derived from bulk sequencing analyses, which are common in the field, may obscure subtype-specific or cell-type-specific m6A patterns. Integrating single-cell sequencing and spatial omics is essential for resolving cell type-specific regulatory patterns and generating more precise, context-dependent evidence for the role of m6A in cervical cancer heterogeneity.
Mao et al (95) discussed the functions, molecular mechanisms and clinical applications of m6A modification in cervical cancer. The authors particularly highlighted its regulatory role in non-coding RNAs, including circRNA, lncRNA and miRNA. Moreover, Hu et al (96) summarized the detailed mechanisms of the m6A regulatory factors on tumor cell proliferation, migration, invasion, cell cycle, apoptosis, chemotherapy resistance and sensitivity to chemotherapy. The novelty of the present review compared with previous works shows in the following ways: Firstly, it systematically dissects the cross-regulatory mechanisms between HPV oncoproteins and m6A modification networks. Secondly, in the present review, bioinformatics tools were used to performed a detailed analysis of 24 immune cell subtypes, revealing the systematic associations between m6A modification-related regulators and the characteristics of the immune microenvironment. Furthermore, the present study brings together several m6A regulators (METTL3/FTO/YTHDF1) within key signaling pathways, including the NEK2-Wnt, BMP4-Hippo and MCT1-lactate axes, to elucidate their synergistic cascade effects in driving therapeutic resistance. Concurrently, it proposes precision strategies such as METTL3 inhibition combined with PD-1 blockade, thereby establishing a distinctive systemic perspective and interventional paradigm for cervical cancer research.
Precision targeting of m6A networks in cervical cancer
m6A modification dynamically orchestrates cervical cancer progression through core regulators, such as METTL3/FTO/YTHDF1-mediated NEK2-Wnt, BMP4-Hippo and MCT1-lactate signaling networks, driving tumor proliferation, metastasis and therapy resistance. Future investigations should prioritize the development of cervical tissue-specific delivery systems for m6A inhibitors and advance the clinical validation of multi-omics biomarkers (such as m6A-circRNA/PD-L1 composite signatures). Systems pharmacology models that integrate epigenetic regulation with immunometabolic pathways may synergize METTL3-targeted agents with PD-1 blockade, potentially overcoming current therapeutic resistance mechanisms. The prognostic value of multidimensional biomarker panels and the therapeutic potential of combination strategies (such as anti-METTL3 + PD-1 inhibitors) offer novel paradigms for clinical translation. In particular, future research should focus on the following: i) Spatiotemporal mapping of the HPV-host m6A interplay; ii) the development of precision tools targeting m6A-dependent networks (subtype-specific proteolysis targeting chimeras and CRISPR-dCas9-based epitranscriptome editing) (97). By integrating these advancements, an epigenetic, metabolic and immune therapeutic framework could redefine precision oncology in cervical cancer. CRISPR-based epitranscriptome editing enables precise m6A modulation of HPV-derived oncogenic transcripts, whereas third-generation liquid biopsy platforms facilitate dynamic monitoring of m6A-modified circRNAs to optimize personalized therapeutic strategies (98); and iii) advancements in liquid biopsy techniques and biomarker-driven clinical trials are revolutionizing cancer diagnostics and therapeutic monitoring. Liquid biopsy platforms for m6A-circRNAs directly decode methylation patterns in biofluids, facilitating dynamic monitoring of therapy and early detection of resistance (70,98–100). The integration of these technologies would provide a closed-loop precision medicine framework that combines targeted epitranscriptomic manipulation with real-time molecular surveillance.
Acknowledgements
Not applicable
Funding Statement
Funding: No funding was received
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
FJ designed the study and checked the literature. MX and FJ confirm the authenticity of all the raw data. MX and FY performed the data analysis, and prepared and wrote the manuscript. HC prepared, edited and read though the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Weaver C, Nam A, Settle C, Overton M, Giddens M, Richardson KP, Piver R, Mysona DP, Rungruang B, Ghamande S, et al. Serum proteomic signatures in cervical cancer: Current status and future directions. Cancers (Basel) 2024;16:1629. doi: 10.3390/cancers16091629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tantengco OAG, Nakura Y, Yoshimura M, Nishiumi F, Llamas-Clark EF, Yanagihara I. Co-infection of human papillomavirus and other sexually transmitted bacteria in cervical cancer patients in the Philippines. Gynecol Oncol Rep. 2022;40:100943. doi: 10.1016/j.gore.2022.100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brotherton JML, Vajdic CM, Nightingale C. The socioeconomic burden of cervical cancer and its implications for strategies required to achieve the WHO elimination targets. Expert Rev Pharmacoecon Outcomes Res. 2025;25:1–20. doi: 10.1080/14737167.2025.2451732. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein A, Gersh M, Skovronsky G, Moss C. The future of cervical cancer screening. Int J Womens Health. 2024;16:1715–1731. doi: 10.2147/IJWH.S474571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nartey Y, Amo-Antwi K, Hill PC, Dassah ET, Asmah RH, Nyarko KM, Agambire R, Konney TO, Yarney J, Damale N, Cox B. Human papillomavirus genotype distribution among women with and without cervical cancer: Implication for vaccination and screening in Ghana. PLoS One. 2023;18:e0280437. doi: 10.1371/journal.pone.0280437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan P, Li X, Feng Y, Cai H, Dong D, Peng Y, Yao X, Guo Y, Ma M, Dong T, Wang R. PD-1 expression status on CD8+ tumour infiltrating lymphocytes associates with survival in cervical cancer. Front Oncol. 2021;11:678758. doi: 10.3389/fonc.2021.678758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braila AD, Poalelungi CV, Albu CC, Damian CM, Dira LM, Banateanu AM, Bogdan-Andreescu CF. The relationship between cervicovaginal infection, human papillomavirus infection and cervical intraepithelial neoplasia in Romanian women. Diseases. 2025;13:18. doi: 10.3390/diseases13010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meghani K, Puri P, Bazzett-Matabele L, Vuylsteke P, Luckett R, Monare B, Chiyapo S, Ketlametswe R, Ralefala TB, Bvochora-Nsingo M, et al. Significance of HIV status in cervical cancer patients receiving curative chemoradiation therapy, definitive radiation alone, or palliative radiation in Botswana. Cancer. 2024;130:2462–2471. doi: 10.1002/cncr.35289. [DOI] [PubMed] [Google Scholar]
- 9.Teshome R, Yang I, Woldetsadik E, Girma E, Higgins M, Wells J. Survival status and predictors among women with advanced stage of cervical cancer. Int J Womens Health. 2024;16:605–617. doi: 10.2147/IJWH.S455235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen M, Yi N, Mijiti B, Zhao S, Shen G. N(6)-methyladenosine (m6A) reader HNRNPA2B1 accelerates the cervical cancer cells aerobic glycolysis. J Bioenerg Biomembr. 2024;56:657–668. doi: 10.1007/s10863-024-10042-x. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, Ju M, Zheng X, Jiang Y, Yu X, Pan B, Luo R, Jia W, Zheng M. Methyltransferase-like 3 promotes cervical cancer metastasis by enhancing cathepsin L mRNA stability in an N6-methyladenosine-dependent manner. Cancer Sci. 2023;114:837–854. doi: 10.1111/cas.15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo J, Zhao J, Tian P, Xu Z, Wang R, Chen W, Wang X, Wan S, Yang Y, Zhang H. BaP/BPDE exposure causes human trophoblast cell dysfunctions and induces miscarriage by up-regulating lnc-HZ06-regulated IL1B. J Hazard Mater. 2024;476:134741. doi: 10.1016/j.jhazmat.2024.134741. [DOI] [PubMed] [Google Scholar]
- 13.Dai M, Huang W, Huang X, Ma C, Wang R, Tian P, Chen W, Zhang Y, Mi C, Zhang H. BPDE, the migration and invasion of human trophoblast cells, and occurrence of miscarriage in humans: Roles of a novel lncRNA-HZ09. Environ Health Perspect. 2023;131:17009. doi: 10.1289/EHP10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Xiao P, Tang J, Wang R, Wang X, Wang F, Ruan J, Yu S, Tang J, Huang R, Zhao X. m6A Regulator-Mediated tumour infiltration and methylation modification in cervical cancer microenvironment. Front Immunol. 2022;13:888650. doi: 10.3389/fimmu.2022.888650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Kong W, Zhao X, Han C, Liu T, Li J, Song D. N6-Methyladenosine-Related lncRNAs as potential biomarkers for predicting prognoses and immune responses in patients with cervical cancer. BMC Genom Data. 2022;23:8. doi: 10.1186/s12863-022-01024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condic M, Ralser DJ, Klumper N, Ellinger J, Qureischi M, Egger EK, Kristiansen G, Mustea A, Thiesler T. Comprehensive analysis of N6-Methyladenosine (m6A) writers, erasers, and readers in cervical cancer. Int J Mol Sci. 2022;23:7165. doi: 10.3390/ijms23137165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Li Z, Kong B, Song C, Cong J, Hou J, Wang S. Reduced m6A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget. 2017;8:98918–98930. doi: 10.18632/oncotarget.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Gu L, Xiao J, Jin F. Knockdown of RBM15 inhibits tumor progression and the JAK-STAT signaling pathway in cervical cancer. BMC Cancer. 2023;23:684. doi: 10.1186/s12885-023-11163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu R, Wei Y, He C, Zhou P, Yang H, Deng C, Liu R, Wu P, Gao Q, Cao C. Integrative analyses of m6A regulators identify that METTL3 is associated with HPV status and immunosuppressive microenvironment in HPV-related cancers. Int J Biol Sci. 2022;18:3874–3887. doi: 10.7150/ijbs.70674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji H, Zhang JA, Liu H, Li K, Wang ZW, Zhu X. Comprehensive characterization of tumor microenvironment and m6A RNA methylation regulators and its effects on PD-L1 and immune infiltrates in cervical cancer. Front Immunol. 2022;13:976107. doi: 10.3389/fimmu.2022.976107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pu M, Xiao X, Lv S, Ran D, Huang Q, Zhou M, Lei Q, Kong L, Zhang Q. METTL3-dependent DLG2 inhibits the malignant progression of cervical cancer by inactivating the Hippo/YAP signaling. Hereditas. 2025;162:9. doi: 10.1186/s41065-025-00365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou PX, Fan Q, Zhang Q, Liu JJ, Wu Q. M6A-induced transcription factor IRF5 contributes to the progression of cervical cancer by upregulating PPP6C. Clin Exp Pharmacol Physiol. 2024;51:e13868. doi: 10.1111/1440-1681.13868. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Li C, Deng Q, Ren X, Wang H. METTL3′s role in cervical cancer development through m6A modification. FASEB J. 2024;38:e23693. doi: 10.1096/fj.202400580. [DOI] [PubMed] [Google Scholar]
- 24.Guo Y, Bai Y, Wang L, Xu Z, Zhang N, Wang W, Zhao H. METTL3 facilitates the progression of cervical cancer by m6A modification-mediated up-regulation of NEK2. Sci Rep. 2024;14:24469. doi: 10.1038/s41598-024-73601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y, Shi H, Zheng W. METTL3 regulates the translation of oncogene Myc through m6A Modification and promotes the occurrence and development of cervical cancer. Discov Med. 2024;36:1902–1910. doi: 10.24976/Discov.Med.202436188.176. [DOI] [PubMed] [Google Scholar]
- 26.Yu T, Wu F, Jia Y, Zhang X, Qi X, Jin Z, Hao T, Zhao J, Liu Z, Wang C, et al. RNA N(6)-methyladenosine modification mediates downregulation of NR4A1 to facilitate malignancy of cervical cancer. Cell Biosci. 2022;12:207. doi: 10.1186/s13578-022-00937-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Xie G, Tian Y, Li W, Wu Y, Chen F, Lin Y, Lin X, Au SWN, Cao J, et al. RNA m6A methylation regulates dissemination of cancer cells by modulating expression and membrane localization of β-catenin. Mol Ther. 2022;30:1578–1596. doi: 10.1016/j.ymthe.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ao K, Yin M, Lyu X, Xiao Y, Chen X, Zhong S, Wen X, Yuan J, Ye M, Zhang J, et al. METTL3-mediated HSPA9 m6A modification promotes malignant transformation and inhibits cellular senescence by regulating exosomal mortalin protein in cervical cancer. Cancer Lett. 2024;587:216658. doi: 10.1016/j.canlet.2024.216658. [DOI] [PubMed] [Google Scholar]
- 29.Rui Y, Zhang H, Yu K, Qiao S, Gao C, Wang X, Yang W, Asadikaram G, Li Z, Zhang K, et al. N(6)-Methyladenosine regulates cilia elongation in cancer cells by modulating HDAC6 expression. Adv Sci (Weinh) 2025;12:e2408488. doi: 10.1002/advs.202408488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Zhong Y, Cao G, Shi H, Liu Y, Li L, Yin P, Chen J, Xiao Z, Du B. METTL3 promotes cell cycle progression via m6A/YTHDF1-dependent regulation of CDC25B translation. Int J Biol Sci. 2022;18:3223–3236. doi: 10.7150/ijbs.70335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Guo X, Li L, Gao Z, Su X, Ji M, Liu J. N(6)-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. Cell Death Dis. 2020;11:911. doi: 10.1038/s41419-020-03071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu HT, Zhao Y, Wang HC, Liu QL. METTL3-mediated m6A methylation of SLC38A1 stimulates cervical cancer growth. Biochem Biophys Res Commun. 2024;716:150039. doi: 10.1016/j.bbrc.2024.150039. [DOI] [PubMed] [Google Scholar]
- 33.Du QY, Huo FC, Du WQ, Sun XL, Jiang X, Zhang LS, Pei DS. METTL3 potentiates progression of cervical cancer by suppressing ER stress via regulating m6A modification of TXNDC5 mRNA. Oncogene. 2022;41:4420–4432. doi: 10.1038/s41388-022-02435-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang YY, Ye LH, Zhao AQ, Gao WR, Dai N, Yin Y, Zhang X. M6A modification regulates tumor suppressor DIRAS1 expression in cervical cancer cells. Cancer Biol Ther. 2024;25:2306674. doi: 10.1080/15384047.2024.2306674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen S, Jin H, Zhang X, Zhang Y, Li X, Yan W, Xie S, Yu B, Hu J, Liu H, et al. LINC00426, a novel m6A-regulated long non-coding RNA, induces EMT in cervical cancer by binding to ZEB1. Cell Signal. 2023;109:110788. doi: 10.1016/j.cellsig.2023.110788. [DOI] [PubMed] [Google Scholar]
- 36.Shen W, Zhu M, Wang Q, Zhou X, Wang J, Wang T, Zhang J. DARS-AS1 recruits METTL3/METTL14 to bind and enhance DARS mRNA m6A modification and translation for cytoprotective autophagy in cervical cancer. RNA Biol. 2022;19:751–763. doi: 10.1080/15476286.2022.2079889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, Ling K, Zhu Y, Deng L, Li Y, Liang Z. circ0000069 promotes cervical cancer cell proliferation and migration by inhibiting miR-4426. Biochem Biophys Res Commun. 2021;551:114–120. doi: 10.1016/j.bbrc.2021.03.020. [DOI] [PubMed] [Google Scholar]
- 38.Shi J, Rui X, Han C, Wang C, Xu L, Jiang X. circRNF13, a novel N(6)-methyladenosine-modified circular RNA, enhances radioresistance in cervical cancer by increasing CXCL1 mRNA stability. Cell Death Discov. 2023;9:253. doi: 10.1038/s41420-023-01557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han X, Xia L, Wu Y, Chen X, Wu X. m6A-modified circSTX6 as a key regulator of cervical cancer malignancy via SPI1 and IL6/JAK2/STAT3 pathways. Oncogene. 2025;44:968–982. doi: 10.1038/s41388-024-03260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji F, Lu Y, Chen S, Lin X, Yu Y, Zhu Y, Luo X. m6A methyltransferase METTL3-mediated lncRNA FOXD2-AS1 promotes the tumorigenesis of cervical cancer. Mol Ther Oncolytics. 2021;22:574–581. doi: 10.1016/j.omto.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen G, Li F, Wang Y, Huang Y, Aizezi G, Yuan J, Ma C, Lin C. New insights on the interaction between m6A modification and non-coding RNA in cervical squamous cell carcinoma. World J Surg Oncol. 2023;21:25. doi: 10.1186/s12957-023-02907-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Ma J, Han S, Li X, Guo H, Liu D. ZFAS1 exerts an oncogenic role via suppressing miR-647 in an m6A-dependent manner in cervical cancer. Onco Targets Ther. 2020;13:11795–11806. doi: 10.2147/OTT.S274492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong Y, Luo G, Zhang S, Chen Y, Hu Y. Transcriptome sequencing analysis reveals miR-30c-5p promotes ferroptosis in cervical cancer and inhibits growth and metastasis of cervical cancer xenografts by targeting the METTL3/KRAS axis. Cell Signal. 2024;117:111068. doi: 10.1016/j.cellsig.2024.111068. [DOI] [PubMed] [Google Scholar]
- 44.Huang C, Liang J, Lin S, Wang D, Xie Q, Lin Z, Yao T. N(6)-methyladenosine associated silencing of miR-193b promotes cervical cancer aggressiveness by targeting CCND1. Front Oncol. 2021;11:666597. doi: 10.3389/fonc.2021.666597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alasar AA, Saglam B, Vatansever IE, Akgul B. Expression patterns of m6A RNA methylation regulators under apoptotic conditions in various human cancer cell lines. Turk J Biol. 2024;48:24–34. doi: 10.55730/1300-0152.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Zeng J, He S, Yang Y, Wang C. METTL14 decreases FTH1 mRNA stability via m6A methylation to promote sorafenib-induced ferroptosis of cervical cancer. Cancer Biol Ther. 2024;25:2349429. doi: 10.1080/15384047.2024.2349429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q, Zhao N, Ding X, Zhao J. METTL14-mediated m6A modification upregulates HOXB13 expression to activate NF-κB and exacerbate cervical cancer progression. Mol Cell Oncol. 2024;11:2423986. doi: 10.1080/23723556.2024.2423986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P, Tang Y, Zhao J, Yang J, Chen Y, Gong Y, Meng S, Shu C. TRIM11 regulated by m6A modification promotes the progression of cervical cancer by PHLPP1 ubiquitination. Neoplasma. 2023;70:659–669. doi: 10.4149/neo_2023_230104N7. [DOI] [PubMed] [Google Scholar]
- 49.Geng F, Fan MJ, Li J, Liang SM, Li CY, Li N. Knockdown of METTL14 inhibits the growth and invasion of cervical cancer. Transl Cancer Res. 2019;8:2307–2315. doi: 10.21037/tcr.2019.09.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Q, Li Z, Luo X, Wang D, Zhou Y, Zhao J, Gao S, Yang Y, Fu W, Kong L, Sun T. piRNA-14633 promotes cervical cancer cell malignancy in a METTL14-dependent m6A RNA methylation manner. J Transl Med. 2022;20:51. doi: 10.1186/s12967-022-03257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang R, Tan W. RBM15-Mediated N6-Methyl adenosine (m6A) modification of EZH2 drives the epithelial-mesenchymal transition of cervical cancer. Crit Rev Eukaryot Gene Expr. 2024;34:15–29. doi: 10.1615/CritRevEukaryotGeneExpr.2024052205. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Li C, Wei Q, Zhang E, Yang Y, Sha L, Wang D. RBM15 knockdown impairs the malignancy of cervical cancer by mediating m6A modification of decorin. Biochem Genet. 2024;63:225–238. doi: 10.1007/s10528-024-10757-x. [DOI] [PubMed] [Google Scholar]
- 53.Quan Y, Zhou M, Li J, Yang Y, Guo J, Tang T, Liu P. The m6A methyltransferase RBM15 affects tumor cell stemness and progression of cervical cancer by regulating the stability of lncRNA HEIH. Exp Cell Res. 2024;436:113924. doi: 10.1016/j.yexcr.2024.113924. [DOI] [PubMed] [Google Scholar]
- 54.Song Y, Wu Q. RBM15 m(6) A modification-mediated OTUB2 upregulation promotes cervical cancer progression via the AKT/mTOR signaling. Environ Toxicol. 2023;38:2155–2164. doi: 10.1002/tox.23852. [DOI] [PubMed] [Google Scholar]
- 55.Feng X, Shu L. The methyltransferase KIAA1429 potentiates cervical cancer tumorigenesis via modulating LARP1 mRNA m6A modification and stability. Histol Histopathol. 2024;40:1095–1103. doi: 10.14670/HH-18-843. [DOI] [PubMed] [Google Scholar]
- 56.Wang M, Wang Z, Zou X, Yang D, Xu K. The regulatory role of KIAA1429 in epithelial-mesenchymal transition in cervical cancer via mediating m6A modification of BTG2. Cytotechnology. 2025;77:34. doi: 10.1007/s10616-024-00694-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei Q, Yang Y, Li C, Wang H. ZC3H13-induced the m6A modification of hsa_circ_0081723 promotes cervical cancer progression via AMPK/p53 pathway. J Obstet Gynaecol Res. 2024;50:2286–2298. doi: 10.1111/jog.16140. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Chen X, Chen H, Zhang Y. ZC3H13 enhances the malignancy of cervical cancer by regulating m6a modification of CKAP2. Crit Rev Immunol. 2023;43:1–13. doi: 10.1615/CritRevImmunol.2023049342. [DOI] [PubMed] [Google Scholar]
- 59.Lin X, Wang F, Chen J, Liu J, Lin YB, Li L, Chen CB, Xu Q. N(6)-methyladenosine modification of CENPK mRNA by ZC3H13 promotes cervical cancer stemness and chemoresistance. Mil Med Res. 2022;9:19. doi: 10.1186/s40779-022-00378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu X, Li R, Ying Y, Zhang W, Wang W. Gene signatures, immune infiltration, and drug sensitivity based on a comprehensive analysis of m6a RNA methylation regulators in cervical cancer. J Transl Med. 2022;20:385. doi: 10.1186/s12967-022-03600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu J, Wang S, Zhang X, Yan W, Liu H, Chen X, Nie Y, Liu F, Zheng Y, Lu Y, Jin H. A genetic variant in the TAPBP gene enhances cervical cancer susceptibility by increasing m6A modification. Arch Toxicol. 2024;98:3425–3438. doi: 10.1007/s00204-024-03820-4. [DOI] [PubMed] [Google Scholar]
- 62.Zou D, Dong L, Li C, Yin Z, Rao S, Zhou Q. The m6A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int. 2019;19:321. doi: 10.1186/s12935-019-1045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen B, Wang L, Li X, Ren C, Gao C, Ding W, Wang H. FTO facilitates cervical cancer malignancy through inducing m6A-Demethylation of PIK3R3 mRNA. Cancer Med. 2024;13:e70507. doi: 10.1002/cam4.70507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu C, Li Y, Dong C, Qu L, Zuo Y. E6E7 regulates the HK2 expression in cervical cancer via GSK3β/FTO signal. Arch Biochem Biophys. 2022;729:109389. doi: 10.1016/j.abb.2022.109389. [DOI] [PubMed] [Google Scholar]
- 65.Azzam SK, Alsafar H, Sajini AA. FTO m6A demethylase in obesity and cancer: Implications and underlying molecular mechanisms. Int J Mol Sci. 2022;23:3800. doi: 10.3390/ijms23073800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang J, Yang J, Zhang Y, Lu D, Dai Y. FTO promotes cervical cancer cell proliferation, colony formation, migration and invasion via the regulation of the BMP4/Hippo/YAP1/TAZ pathway. Exp Cell Res. 2023;427:113585. doi: 10.1016/j.yexcr.2023.113585. [DOI] [PubMed] [Google Scholar]
- 67.Wang T, Li W, Ye B, Zhang S, Lei X, Zhang D. FTO-stabilized lncRNA HOXC13-AS epigenetically upregulated FZD6 and activated Wnt/β-catenin signaling to drive cervical cancer proliferation, invasion, and EMT. J BUON. 2021;26:1279–1291. [PubMed] [Google Scholar]
- 68.Wang A, Jin C, Wang Y, Yu J, Wang R, Tian X. FTO promotes the progression of cervical cancer by regulating the N6-methyladenosine modification of ZEB1 and Myc. Mol Carcinog. 2023;62:1228–1237. doi: 10.1002/mc.23559. [DOI] [PubMed] [Google Scholar]
- 69.Huo FC, Zhu ZM, Du WQ, Pan YJ, Jiang X, Kang MJ, Liu BW, Mou J, Pei DS. HPV E7-drived ALKBH5 promotes cervical cancer progression by modulating m6A modification of PAK5. Pharmacol Res. 2023;195:106863. doi: 10.1016/j.phrs.2023.106863. [DOI] [PubMed] [Google Scholar]
- 70.Liang L, Zhu Y, Li J, Zeng J, Wu L. ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J Exp Clin Cancer Res. 2022;41:261. doi: 10.1186/s13046-022-02462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu S, Liu L, Xu H, Zhu Q, Tan M. The involvement of MALAT1-ALKBH5 signaling axis into proliferation and metastasis of human papillomavirus-positive cervical cancer. Cancer Biol Ther. 2023;24:2249174. doi: 10.1080/15384047.2023.2249174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhen L, Pan W. ALKBH5 inhibits the SIRT3/ACC1 axis to regulate fatty acid metabolism via an m6A-IGF2BP1-dependent manner in cervical squamous cell carcinoma. Clin Exp Pharmacol Physiol. 2023;50:380–392. doi: 10.1111/1440-1681.13754. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–4921. [PMC free article] [PubMed] [Google Scholar]
- 74.Xiong J, He L, Chai X, Zhang Y, Sun S. YTHDF1 boosts the lactate accumulation to potentiate cervical cancer cells immune escape. Cell Death Dis. 2024;15:843. doi: 10.1038/s41419-024-07128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H, Luo Q, Kang J, Wei Q, Yang Y, Yang D, Liu X, Liu T, Yi P. YTHDF1 aggravates the progression of cervical cancer through m6A-mediated up-regulation of RANBP2. Front Oncol. 2021;11:650383. doi: 10.3389/fonc.2021.650383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, Lin S, Wang H. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11:2578. doi: 10.1038/s41467-020-16306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong S, Guo Q, Chen X, Luo X, Long Y, Chong T, Ye M, He H, Lu A, Ao K, et al. The inhibition of YTHDF3/m6A/LRP6 reprograms fatty acid metabolism and suppresses lymph node metastasis in cervical cancer. Int J Biol Sci. 2024;20:916–936. doi: 10.7150/ijbs.87203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Du H, Zou NY, Zuo HL, Zhang XY, Zhu SC. YTHDF3 mediates HNF1alpha regulation of cervical cancer radio-resistance by promoting RAD51D translation in an m6A-dependent manner. FEBS J. 2023;290:1920–1935. doi: 10.1111/febs.16681. [DOI] [PubMed] [Google Scholar]
- 79.Zhang C, Guo C, Li Y, Ouyang L, Zhao Q, Liu K. The role of YTH domain containing 2 in epigenetic modification and immune infiltration of pan-cancer. J Cell Mol Med. 2021;25:8615–8627. doi: 10.1111/jcmm.16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sui H, Shi C, Yan Z, Chen J, Man L, Wang F. LRRC75A-AS1 drives the epithelial-mesenchymal transition in cervical cancer by binding IGF2BP1 and inhibiting SYVN1-mediated NLRP3 ubiquitination. Mol Cancer Res. 2024;22:1075–1087. doi: 10.1158/1541-7786.MCR-23-0478. [DOI] [PubMed] [Google Scholar]
- 81.Yu B, Li X, Yan W, Ding B, Zhang X, Shen S, Xie S, Hu J, Liu H, Chen X, et al. Post-transcriptional regulation of tumor suppressor gene lncRNA CARMN via m6A modification and miRNA regulation in cervical cancer. J Cancer Res Clin Oncol. 2023;149:10307–10318. doi: 10.1007/s00432-023-04893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu C, Liu T, Han C, Xuan Y, Jiang D, Sun Y, Zhang X, Zhang W, Xu Y, Liu Y, et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m6A-MYC expression. Int J Biol Sci. 2022;18:507–521. doi: 10.7150/ijbs.67770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ji F, Lu Y, Chen S, Yu Y, Lin X, Zhu Y, Luo X. IGF2BP2-modified circular RNA circARHGAP12 promotes cervical cancer progression by interacting m6A/FOXM1 manner. Cell Death Discov. 2021;7:215. doi: 10.1038/s41420-021-00595-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han C, Hu C, Liu T, Sun Y, Hu F, He Y, Zhang J, Chen J, Ding J, Fan J, et al. IGF2BP3 enhances lipid metabolism in cervical cancer by upregulating the expression of SCD. Cell Death Dis. 2024;15:138. doi: 10.1038/s41419-024-06520-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou T, Xiao Z, Lu J, Zhang L, Bo L, Wang J. IGF2BP3-mediated regulation of GLS and GLUD1 gene expression promotes treg-induced immune escape in human cervical cancer. Am J Cancer Res. 2023;13:5289–5305. [PMC free article] [PubMed] [Google Scholar]
- 86.Sun X, Ye G, Li J, Shou H, Bai G, Zhang J. Parkin regulates IGF2BP3 through ubiquitination in the tumourigenesis of cervical cancer. Clin Transl Med. 2023;13:e1457. doi: 10.1002/ctm2.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu YY, Xia M, Chen ZB, Liao YD, Zhang CY, Yuan L, Pan YW, Huang H, Lu HW, Yao SZ. HNRNPC mediates lymphatic metastasis of cervical cancer through m6A-dependent alternative splicing of FOXM1. Cell Death Dis. 2024;15:732. doi: 10.1038/s41419-024-07108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang B, Mao Z, Ye J, Jiao X, Zhang T, Wang Q, Han S, Zhang Y, Wang C, Dong T, Cui B. Glycolysis induced by METTL14 is essential for macrophage phagocytosis and phenotype in cervical cancer. J Immunol. 2024;212:723–736. doi: 10.4049/jimmunol.2300339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liang Y, Diao W, Yang X, Tao Y, Hong L, Li W. Regulator of calcineurin 3 as a novel predictor of diagnosis and prognosis in pan-cancer. Croat Med J. 2024;65:356–372. doi: 10.3325/cmj.2024.65.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hanzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 92.Shi YL, Liu MB, Wu HT, Han Y, He X. GLTP is a potential prognostic biomarker and correlates with immunotherapy efficacy in cervical cancer. Dis Markers. 2022;2022:9109365. doi: 10.1155/2022/9109365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, Shen Q, Xu P, Zeng L, Zhou Y, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benak D, Alanova P, Holzerova K, Chalupova M, Opletalova B, Kolar F, Pavlinkova G, Hlavackova M. Epitranscriptomic regulation of HIF-1. Bidirectional regulatory pathways. Mol Med. 2025;31:105. doi: 10.1186/s10020-025-01149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mao Z, Wang B, Zhang T, Cui B. The roles of m6A methylation in cervical cancer: Functions, molecular mechanisms, and clinical applications. Cell Death Dis. 2023;14:734. doi: 10.1038/s41419-023-06265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu J, Wang S, Li X. A comprehensive review of m6A research in cervical cancer. Epigenomics. 2024;16:753–773. doi: 10.2217/epi-2024-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao J, Li B, Ma J, Jin W, Ma X. Photoactivatable RNA N(6)-methyladenosine editing with CRISPR-Cas13. Small. 2020;16:e1907301. doi: 10.1002/smll.201907301. [DOI] [PubMed] [Google Scholar]
- 98.Lo N, Xu X, Soares F, He HH. The basis and promise of programmable RNA editing and modification. Front Genet. 2022;13:834413. doi: 10.3389/fgene.2022.834413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He F, Guo Q, Jiang GX, Zhou Y. Comprehensive analysis of m6A circRNAs identified in colorectal cancer by MeRIP sequencing. Front Oncol. 2022;12:927810. doi: 10.3389/fonc.2022.927810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Song C, Zhao C, Nong Y, Lin Y, Huang A, Xi S, Wei X, Zeng C, Qin Y, Zhu Q. Exploring the accuracy of third-generation Nanopore Sequencing technology for detecting mycobacterium tuberculosis in patients with diabetes mellitus. Diagn Microbiol Infect Dis. 2024;110:116392. doi: 10.1016/j.diagmicrobio.2024.116392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated in the present study may be requested from the corresponding author.