Abstract
Toll/IL-1 receptor family members are central components of host defense mechanisms in a variety of species. One well conserved element in their signal transduction is Ser/Thr kinases, which couple early signaling events in a receptor complex at the plasma membrane to larger signalosomes in the cytosol. The fruit fly Drosophila melanogaster has one member of this family of kinases, termed Pelle. The complexity of this pathway is vastly increased in vertebrates, and several Pelle homologs have been described and termed IL-1 receptor-associated kinase (IRAK). Here we report the identification of a novel and distinct member of the IRAK family, IRAK-4. IRAK-4 is the closest human homolog to Pelle. Endogenous IRAK-4 interacts with IRAK-1 and TRAF6 in an IL-1-dependent manner, and overexpression of IRAK-4 can activate NF-κB as well as mitogen-activated protein (MAP) kinase pathways. Most strikingly, and in contrast to the other IRAKs, IRAK-4 depends on its kinase activity to activate NF-κB. In addition, IRAK-4 is able to phosphorylate IRAK-1, and overexpression of dominant-negative IRAK-4 is blocking the IL-1-induced activation and modification of IRAK-1, suggesting a role of IRAK-4 as a central element in the early signal transduction of Toll/IL-1 receptors, upstream of IRAK-1.
IL-1 receptor-associated kinases (IRAKs) are important mediators in the signal transduction of Toll/IL-1 receptor (TIR) family members (1). All TIRs with known function are involved in host defense mechanisms, either by the recognition of pathogens or as receptors for proinflammatory cytokines (2). They play a crucial role in the switch from innate to adaptive immunity in mammals, and the signaling cascades initiated by these receptors are implicated in a number of human diseases (3).
Remarkably, the signal transduction mechanisms used by TIRs are at least in part conserved from invertebrates to vertebrates (2, 4), and to our knowledge, the early signal transduction events triggered by all mammalian members of the TIR family are basically identical. The first step is the ligand-induced formation of a receptor complex. This complex can consist of a receptor and a coreceptor, such as IL-1RI and IL-1RAcP. Alternatively, several chains of one family member form a complex, as was reported for TLR4 (5). In either case, the close proximity of the TIR domains of the individual receptor chains allows a homotypic interaction with the TIR domain of the adaptor molecules MyD88 (6, 7) or Mal (8). MyD88 can then in turn recruit an IRAK via a death domain–death domain interaction. An IL-1-induced, IL-1 receptor-associated kinase activity was first described in 1994 (9), and the first, prototypic family member, IRAK-1, was cloned in 1996 (10). IRAK-1 is activated at the receptor complex, becomes rapidly phosphorylated, and leaves the receptor complex to interact with the adaptor TRAF6 (11). The IRAK/TRAF6 interaction is a key step in the assembly of a multiprotein signalosome, which includes the mitogen-activated protein kinase kinase kinase (MAPKKK) TAK1. This complex activates a number of downstream signaling cascades, including IκB kinases (IKKs), p38, and Jun-N-terminal kinases (JNKs), leading to the activation of transcription factors such as NF-κB and AP-1 (for review see ref. 2).
Surprisingly, mice deficient in IRAK-1 are severely compromised in their ability to respond to IL-1, but the response is not completely abolished (12, 13). This observation was explained by redundancy in the IRAK family: Two additional members were identified in humans, termed IRAK-2 and IRAK-M (6, 14, 15). Both proteins behave much like IRAK-1 in overexpression studies and they can compensate for the loss of IRAK-1 in a mutant 293 cell line (14, 16). The most striking difference among the three proteins is that only IRAK-1 has potent kinase activity. The function of the kinase activity of IRAK-1 is unclear, however, and seems to be dispensable for IL-1-induced NF-κB activation, at least under overexpression conditions (16–18).
Here we describe the identification and characterization of IRAK-4, a novel member of the IRAK family with unique functional properties. IRAK-4 shares the domain structure of the other IRAKs and it is able to activate similar signal transduction pathways, namely NF-κB and MAPK pathways. It rapidly and transiently associates with IRAK-1 and TRAF6 in an IL-1-dependent manner but it is not functionally redundant with IRAK-1. Most strikingly, IRAK-4 is an active protein kinase and requires its kinase activity to activate NF-κB. Finally, we present evidence that IRAK-4 might act upstream of IRAK-1 as an IRAK-1 activator.
Methods
Biological Reagents and Cell Culture.
Recombinant human IL-1β and tumor necrosis factor α (TNFα) were purchased from BioSource International (Camarillo, CA). The anti-Flag mAbs M2 and M2 cross-linked to Sepharose beads (M2 beads) were purchased from Sigma–Aldrich. Rabbit anti-myc polyclonal Abs were from Santa Cruz Biotechnology. The 293 cells and 293 sublines were maintained in DMEM supplemented with 10% FCS. Polyclonal antisera against IRAK-4 were raised in rabbits from Zymed, using IRAK-4 expressed in Escherichia coli and purified under denaturing conditions as described elsewhere (10). His-tagged recombinant IRAK-4 was expressed in insect cells by using a GIBCO bac-to-bac baculovirus expression system, and Flag-tagged recombinant IRAK-1 and NIK were expressed in insect cells by using the Baculogold system from PharMingen (following the manufacturer's recommendations). Antisera against IRAK-1 and TRAF6 were described (10, 11).
Cloning and Expression Vectors.
Human IRAK-4 cDNA was obtained by PCR with a universal cDNA library as template (CLONTECH). Murine IRAK-4 was isolated from a murine kidney cDNA phage library (CLONTECH) under low stringency conditions with a probe derived from full-length human IRAK-4 according to standard procedures. Mammalian expression vectors encoding Flag- and myc-tagged IKKβ K44A, IRAK-1, IRAK-1 K239S, MyD88, and TRAF6 have been described elsewhere (14, 19). Expression vectors for N-terminal Flag- or myc-tagged IRAK-4, N-IRAK-4 (amino acid 1–191) were constructed by inserting PCR-generated cDNA fragments in the mammalian expression pRK7 (20). IRAK-4 KK213AA was constructed with the Quick-Change site-directed mutagenesis kit (Stratagene).
Reporter Assays.
Cells (3 × 105) were seeded into 6-well plates. For experiments with the NF-κB-dependent endothelial leukocyte adhesion molecule (ELAM) promoter, cells were transfected on the following day by the calcium phosphate precipitation method with 0.1 μg pELAM-luc/1 μg pRSV-β-gal and the indicated amounts of expression constructs. The PathDetect transreporting systems (Stratagene) were used according to the manufacturer's protocol. After 24 h, the cells were left untreated or stimulated with IL-1β (50 ng/ml) or tumor necrosis factor α (100 ng/ml) for 6–8 h before harvest. Luciferase activity and β-galactosidase activity were determined with the Luciferase Assay System (Promega) and chemiluminescence reagents from Tropix (Bedford, MA), respectively.
Immunoprecipitations of Overexpressed Proteins and Immunoblotting.
For coprecipitation of transfected proteins, 3 × 106 293 cells were plated on 10-cm dishes and transfected on the following day by the calcium phosphate precipitation method. Lysis and immunoprecipitation were performed as described elsewhere (14).
Immunoprecipitation of Endogenous Proteins.
Suspension cultures of 293RI cells (2 × 108 cells per sample) were incubated with or without 100 ng/ml of IL-1β for the indicated amount of time. Cells were collected by centrifugation (10 min at 600 × g), and cell lysates were prepared as described above. The lysates were divided into two aliquots. One aliquot was incubated with 50 μl of Protein A Sepharose (Amersham Pharmacia) and 2 μl of anti-IRAK-4 rabbit serum, the other one with 50 μl of Protein A Sepharose and 2 μl of preimmune serum. After overnight incubation, the immunoprecipitates were washed extensively, and immunoblotting was performed as described above.
In Vitro Kinase Assays.
Recombinant kinases and synthetic peptides were incubated in 20 μl of kinase buffer [20 mM Tris (pH 7.6)/1 mM DTT/20 mM MgCl2/20 mM β-glycerophosphate/20 mM p-nitrophenylphosphate/1 mM EDTA/1 mM sodium orthovanadate, protease inhibitors (Complete, Roche Molecular Biochemicals)/20 μM ATP] with 10 μCi [γ-32P]ATP for 20 min at 30°C. Boiling in 20 μl of SDS sample buffer stopped the kinase reaction. Proteins and peptides were separated by way of SDS/PAGE, and gels were dried and exposed to x-ray film.
Results and Discussion
Identification of IRAK-4.
In an attempt to identify novel members of the IRAK family, we performed database searches at National Center for Biotechnology Information. A human cDNA sequence was found that encodes a polypeptide sharing significant but previously unrecognized homology with IRAK-1 (NY-REN-64 antigen, GenBank accession no. AF155118) (21). We obtained full-length cDNA clones from a cDNA library by means of PCR. DNA sequencing of several independently obtained clones revealed five amino acid substitutions compared with NY-REN-64 (A81V, V432G, L437R, R444S, and Q451H). The new sequence was deposited at GenBank and termed IRAK-4 (accession no. AF445802).
The IRAK-4 cDNA encodes a protein with 460 amino acids and a calculated molecular mass of 52 kDa. Analysis of the deduced protein sequence revealed an N-terminal death domain (22) and a central kinase domain, similar to the domain structures of IRAK-1, IRAK-2, IRAK-M, and Pelle (14). Similar to Pelle, but in contrast to the other family members, which contain a long, unique C-terminal domain, the IRAK-4 kinase domain is followed only by a few amino acids (Fig. 1). The overall sequence identity shared between the newly identified protein and the existing IRAK-like molecules is between 30–40% (Table 1). IRAK-4 is an active protein kinase and undergoes auto- and/or crossphosphorylation upon activation by overexpression (data not shown), similar to IRAK-1 and Pelle, but unlike IRAK-2 and IRAK-M, which bear inactivating mutations in their kinase domains (14).
Figure 1.
Protein sequences of human IRAK-4, murine IRAK-4, human IRAK-1, and Drosophila melanogaster Pelle. Dark and light shading indicates identical and similar residues, respectively.
Table 1.
Sequence similarities among hu-IRAK-4 and IRAK/Pelle family members
| Homology with | Sequence identity | Sequence similarity |
|---|---|---|
| hu-IRAK-1 | 36% | 48% |
| hu-IRAK-2 | 28% | 39% |
| hu-IRAK-M | 31% | 41% |
| dm-Pelle | 30% | 40% |
Sequence comparisons were performed with the gap program of the Wisconsin GCG package. Numbers indicate percentage of sequence identity or similarity. hu, human; dm, D. melanogaster.
Murine IRAK-4 was obtained from a kidney cDNA phage library with a probe derived from full-length human IRAK-4 (GenBank accession no. AF445803). m-IRAK-4 encodes a 459-aa protein with a calculated molecular mass of 51 kDa, which shares 87% similarity and 84% identity with human IRAK-4 protein (Fig. 1).
Expression Pattern of IRAK-4.
Analysis of multiple-tissue Northern blots with a probe derived from full-length human IRAK-4 revealed two mRNA species with sizes of ≈3 and 4.4 kb. The strongest hybridization signal was observed in kidney and liver samples (data not shown). A broader analysis of a tissue cDNA panel by reverse transcription–PCR revealed a low level of expression in a wide variety of tissues (data not shown).
Interaction of IRAK-4 with Components of the IL-1R Complex.
To study the functional properties of IRAK-4, we performed coprecipitation studies with known adaptor proteins of the Toll/IL-1R system. Similar to IRAK-1, only kinase-inactive IRAK-4 (IRAK-4 KK213AA) is able to interact with MyD88 upon overexpression in 293 cells, whereas no interaction is detectable between wild-type IRAK-1 or IRAK-4 and MyD88 (Fig. 2A). Both kinase-inactive IRAK-1 and IRAK-4 coprecipitated with coexpressed TRAF6, although a similar interaction between wild-type IRAK-4 and TRAF6 was not observed. This behavior differs from IRAK-1; wild-type IRAK-1 shows a weak but reproducible association with TRAF6 under overexpression conditions (Fig. 2B).
Figure 2.
Interaction of IRAK-4 with components of the IL-1R complex. (A) Coprecipitation of MyD88 with IRAK-4 and IRAK-1. The 293 cells were transfected with expression plasmids coding for myc-tagged IRAK-1 or IRAK-4 (each wild-type and kinase-inactive mutant) alone or together with Flag-tagged MyD88. MyD88 was immunoprecipitated (IP) with anti-Flag mAb, and coprecipitating IRAKs were detected with anti-myc antiserum (Top). Western blot analysis (WB) of the same blot with mAb to the Flag epitope shows that similar amounts of MyD88 were IP (Middle). The lysates of the transfected cells were immunoblotted with myc antiserum to monitor the expression of IRAKs (Bottom). (B) Coprecipitation of TRAF6 with IRAK-4 and IRAK-1. The 293 cells were transfected with expression plasmids coding for myc-tagged IRAK-1 or IRAK-4 alone or together with Flag-tagged TRAF6. IP and WB were performed as described in A. (C) IL-1-induced coprecipitation of IRAK-4 with TRAF6 and IRAK-1. Untransfected 293RI cells (2 × 108 cells per sample) were stimulated for the indicated amount of time with 100 ng/ml of IL-1β. Cell lysates were IP with either IRAK-4 antiserum or preimmune serum. Coprecipitating IRAK-1 and TRAF6 were detected with polyclonal antisera (Upper). In a second Western blot, the same membrane was used to show equal precipitation of IRAK-4.
We also studied the ability of IRAK-4 to heterodimerize with other IRAK-family members. Although IRAK-1, IRAK-2, and IRAK-M form heterodimers when coexpressed (14), IRAK-4 does not stably associate with the other family members under overexpression conditions in 293 cells. An interaction between IRAK-1 and IRAK-4, however, can be detected in a yeast two-hybrid system (data not shown), suggesting that the direct interaction is weak or transient.
Given that IRAK-4 is able to interact with MyD88 and TRAF6 upon overexpression, we investigated whether endogenous IRAK-4 is involved in IL-1 signaling in untransfected cells. We stimulated 293RI cells (293 cells stably expressing IL-1RI; ref. 10) with IL-1 for 2, 10, and 30 min, lysed the cells, and immunoprecipitated IRAK-4. Coprecipitating proteins were detected by Western blotting (Fig. 2C). Both IRAK-1 and TRAF6 associate with endogenous IRAK-4 in a rapid, transient, and IL-1-dependent manner, strongly supporting a role for IRAK-4 in the signaling of the TIR family. This interaction is already detectable after 2 min of IL-1 stimulation and peaks around 10 min. After 30 min of stimulation the interaction between TRAF6 and IRAK-4 is no longer detectable. Even though IRAK-4 and IRAK-1 do not seem to be able to form a stable, direct interaction under overexpression conditions (see above), they can be readily detected in an IL-1-induced protein complex consisting of at least three members.
One hallmark of the IL-1-induced activation of IRAK-1 is its rapid modification, which is reflected by a shift of the apparent molecular mass from about 80 kDa to more than 100 kDa (10) and which precedes its degradation (23). The apparent molecular mass of the IRAK-1 species detected in this experiment is about 80 kDa, suggesting that it is unmodified IRAK-1 and that the immunoprecipitated complex is involved in an early step in the IL-1 signal transduction, before full activation of IRAK-1.
Taken together these data indicate that IRAK-4 is indeed a component of the IL-1 signal transduction cascade. It is able to interact with the same adaptor molecules as the other IRAK family members and might therefore be involved in similar signal transduction pathways, but these interactions differ in their nature from IRAK-1, pointing toward a distinct role of IRAK-4 in cytokine signaling.
Activation of Erk, JNK, and NF-κB Pathways.
We performed reporter assays to elucidate the function of IRAK-4 in TIR signaling. First we analyzed the ability of IRAK-4 to activate MAPK pathways, in particular Erk and JNK-dependent transcription-factor activation. For this analysis, we used a transreporter system, in which the activation domains (ADs) of the transcription factors Elk1 or cJun were fused to the GAL4 DNA binding domain. Activation of the Erk or JNK pathways leads to phosphorylation of the Elk1-AD or cJun-AD, respectively, which in turn will switch on a GAL4-dependent luciferase construct. Known activators of these pathways were used as positive controls (MEK1 for Elk1 activation and MEKK1 for cJun activation). Transient overexpression of IRAK-4 activated both pathways in a dose-dependent manner (Fig. 3 A and B).
Figure 3.
Activation of Elk1, cJun, and NF-κB by IRAK-4. (A) Elk1 activation. The 293 cells were transfected with the indicated amounts of expression plasmids for IRAK-4 and MEK1 (positive control), pRSVβgal for normalization, the GAL4-dependent reporter construct pFR-luc and the transactivator plasmid pFA2-Elk1, a fusion of the DNA binding domain of GAL4 and the transactivation domain of Elk1. The y axis represents the normalized fold of luciferase activity induction relative to cells transfected with a transactivator plasmid lacking the transactivation domain. (B) cJun activation. The experiment was performed as described above, with expression plasmids for MEKK as positive control and pFA2-cJun as transactivator. (C) Activation of NF-κB. The 293 cells were transfected with an NF-κB-dependent ELAM-luc reporter gene construct, pRSVβgal for normalization, and the indicated amounts of wild-type IRAK-4, kinase-inactive IRAK-4 (IRAK-4 KK213AA), and truncated IRAK-4 (amino acids 1–191). Twenty-four hours after transfection, luciferase and β-galactosidase activity were determined. The y axis represents the normalized fold of luciferase activity induction relative to cells transfected with empty vector.
To study the activation of NF-κB, 293 cells were transiently transfected with an NF-κB-dependent ELAM-luciferase reporter construct and increasing amounts of wild-type IRAK-4, kinase-inactive IRAK-4 (IRAK-4 KK213AA), or the IRAK-4 N terminus (amino acids 1–191) (Fig. 3C). Only overexpression of full-length wild-type IRAK-4 activates an NF-κB-dependent reporter construct. In striking contrast to the other IRAKs, kinase-inactive IRAK-4 cannot activate this reporter construct, suggesting that IRAK-4 needs its kinase activity to activate NF-κB. It should be noted, that the amount of NF-κB activation induced by IRAK-4 overexpression is moderate compared with IRAK-1, although the physiological relevance of this observation is unclear.
Inhibition of IL-1-Induced NF-κB Activation and IL-1-Induced IRAK-1 Activation.
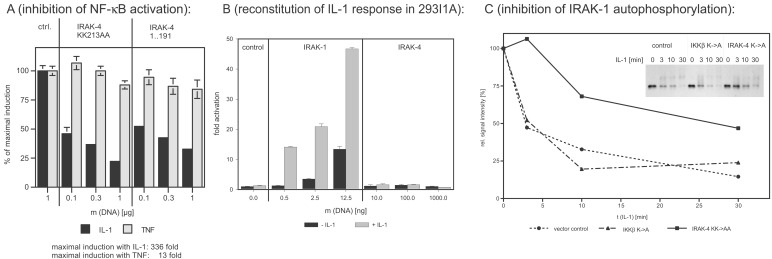
Earlier studies showed that the death-domain-containing N termini of IRAK-1, IRAK-2, and IRAK-M have a dominant-negative effect on IL-1-induced NF-κB activation upon overexpression in 293RI cells (14), whereas kinase-inactive full-length constructs were not only not dominant negative, but were in fact activators of NF-κB (unpublished results and ref. 17). Like analogous IRAK-1 mutants, truncated IRAK-4 (amino acids 1–191) inhibits IL-1-induced NF-κB activation in a dose-dependent manner in 293RI cells (7), most likely because of the interaction of the death domain of the truncated kinases with the death domain of MyD88, and has no significant effect on tumor necrosis factor α-induced NF-κB activation. But in sharp contrast to kinase-inactive IRAK-1, IRAK-4 KK213AA specifically inhibits IL-1-induced NF-κB activation in a dominant-negative fashion (Fig. 4A).
Figure 4.
Dominant-negative effects of IRAK-4 mutants and reconstitution experiments in IRAK-1-deficient 293 cells. (A) Inhibition of IL-1-induced NF-κB activation. The 293RI cells were transfected with an NF-κB-dependent ELAM-luc reporter gene construct, pRSVβgal for normalization, and the indicated amounts of kinase-inactive IRAK-4 (IRAK-4 KK213AA) and truncated IRAK-4 (amino acids 1–191). Twenty-four hours after transfection, cells were stimulated with 50 ng/ml of IL-1β or 100 ng/ml of tumor necrosis factor α for 6 h, and luciferase and β-galactosidase activity were determined. The y axis represents the percentage of NF-κB activation relative to cells transfected with empty vector. (B) Reconstitution of IL-1 response in 293I1A. IRAK-1-deficient 293 cells [293I1A(16)] were transfected and stimulated as described for Fig. 4A. The y axis represents the fold of NF-κB activation relative to cells transfected with empty vector. (C) Inhibition of IL-1-induced IRAK-1 activation. The 293RI cells were transfected with control vector, kinase-inactive IKKβ (negative control), or kinase-inactive IRAK-4 for 24 h. After stimulation with 100 ng/ml of IL-1β for the indicated amount of time, the cells were lysed, and IRAK-1 was immunoprecipitated with the mAb 2A9. The activation level of IRAK-1 was assessed by determining the shift in apparent molecular weight and subsequent degradation of IRAK-1 by Western blotting (Inset). The signal intensity was quantified by densitometry and plotted as a function of incubation time.
The distinct properties of IRAK-4 are also reflected by the fact that it is not able to compensate for the loss of IRAK-1: In an IRAK-1- deficient 293 cell line (293I1A), the IL-1-induced NF-κB activation is completely abolished (16) but can be restored by the introduction of IRAK-1, IRAK-2, or IRAK-M (14). In this cell line, transient expression of IRAK-4 does not activate NF-κB and it does not restore the IL-1 response (Fig. 4B), suggesting that the function of IRAK-4 is not redundant with or downstream of IRAK-1, but that IRAK-4 might act upstream of IRAK-1.
This interpretation is supported by Fig. 4C. The overexpression of IRAK-4 KK213AA inhibits the IL-1-induced modification and degradation of IRAK-1, which is a hallmark of IRAK-1 activation. For this study we transfected 293RI cells transiently with IRAK-4 KK213AA or, as a negative control, with a kinase-inactive version of the downstream effector IKKβ (IKKβ K44A) (19). After IL-1 stimulation for 3, 10, or 30 min, the cells were lysed, and IRAK-1 was immunoprecipitated and analyzed by Western blotting (Fig. 4C, Inset shows the result of the Western blot and the graph shows the densitometric analysis of the same blot). In control-transfected cells, the IRAK-1 protein amount is reduced by 50% within 3 min of IL-1 stimulation, and after 10–30 min, 75% of the protein is degraded. Expression of IRAK-4 KK213AA prevents these events: After 3 min only a very small fraction of IRAK-1 is modified, and even after 30 min only 50% of the protein is degraded (the still considerable amount of IRAK-1 activation and degradation is because of the fact that the transfection efficiency in this transient expression system is only between 50–80%).
Phosphorylation of IRAK-1 by IRAK-4.
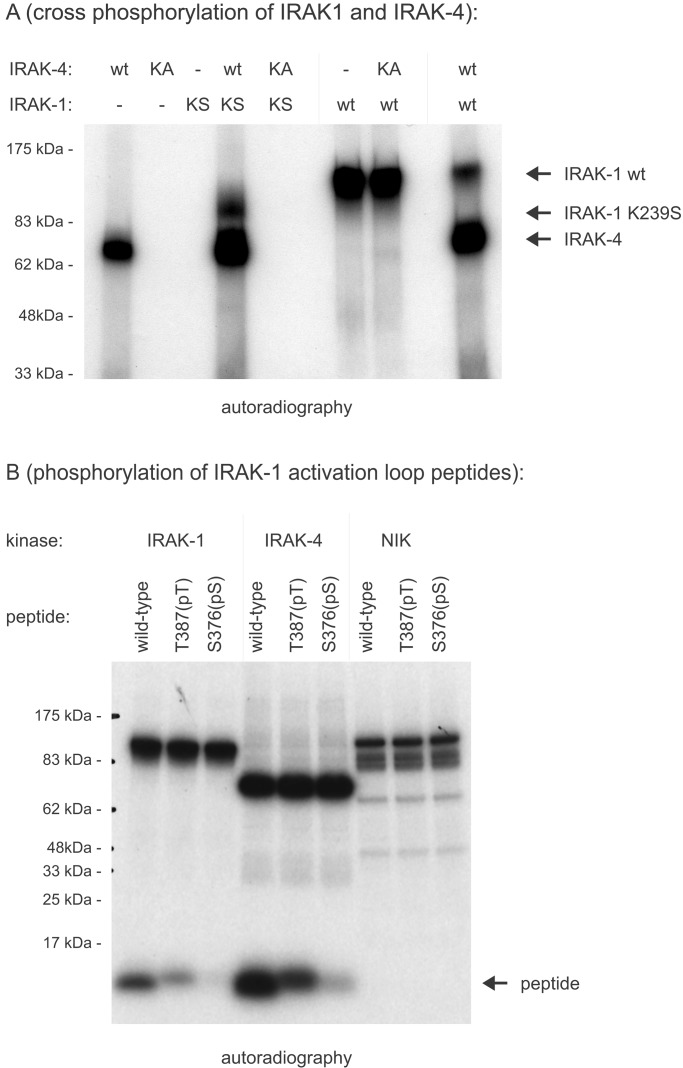
Reconstitution experiments with kinase-inactive IRAK-1 in IRAK-1-deficient 293RI cells showed an IL-1-dependent but IRAK-1-independent phosphorylation of IRAK-1, leading to the postulation of an IRAK-1 kinase (16). Here we show that recombinant IRAK-1 is a substrate for IRAK-4, whereas IRAK-1 is not able to phosphorylate IRAK-4 in vitro (Fig. 5A). In contrast to IRAK-1 autophoshorylation, phosphorylation by IRAK-4 does not lead to a dramatic shift in electrophoretic mobility, showing a qualitative difference between these two phosphorylation events.
Figure 5.
Phosphorylation of the IRAK-1 activation loop. (A) Crossphosphorylation of IRAK-1 and IRAK-4. Full-length IRAK-1 and IRAK-4 expressed in insect cells were tested for their ability to phosphorylate each other in vitro. One hundred nanograms of wild-type protein (wt) were incubated with kinase-inactive mutants (KA, IRAK-4 KK213AA; KS, IRAK-1 K239S) in the presence of [γ-32P]ATP. (B) Phosphorylation of IRAK-1 activation loop peptides. Synthetic peptides were derived from the IRAK-1 activation loop (amino acids 359–389). The substrate quality of wild-type peptides was compared with peptides, in which either Thr-387 or Ser-376 was replaced with phospho-threonine or phospho-serine, respectively. After incubation in an in vitro kinase assay with recombinant IRAK-1, recombinant IRAK-4, and recombinant NIK (negative control), phosphorylation of peptides was determined by incorporation of radioactive phosphate after SDS/PAGE and autoradiography.
In addition, it was shown that phosphorylation of Thr-387 in the IRAK-1 activation loop (amino acids 358–389) is critical for the activation of IRAK-1 kinase activity, because substitution with alanine leads to a dramatic decrease of IRAK-1 kinase activity (C. Kollewe, unpublished data). By using peptides derived from the IRAK-1 activation loop, we found that at least in vitro the IRAK-1 activation loop is a good substrate for IRAK-4, and that T387 and S376 are the main sites of phosphorylation by both IRAK-1 and IRAK-4. The unrelated kinase NIK (24) was not able to phosphorylate IRAK-1 activation-loop peptides (Fig. 5B). This observation is consistent with a model in which IRAK-4 acts upstream of IRAK-1 and in which it would initiate the activation of the kinase activity of IRAK-1 by phosphorylating critical residues in the IRAK-1 activation loop. In a positive feedback loop, this activated IRAK-1 would then be able to crossphosphorylate other IRAK-1 molecules as well as IRAK-2 and IRAK-M (14), resulting in rapid and full activation of all IRAK family members.
It should be noted, however, that the physiological role of the kinase activity of IRAK-1 is still unclear. The time course of this activity correlates both with initiation of the IL-1 response and with the degradation of IRAK-1, and therefore it was speculated that the phosphorylation might be a negative feedback loop, leading to the termination of the IL-1 response (23). Another hypothesis is based on the observation that only kinase-inactive IRAK-1 can interact with MyD88, and that the phosphorylation of IRAK-1 is terminating the interaction between these two molecules, allowing IRAK-1 to leave the IL-1 receptor complex efficiently and to interact with TRAF6 in the cytosol (7). It is conceivable that both theories are correct, and that the activation of the kinase activity of IRAK-1 on the one hand is necessary to initiate the IL-1 response, but on the other hand is also limiting the duration of this response.
Recently, two novel adaptor molecules were identified, termed Tollip and Mal (8, 25). Both molecules are implicated in the early signal transduction events of members of the TIR family and both have been reported to be able to interact with IRAK family members at least under overexpression conditions. It is tempting to speculate that the exact molecular composition of signaling complexes of TIR family members is variable and depending on the cell type and activation state of the cell studied, and it can even be speculated that the exact function of one and the same molecule in this complex might depend on the cellular and molecular context.
We are proposing the following model for IRAK-4 function in TIR signaling: IRAK-4 is the mammalian homolog of Drosophila melanogaster Pelle. Like Pelle, and unlike the other mammalian IRAKs, IRAK-4 needs its kinase activity for its function (26, 27). IRAK-4 is acting upstream of the other IRAKs and might function as an IRAK-1 kinase, triggering a cascade of phosphorylation events. This model is supported by the characterization of IRAK-4 knockout mice by Wen-Chen Yeh and colleagues (unpublished observations): In contrast to mice lacking IRAK-1, in which the response to TIR triggering is reduced but not completely abolished, presumably because of the redundancy among IRAK-1, IRAK-2, and IRAK-M, the phenotype of IRAK-4 knockout mice is much more severe, and signaling through TIRs is virtually abolished.
Acknowledgments
We thank Tim Hoey and Nigel Walker for helpful comments on the manuscript, Linda Huang for peptide synthesis, and Miki Rich for DNA-sequencing.
Abbreviations
- IRAK
IL-1 receptor-associated kinase
- TIR family
Toll/IL-1 receptor family
- ELAM
endothelial leukocyte adhesion molecule
Footnotes
References
- 1. Kollewe, C. & Martin, M. U. (2002) Signal Trans., in press.
- 2. Wesche, H. & Martin, M. U. (2002) Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 3.Dinarello C A. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 4.Fallon P G, Allen R L, Rich T. Trends Immunol. 2001;22:63–66. doi: 10.1016/s1471-4906(00)01800-7. [DOI] [PubMed] [Google Scholar]
- 5.Ulevitch R J, Tobias P S. Curr Opin Immunol. 1999;11:19–22. doi: 10.1016/s0952-7915(99)80004-1. [DOI] [PubMed] [Google Scholar]
- 6.Muzio M, Ni J, Feng P, Dixit V M. Science. 1997;278:1612–1615. [Google Scholar]
- 7.Wesche H, Henzel W J, Shillinglaw W, Li S, Cao Z. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald K A, Palsson-McDermott E M, Bowie A G, Jefferies C A, Mansell A S, Brady G, Brint E, Dunne A, Gray P, Harte M T, et al. Nature (London) 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 9.Martin M, Bol G F, Eriksson A, Resch K, Brigelius-Flohe R. Eur J Immunol. 1994;24:1566–1571. doi: 10.1002/eji.1830240717. [DOI] [PubMed] [Google Scholar]
- 10.Cao Z, Henzel W J, Gao X. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 11.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D V. Nature (London) 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 12.Kanakaraj P, Schafer P H, Cavender D E, Wu Y, Ngo K, Grealish P F, Wadsworth S A, Peterson P A, Siekierka J J, Harris C A, Fung-Leung W P. J Exp Med. 1998;187:2073–2079. doi: 10.1084/jem.187.12.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas J A, Allen J L, Tsen M, Dubnicoff T, Danao J, Liao X C, Cao Z, Wasserman S A. J Immunol. 1999;163:978–984. [PubMed] [Google Scholar]
- 14.Wesche H, Gao X, Li X, Kirschning C J, Stark G R, Cao Z. J Biol Chem. 1999;274:19403–19410. doi: 10.1074/jbc.274.27.19403. [DOI] [PubMed] [Google Scholar]
- 15.Aravind L, Dixit V M, Koonin E V. Science. 2001;291:1279–1284. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Commane M, Burns C, Vithalani K, Cao Z, Stark G R. Mol Cell Biol. 1999;19:4643–4652. doi: 10.1128/mcb.19.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knop J, Martin M U. FEBS Lett. 1999;448:81–85. doi: 10.1016/s0014-5793(99)00322-1. [DOI] [PubMed] [Google Scholar]
- 18.Maschera B, Ray K, Burns K, Volpe F. Biochem J. 1999;339:227–231. [PMC free article] [PubMed] [Google Scholar]
- 19.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 20.Schall T J, Lewis M, Koller K J, Lee A, Rice G C, Wong G H, Gatanaga T, Granger G A, Lentz R, Raab H, et al. Cell. 1990;61:361–370. doi: 10.1016/0092-8674(90)90816-w. [DOI] [PubMed] [Google Scholar]
- 21.Scanlan M J, Gordan J D, Williamson B, Stockert E, Bander N H, Jongeneel V, Gure A O, Jager D, Jager E, Knuth A, et al. Int J Cancer. 1999;83:456–464. doi: 10.1002/(sici)1097-0215(19991112)83:4<456::aid-ijc4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Feinstein E, Kimchi A, Wallach D, Boldin M, Varfolomeev E. Trends Biochem Sci. 1995;20:342–344. doi: 10.1016/s0968-0004(00)89070-2. [DOI] [PubMed] [Google Scholar]
- 23.Yamin T T, Miller D K. J Biol Chem. 1997;272:21540–21547. doi: 10.1074/jbc.272.34.21540. [DOI] [PubMed] [Google Scholar]
- 24.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 25.Burns K, Clatworthy J, Martin L, Martinon F, Plumpton C, Maschera B, Lewis A, Ray K, Tschopp J, Volpe F. Nat Cell Biol. 2000;2:346–351. doi: 10.1038/35014038. [DOI] [PubMed] [Google Scholar]
- 26.Edwards D N, Towb P, Wasserman S A. Development (Cambridge, UK) 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- 27.Towb P, Bergmann A, Wasserman S A. Development (Cambridge, UK) 2001;128:4729–4736. doi: 10.1242/dev.128.23.4729. [DOI] [PubMed] [Google Scholar]