Abstract
The exon-1 peptide of huntingtin has 51 Gln repeats and produces the symptoms of Huntington's disease in transgenic mice. Aggregation of the yeast Sup35 protein into prions has been attributed to its glutamine-rich and asparagine-rich domain. Here, we show that poly-l-asparagine forms polar zippers similar to those of poly-l-glutamine. In solution at acid pH, the glutamine-rich and asparagine-rich 18-residue Sup35 peptide, rendered soluble by the addition of two aspartates at the amino end and two lysines at the carboxyl end, gives a β-sheet CD spectrum; it aggregates at neutral pH. A poly-alanine peptide D2A10K2 gives an α-helical CD spectrum at all pHs and does not aggregate; a peptide with the sequence of the C-terminal helix of the α-chain of human hemoglobin, preceded by two aspartates and followed by two lysines, exhibits a random coil spectrum and does not aggregate either. Alignment of several β-strands with the sequence of the 42-residue Alzheimer's amyloid β-peptide shows that they can be linked together by a network of salt bridges. We also asked why single amino acid replacements can so destabilize the native structures of proteins that they unfold and form amyloids. The difference in free energy of a protein molecule between its native, fully ordered structure and an amorphous mixture of randomly coiled chains is only of the order of 10 kcal/mol. Theory shows that destabilization of the native structure by no more than 2 kcal/mol can increase the probability of nucleation of disordered aggregates from which amyloids could grow 130,000-fold.
Intracellular inclusions are now recognized as pathogenic. Many of them have one important feature in common: they consist of normally soluble proteins with a variety of complex three-dimensional structures that are apparently stable until some trigger converts them to bundles of fibers uniformly made up of β-pleated sheets, with their strands running normal to the fiber axis. The inclusions comprise the aggregates formed by proteins with extended Gln repeats, the amyloids formed by many unrelated proteins, the yeast, animal, and human prions, the neurofibrillar tangles and amyloid plaques of Alzheimer's disease, synuclein of Parkinson's disease, and the aggregates caused by expansion of trinucleotide repeats. The question arises whether the conversion of these chemically and structurally unrelated proteins to one common structure has a common cause. It has led us to study the formation of yeast prions, to compare the structures of Asn and Ala repeats and of the exon-1 fragment of huntingtin with those of the Gln repeats determined earlier, to investigate the possible chemical bonds between the strands of the Alzheimer's amyloid β-peptide (Aβ), and, finally, to consider the general thermodynamics of protein aggregation. We found that small reductions in the stability of native, folded proteins can lead to large rises in the probability of nucleation of the unfolded, denatured form.
It has been shown that poly-l-glutamine aggregates into β-pleated sheets by forming hydrogen bonds between both the side-chain and main-chain amides, turning them into polar zippers. The exon-1 fragment of huntingtin contains the Gln repeat followed by a repeat of prolines and 24 mixed residues. The yeast prion protein Sup35 is a transcription termination factor that has a glutamine-rich and asparagine-rich N-terminal region that causes its aggregation (1–3). Poly-l-glutamine aggregates into β-pleated sheets held together by hydrogen bonds (4). Model building suggested that poly-l-asparagine might do the same. CD spectra of poly-l-asparagine confirmed this possibility; they showed that the exon-1 fragment of huntingtin and the 24-residue glutamine-rich and asparagine-rich peptide of Sup35 do indeed aggregate into pleated β-sheets. We then wondered whether other peptides also would form pleated β-sheets under our conditions, but a 15-residue peptide with the sequence of the C-terminal α-helix of the α-chain of human hemoglobin gave a random coil spectrum.
The nuclei of skeletal muscle fibers of patients with oculopharyngeal muscular dystrophy contain filamentous inclusions (5). They are linked to expansion of an Ala repeat in the polyadenine-binding protein 2, but a synthetic ten-mer of l-alanine gave an α-helical rather than a β-sheet spectrum and provided no clue to the cause of the aggregation, which we also found elsewhere. The 42-residue Aβ peptide contained in the extracellular plaques of Alzheimer disease forms amyloid-like filaments (6). When we searched for the cause of their aggregation, we found that the sequence of this peptide can be aligned, so that neighboring β-strands are held together by networks of strong hydrogen bonds between oppositely charged ions.
Materials and Methods
Peptides were assembled by using a continuous-flow peptide synthesizer on polyethylene glycol-polystyrene resins with fluorenylmethoxycarbonyl chemistry. Peptides were purified by using preparative reverse-phase HPLC and their identities confirmed by analytical reverse-phase HPLC, time-of-flight mass spectrometry, and amino acid analysis. Far-UV CD measurements were made on a Jobin-Yvon CD6 Circular Dichrograph (Jobin-Yvon, Longjumeau, France) which was calibrated with a 0.06% (wt/vol) solution of d-10-camphorosulphonate. Peptides were freshly prepared as 1 mg/ml working solutions in 0.1% trifluoroacetic acid, pH 2.0; 300 μl samples were analyzed in a 0.1-cm path length cell at 20°C. Data were collected in the far UV (190–250 nm) at 0.5 nm intervals with 2 s intervals. Five such runs were averaged, calculated net of a solvent baseline and factor 3 smoothed. The 1 mg/ml D2A10K2 peptide (822 μM) and the hemoglobin peptide (492 μM) exceeded the maximum voltage for the machine to 200 nm and were, therefore, diluted four-fold for assay. Data for all peptides were expressed as mean residue ellipticity ([θ]MR) in degrees cm2/dmol−1. Additions of 1–10 μl of 0.1 N NaOH were added to 0.5 ml samples of peptide to raise the pH to the respective values. Yeast prion peptide concentrations were confirmed from absorbance measurements at 280 nm, based on the tyrosine content (7); A280 = 1.0 cm−1 is equivalent to 390.6 μM, 1.027 mg/ml. Yeast prion peptide was diluted to 0.25 mg/ml (96 μM) for measurements of light scattering on a Perkin–Elmer LS50B luminescence spectrometer. Measurements were made at 20°C on 400 μl samples in 1-cm path length fluorescence cuvettes with excitation and emission wavelengths of 520 nm and with 2.5 nm slit widths. Hemoglobin peptide (1 mg/ml) was used as a control. The pH of samples was adjusted with 1–10 μl of 1 M stocks of sodium phosphate pH 7.0, or Tris pH 8.0, or unbuffered Tris pH 10.0. Measurements were taken within 30 s of mixing and again at 2 h.
Results and Discussion
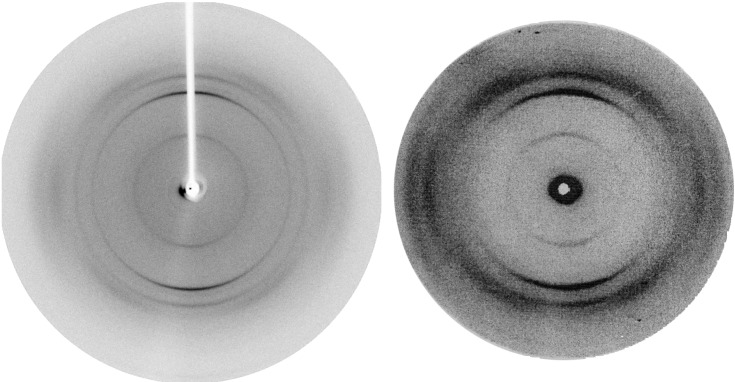
Figure 1 shows the x-ray diffraction patterns of fibers of the exon-1 protein of huntingtin together with those of poly-l-glutamine (D2Q15K2) obtained earlier. Both exhibit one meridional reflection at 4.75 Å together with its second order and another at 8.3 Å together with its second and third orders. The pictures were taken with the x-ray beam parallel to a thin film of the fibers, so that only fibers parallel to the plane of the film and roughly normal to the beam would have contributed. An electron diffraction picture (not shown) taken with the electron beam normal to the plane of the film showed the two sets of reflections at right angles to each other, indicating that they came from different fibers. The accompanying paper presents a structure for the fibers with the 4.75-Å repeat (8). The interpretation of the one with the 8.3 Å is still unclear. The Gln repeat in the exon-1 protein of huntingtin is followed by long repeats of prolines. The identity of its diffraction pattern with that of poly-l-glutamine shows that the prolines are not part of the ordered fiber structures.
Figure 1.
X-ray fiber diffraction diagrams of the peptide D2Q15K2 (Left) and of the exon-1 protein of huntingtin with 51 Gln repeats.
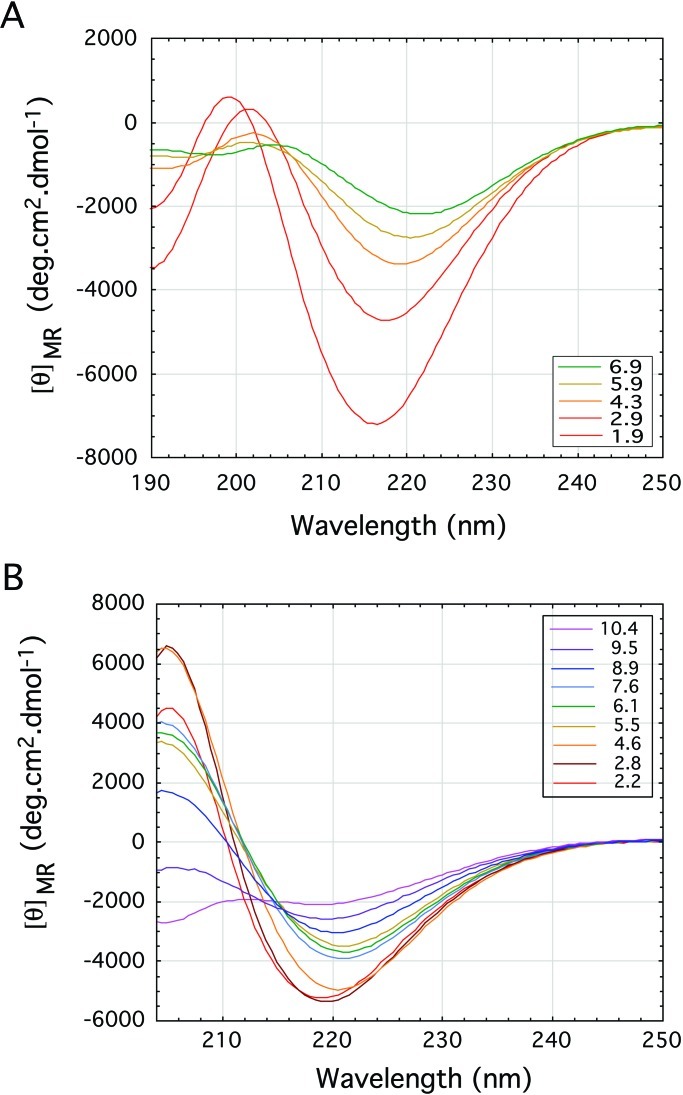
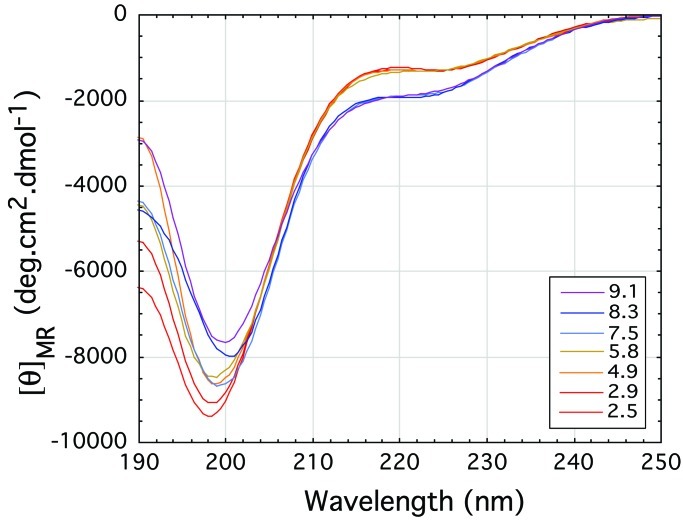
Poly-l-glutamine forms β-pleated sheets in solution. We now wondered whether the glutamine-rich and asparagine-rich regions of the Sup35 protein of yeast prions and poly-l-asparagine alone do so too and measured their CD spectra. To make them soluble in water, we attached two aspartates to the amino ends and two lysines to the carboxyl ends of both peptides. Figure 2 illustrates the molar ellipticities as a function of pH of solutions of D2N15K2 and of the yeast prion Sup35 peptide with the sequence H-Asp-Asp-Asn-Asn-Gln-Gln-Asn-Tyr-Gln-Gln-Tyr-Ser-Gln-Asn-Gly-Asn-Gln-Gln-Gln-Gly-Lys-Lys-OH (9).
Figure 2.
Far-UV CD spectra plotting the mean residue ellipticity against wavelength for (A) the poly-l-asparagine peptide D2N15K2 and (B) the yeast prion protein Sup35 peptide. The spectra are typical of pure β-sheet at pH 2 but suggest a decrease of β-sheet with increasing pH to neutral pH. The signal for the yeast prion protein Sup35 peptide eventually becomes that of a random coil at the highest pH of 10.4.
Both peptides show strong β-pleated sheet spectra at low pH that get
weaker with increasing pH. At pH 10.4, the yeast peptide shows the
beginning of a random coil spectrum. The pH dependence of the CD
spectra of the poly-l-asparagine and yeast prion peptides
must be caused by the N-terminal aspartates. At low pH, they would be
uncharged, but at higher pH, the charged carboxylates would compete
with the amide carbonyls for hydrogen bonding to the amide
NH2s. At neutral pH, attraction between the
terminal aspartates and lysines also would promote aggregation that
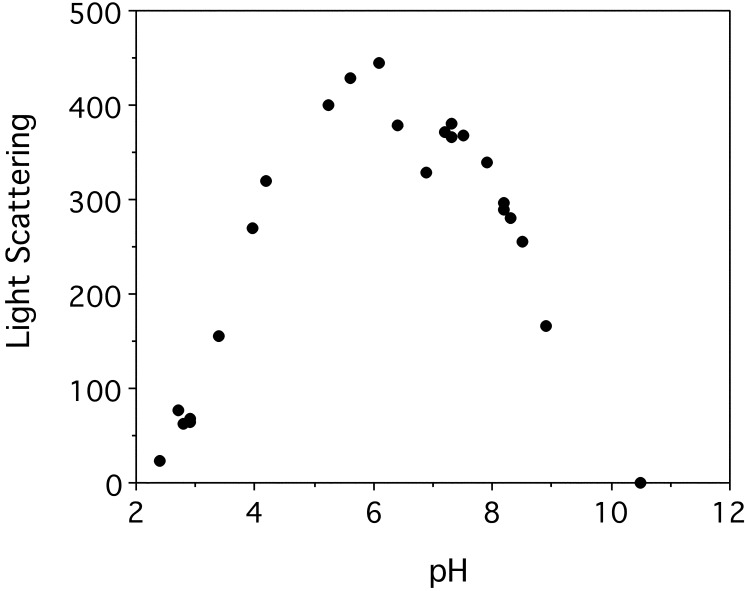
weakens the spectra. Light scattering confirmed this possibility
by showing that the Sup35 peptide is soluble at both low pH and high pH
and aggregates at neutral pH (Fig. 3).
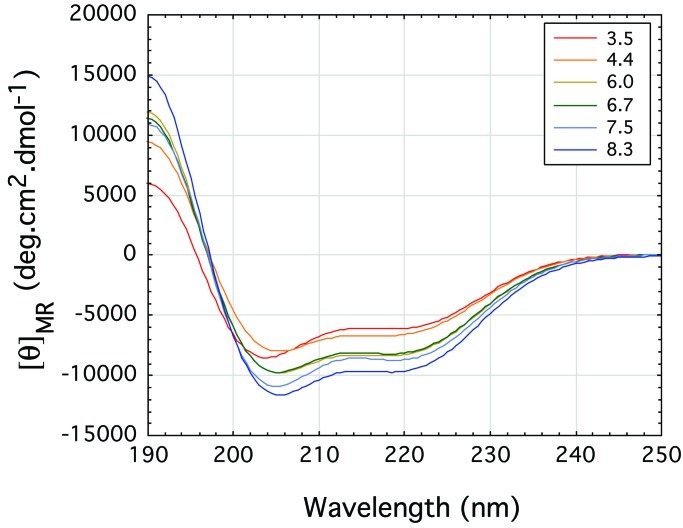
The CD spectra of
D2A10K2
showed that it is α-helical at all pHs. The spectra became stronger
rather than weaker with increasing pH, with no sign of aggregation
(Fig. 4). The hemoglobin peptide with the
sequence
H-Asp-Asp-Ala-Val-His-Ala-Ser-Leu-Asp-Lys-Phe-Leu-Ala-Ser-Val-Ser-Thr-Lys-Lys-OH
(10) exhibits a random coil spectrum (Fig.
5); its light scattering at pH 7.5 was no
more than 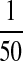 of that of the Sup35 peptide, which shows that
the aspartates and lysines by themselves are not sufficient to cause
aggregation.
of that of the Sup35 peptide, which shows that
the aspartates and lysines by themselves are not sufficient to cause
aggregation.
Figure 3.
Light-scattering experiments on the yeast prion protein Sup35 peptide indicate aggregation that peaks between pH 5.5 and 7.5. This effect is negligible below pH 3.0 and above pH 9.0. (The signal from the hemoglobin peptide rose to the equivalent of eight light-scattering units at pH 7–7.5, too small to show on the plot.)
Figure 4.
The polyalanine peptide D2A10K2 folds to give an α-helix type far-UV CD spectrum. The plot for α-helix at pH 3.5 is weaker than at pH 4.4 and increases slightly with pH to pH 8.3.
Figure 5.
The C-terminal α-helix peptide of hemoglobin exhibits a random coil structure in the far-UV CD. There are no effects on folding with changes in pH.
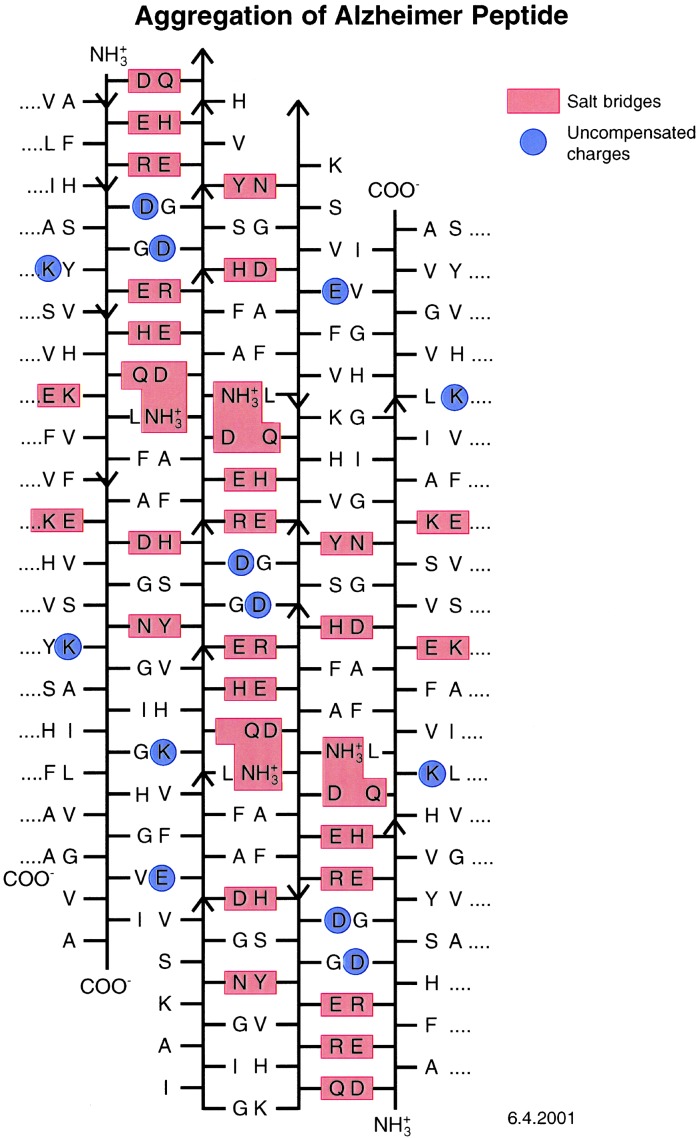
Having found that amino acids with side chains capable of linking neighboring β-strands together by hydrogen bonds tend to make peptides aggregate into amyloid fibers, we tried to find out whether such crosslinks also might contribute to the aggregation of the 42-residue Alzheimer's Aβ peptide. Figure 6 shows four β-strands, some parallel and others antiparallel, lined up so that they are linked together by a network of hydrogen bonds between oppositely charged ions. These β-strands are marked by the red boxes that also mark a tyrosine hydrogen bonded to an aspartate. The blue circles mark uncompensated charges. The structure contains 30 hydrogen-bonded crosslinks and 13 uncompensated charges, but their destabilizing effects could be screened by hydration. The effects of a variety of amino acid substitutions on the aggregation of the Aβ peptide are consistent with this scheme (Table 1). M.F.P. et al. (8) propose that amyloid fibers consist of cylindrical β-sheets. Their paper shows only a parallel β-sheet, but there is no reason why the sheets should not be made of a mixture of parallel and anti-parallel β-strands or only of anti-parallel strands, provided their terminal residues are separated so as not to cause repulsion.
Figure 6.
Alignment of the sequence of the 42-residue peptide of the plaques (10) of Alzheimer's disease. The red boxes mark salt bridges between acids and bases and a hydrogen bond between a Tyr and an Asn; the blue circles mark uncompensated changes.
Table 1.
Effects of amino acid substitutions on aggregation of Aβ (Alzheimer) peptide
| Proposed interactions between β-strands | Substitution | Effect of substitution |
|---|---|---|
| Lys-16–Glu-23 | Lys→Ala | no β-sheets at any pH |
| His-14–Val-24 | His→Ala | increased fibre formation |
| His-14–Val-24 | His→Asp | stabilises fibres at pH 2.5 |
| no β-sheets at pH 7.8 | ||
| Glu-11–Arg-5 | Glu→Ala | reduced interaction with wild type |
| Val-18-–Phe-20 | Val→Lys | reduced interaction with wild type |
| Arg-5–Glu-11 | Arg→Glu | no interaction with wild type |
| His-13–Glu-3 | His→Leu | reduced interaction with wild type |
Inclusion of long Gln repeats in whatever protein gives rise to neurodegenerative disease, because these repeats cause the proteins to form intraneural aggregates. The only other cause of extension of a single amino acid repeat causing an inherited neurodegenerative disease is that of oculopharyngeal muscular dystrophy, in which expansion of an Ala repeat in the polyadenine-binding protein 2 by a single Ala residue from its normal six to seven Alas causes nuclear filamentous inclusions. Similar inclusions were found in cultures of COS-7 cells in which the same protein with expanded Alas was expressed (11). It seemed surprising that extension of an Ala repeat by a single residue should have such a drastic effect. To find out whether Ala repeats tend to aggregate, we made the peptide D2A10K2 and measured its CD spectra in solution as a function of pH (Fig. 4). The spectra show that the peptide forms α-helices at all pHs, with no sign of aggregation. α-Helices can adhere to each other by forming coiled coils, but it is most unlikely that expansion from six to seven Alas would have this effect. The following is a more likely interpretation: Ala repeats are hydrophobic and would, therefore, occupy internal positions in the protein. An additional Ala would be a misfit that lowers the free energy barrier to unfolding of the protein. The ease with which this can happen is illustrated by mutations that make transthyretin form amyloids. Transthyretin is a protein of 127-aa residues folded into tightly packed β-sheets. Replacement of Val-122 by Ile is sufficient to cause amyloid disease. The δ-methyls of the Val are in van der Waals contact with a Phe and 4 Å from a Tyr. The additional carbon of the Ile would push the Val away by no more than 1.5 Å from the bulk of the protein, yet that is enough to loosen the entire structure (12). The additional Ala in the polyadenine-binding protein may have a similar destabilizing effect.
The thermodynamics of protein unfolding help to explain why replacements, additions, or deletions of single amino acid residues lead to aggregation. ΔGN−D, the difference between the free energies of the native, folded and the denatured, unfolded states of typical proteins is only between 5 and 15 kcal/mol of protein (13), compared with the energy of a single carbon–carbon bond of 58.6 kcal/mol. If 10 kcal is taken as an average, then the concentration ratio of folded protein molecules in equilibrium with unfolded ones at 37°C can be derived as follows:
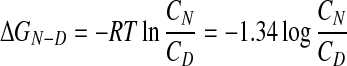 |
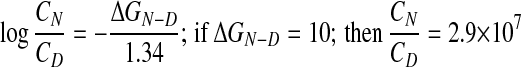 |
Replacement of single amino acid residues can destabilize native structures by between 1 and 2 kcal/mol protein. This amount would reduce the concentration ratio to 5.2 × 106 and 9.3 × 105 respectively, which means that the concentration of denatured molecules will be raised 5.5- or 31-fold. If there is only one single native structure where every atom has its place and its denatured aggregates are amorphous mixtures of randomly coiled chains, then the critical free energy required to create a spherical nucleus of critical radius is
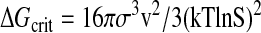 |
where σ is the surface energy of the precipitate, v is the molecular volume, and S is the supersaturation ratio. The probability of this critical energy being reached is
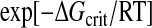 |
If we substitute concentration ratios for supersaturation ratios, then a 5.5-fold rise in concentration of the unfolded protein would increase the probability of nucleation of an amorphous aggregate 14-fold, and a 31-fold increase would raise it 130,000-fold. This result may help to explain the extreme sensitivity of most proteins to any factors that destabilize their native structures (14). Aggregation is reversible: chains dissociating from the amorphous aggregates could refold into either the native structure or into amyloid fibers.
Asparagine-rich and glutamine-rich regions are not confined to Sup35. Michelitsch and Weissman (15) found them in 1.5% of the proteins of the yeast Saccharomyces cerevisiae, 3.5% or 472 of the proteins of Drosophila, and 1% of those of the nematode worm Caenorhabditis elegans. Those regions are highly conserved in eukaryotic microorganisms other than yeasts. Their polar zipper action suggests that they mediate interaction between different proteins. Although long repeats of Gln occur in many species, long repeats of Asn have been found only in the slime mould Dictyostelium discoideum. Despite their obvious importance, no one has so far investigated the normal function of any of these repeats.
Conclusions
The exon-1 peptide of huntingtin with 51 Gln repeats polymerizes into fibers that give the same x-ray diffraction diagram as poly-l-glutamine, which forms cylindrical β-sheets with the β-strands normal to the fiber axis (8). The peptide gives rise to symptoms in transgenic mice similar to those of Huntington's disease of humans. The identity of its diffraction pattern with that of poly-l-glutamine indicates that this structure is itself the pathogen.
The asparagine-rich and glutamine-rich regions of Sup35 can form polar zippers, because poly-l-asparagine forms β-sheets in solution just like poly-l-glutamine.
The 42-residue Aβ protein of Alzheimer plaques is prone to form amyloid fibers for several reasons. Alignment of several strands of Aβ shows that they can form networks of strong hydrogen bonds between ionized side chains of opposite charge. According to secondary structure prediction, residues 16–23 of Aβ are intrinsically apt to form β-structures (16). Finally, a 42-residue peptide can spontaneously form a two-turn helical β-sheet that forms the nucleus for growth of a helical fiber.
The sensitivity of globular proteins to amino acid replacements that cause only small changes in the free energy difference between the correctly folded and the denatured, random coil state arises because the free energy difference between the two states is only 5–15 kcal/mol. In consequence, a reduction of that difference by only 1–2 kcal may lead to very large rises in the probability of nucleation of random aggregates.
Acknowledgments
We thank the Medical Research Council and the Wellcome Trust for support.
Abbreviation
- Aβ protein
amyloid β-peptide
References
- 1.Williams R B. Science. 1994;264:566–569. [Google Scholar]
- 2.Patino M M, Liu J-J, Glover J R, Lindquist S. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 3.DePace A H, Santoso A, Milner O, Weissman J S. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 4.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brais B, Bouchard J P, Xie Y G, Rochefort D L, Chretien N, Tome F M S, Lafreniere R G, Rommens J M, Uyama E, Nohira O, et al. Nat Genet. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 6.Kirschner D A, Inouye H, Duffy L K, Sinclair A, Lind M, Selhoe D J. Proc Natl Acad Sci USA. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill, S. C. & von Hippel, P. H. v. (1989) Anal. Biochem.182, 319–326. [DOI] [PubMed]
- 8.Perutz M F, Finch J T, Berriman J, Lesk A. Proc Natl Acad Sci USA. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoso A, Chien P, Osherovich L Z, Weissman J S. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 10.Fermi G, Perutz M F. Atlas of Molecular Structures in Biology 2: Hemoglobins and Myglobin. Oxford: Clarendon; 1981. [Google Scholar]
- 11.Shanmugam V, Diou P, Rochefort D, Laganière J, Brais B. Ann Neurol. 2000;48:798–800. [PubMed] [Google Scholar]
- 12.Peterson S A, Klabunde T, Lashuel M A, Purkey H, Sachettini J C, Kelly J W. Proc Natl Acad Sci USA. 1998;95:12956–12960. doi: 10.1073/pnas.95.22.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creighton T E. Proteins, Structures and Molecular Principles. New York: Freeman; 1983. p. 299. [Google Scholar]
- 14.Perutz M F, Windle A H. Nature (London) 2001;412:143–144. doi: 10.1038/35084141. [DOI] [PubMed] [Google Scholar]
- 15.Michelitsch M D, Weissman J S. Proc Natl Acad Sci USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallberg Y, Gustafsson M, Personn B, Thyboerg J, Johansson J. J Biol Chem. 2001;276:12945–12950. doi: 10.1074/jbc.M010402200. [DOI] [PubMed] [Google Scholar]